Rust fungi (Basidiomycetes of the order Uredinales) are obligate biotrophs that grow and reproduce only in living plant tissue. There are on the order of 5000 or more species of rust fungi that collectively cause disease on most crops, ornamentals, and many other plants. For example, rusts caused by Puccinia species are some of the most important diseases of wheat and other small grain crops worldwide. A new wheat rust epidemic is currently building in East Africa with the appearance of a highly virulent strain of Puccinia graminis tritici called Ug99, which is perceived as a threat to global wheat production and has led to the establishment of a Global Rust Initiative (http://www.globalrust.org/index.html). Common maize rust caused by Puccinia sorghi is a major disease problem of maize, particularly in tropical and subtropical regions such as South Africa and India. The Asian soybean rust caused by Phakopsora pachyrhizi has recently spread to Africa and the western hemisphere, including the U.S., and is now a major concern in most of the soybean-growing regions of the world.
Rust fungi have extremely complex life cycles, involving up to five different spore-producing stages. Many rusts are heteroecious, requiring two phylogenetically distinct host plants to complete their life cycle. For example, the wheat rust Puccinia graminis alternates between wheat as the primary host and barberry as the alternate host, and Melampsora epitea willow-conifer rusts alternate between a coniferous primary host, such as hemlock or tamarack, and a willow alternate host. Some rusts, such as the flax rust Melampsora lini, are autoecious and complete their life cycle on a single host plant.
Rust fungi are host specific and will develop compatible or incompatible associations with their host plants in a gene-for-gene manner, depending on the presence or absence of avirulence (Avr) genes in the pathogen and corresponding resistance (R) genes in the host. The interaction of M. lini with flax (Linum usitatissimum) has a central place in the history of plant pathology, as studies with this pathosystem led to the development of the gene-for-gene hypothesis of plant–pathogen interactions, one of the most important principles of plant pathology (Flor, 1942, 1955).
During infection of a host plant, rust fungi form haustoria, specialized infection structures that penetrate the plant cell wall and form invaginations in the plasma membrane that are believed to form the major sites of nutrient uptake from the host cell (Hahn and Mendgen, 2001). It is also thought that signals emanating from haustoria suppress host defense responses and facilitate disease in sensitive host plants (Panstruga, 2003; Vogele and Mendgen, 2003) or trigger a hypersensitive response (HR) leading to disease resistance in resistant hosts (i.e., interaction between Avr factors and host R gene products; Heath, 1997).
In this issue of The Plant Cell, Catanzariti et al. (pages 243–256) identify a number of avirulence elicitors (Avr proteins) encoded by genes expressed in haustoria of the flax rust M. lini (see figure). The previously identified AvrL567 proteins from flax rust are expressed in haustoria and contain secretory pathway signal peptides, suggesting that they are secreted into the extrahaustorial matrix (Dodds et al., 2004). In addition, all 19 of the flax R genes identified to date encode predicted cytoplasmic TIR-NBS-LRR proteins, suggesting that the corresponding rust Avr genes encode secreted proteins that somehow gain entry into the host plant cytoplasm. Therefore, the authors searched for haustorially expressed secreted proteins (HESPs) by screening a flax rust haustorium-specific cDNA library for putative secreted peptides.
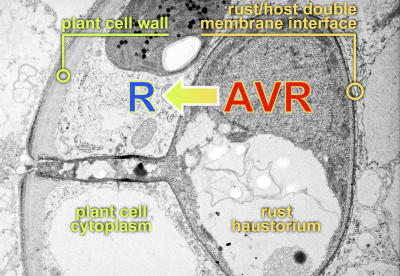
Figure 1.
Electron Micrograph of a Stem Rust Haustorium in a Wheat Mesophyll Cell.
The rust haustorium develops after penetration of the host cell wall by a fungal hypha but remains external to the host cell. Host-derived nutrients cross the host and then fungal plasma membranes to enter the haustorium. There is evidence that proteins, including Avr proteins, are secreted by the fungus across the haustorial membrane by type I secretion and are then taken up by the plant cell using as yet undefined transport mechanisms. Host resistance results from recognition of Avr proteins by host cytoplasmic R proteins. In the absence of R protein recognition, it is believed that Avr proteins are effectors of pathogen virulence. (Figure courtesy of Peter Dodds, Jeff Ellis, and Tony Pryor.)
Among 21 HESPs identified, four cosegregated with the known flax rust avirulence loci AvrL567, AvrM, AvrP4, and AvrP123. AvrL567, previously cloned by Dodds et al. (2004), encodes a novel small secreted protein that induces an HR dependent on the presence of the cytoplasmic NBS-LRR L5, L6, or L7 resistance proteins in the host plant. Catanzariti et al. focused on characterization of AvrM and AvrP4 and to a lesser extent AvrP123. Transient expression of AvrM, AvrP4, and AvrP123 induced HR-like responses in flax leaves dependent on the presence of functional corresponding resistance alleles M, P4, and P1/P2, respectively, indicating that the genes encode functional avirulence proteins.
AvrP123 is a complex locus corresponding to flax resistance genes P1, P2, and P3. The authors identified an HESP corresponding to one gene at this locus, which they named AvrP123-A, and genomic DNA gel blot analysis suggested the presence of several additional homologs. The gene was found to encode a small Cys-rich secreted protein that contains a sequence characteristic of Kazal Ser protease inhibitors, suggesting a possible function in pathogenesis of M. lini as an inhibitor of host proteases. AvrP4 was also found to encode a small Cys-rich secreted protein and may have a Cys knot structure similar to that found in some inhibitors of receptors or proteases. AvrP4 corresponded to a single gene, present in different rust strains as an avirulence (Avr) or virulence (avr) allele.
AvrM was found to be a complex locus that encodes at least six predicted homologous proteins. Five of the sequences segregated with the avirulence allele and were labeled AvrM-A, -B, -C, -D, and -E, whereas the sixth corresponded to the virulence allele (avrM). The AvrM proteins showed no similarities to other known proteins or peptide motifs. The predicted avrM (virulence) protein was unique among the six in having two in-frame deletions and 10 unique amino acids not shared by the AvrM proteins. Analysis of genomic sequence surrounding the set of AvrM genes suggests that they evolved from large duplication events.
Nucleotide variation at both the AvrM and AvrP4 loci showed evidence for diversifying selection acting on these genes. Sequence analysis showed an excess of nonsynonymous (i.e., leading to amino acid changes) over synonymous changes between the virulence allele avrM compared with the five AvrM avirulence alleles and an excess of nucleotide substitutions in the coding regions of AvrM alleles compared with flanking DNA. An excess of nucleotide substitutions was also found between the AvrP4 gene and the virulent avrP4 allele compared with that of flanking DNA. These results suggest that positive selection has acted on the divergence of AvrP4 and avrP4 alleles and on the accumulation of sequence differences between AvrM homologs and the avrM allele.
The authors conducted experiments to determine whether the signal peptides of AvrM and AvrP4 function as secretion signals in the plant to direct the protein through the endoplasmic reticulum (ER). They show that addition of the HDEL ER retention signal to the C terminus inhibited the necrotic response induced by the full-length AvrM-A protein. Importantly, addition of the chemically similar peptide HDDL, which does not function as an ER retention signal, did not affect AvrM-A recognition. These results showed that the predicted signal peptide is functional in plants, and when expressed in plants, the AvrM protein is secreted and reenters the cytoplasm from the apoplast. Addition of either HDEL or HDDL signals abolished recognition of AvrP4 in planta, indicating that the presence of these additional amino acids prevented recognition of this protein irrespective of ER retention. However, truncated AvrP4 lacking the signal peptide yielded significantly reduced recognition (as judged by the occurrence of a necrotic response) in flax carrying the corresponding resistance P4 gene, whereas replacement of the predicted AvrP4 signal peptide with a 44–amino acid plant secretion signal sequence restored recognition, suggesting a requirement for secretion for full recognition of AvrP4. It also may be important to acknowledge that AvrP4 lacking the signal peptide was still capable of causing a degree of necrosis (although much less than that of the secreted AvrP4), indicating that there was some recognition of the nonsecreted peptide, and this must have occurred in the host cytoplasm.
In addition, the Cys-rich AvrP4 contains six Cys residues in the last 28 amino acids that comply with the spacing consensus of inhibitor Cys knot structures, which is similar to the Cladosporium fulvum AVR9 protein. Several other Cys-rich avirulence proteins are subject to posttranslational processing, including C. fulvum AVR9 (Van den Ackerveken et al., 1993) and Fusarium oxysporum SIX1 (Rep et al., 2004). The authors suggest that passage of AvrP4 through the oxidizing environment of the ER may be required for the formation of disulfide bonds. They further show that mutation of two Cys residues in the last 28 amino acids caused a loss of recognition, as expected if these residues participate in structurally important disulfide bonds.
The work of Catanzariti et al. represents an important contribution to the study of the pathogenicity mechanism of biotrophic parasites in general and rust fungi in particular. It also adds to our understanding of the development of resistance to rust diseases in plants. Taken together, the data strongly suggest the existence of a transport mechanism in the host plasma membrane for secreted Avr proteins to gain entry to the host cell. In a recent study of the rust fungi Uromyces fabae and Uromyces striatus, which infect Vicia faba and Medicago truncatula, respectively, Kemen et al. (2005) reported the identification of haustorially expressed novel proteins (Uf-RTP1p and the homologous Us-RPT1p) that localized to the extrahaustorial matrix and entered the host cell cytoplasm by an unknown mechanism, which further supports this hypothesis. Catanzariti et al. discuss two possibilities for this translocation mechanism: a specialized translocation apparatus produced by the rust fungus itself or a host plant transport mechanism. Their experiments with transient expression of the M. lini proteins suggest that the secreted proteins are able to enter the host cytoplasm in the absence of the pathogen. In addition, the described method of screening fungal haustorium ESTs to search for secreted peptides may be applicable to other biotrophic pathogens, such as other rust species as well as downy and powdery mildews that cause severe diseases on a variety of monocotyledonous and dicotyledonous crop species.
References
- Catanzariti, A.-M., Dodds, P.N., Lawrence, G.J., Ayliffe, M.A., and Ellis, J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A., Ayliffe, M.A., and Ellis, J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1942). Inheritance of pathogenicity in Melampsora lini. Phytopathology 32 653–696. [Google Scholar]
- Flor, H.H. (1955). Host-parasite interactions in flax rust—Its genetics and other implications. Phytopathology 45 680–685. [Google Scholar]
- Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr. Opin. Plant Biol. 4 322–327. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. (Lond.) 80 713–720. [Google Scholar]
- Kemen, E., Kemen, A.C., Rafiqi, M., Hempel, U., Mendgen, K., Hahn, M., and Voegele, R.T. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant-Microbe Interact. 18 1130–1139. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6 320–326. [DOI] [PubMed] [Google Scholar]
- Rep, M., van der Does, H.C., Meijer, M., van Wijk, R., Houterman, P.M., Dekker, H.L., de Koster, C.G., and Cornelissen, B.J.C. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53 1373–1383. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken, G.F.J.M., Vossen, P., and de Wit, P.J.G.M. (1993). The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 103 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogele, R.T., and Mendgen, K. (2003). Rust haustorium: Nutrient uptake and beyond. New Phytol. 159 93–100. [DOI] [PubMed] [Google Scholar]