Abstract
Gibberellic acid (GA) promotes germination, stem/hypocotyl elongation, and leaf expansion during seedling development. Using activation-tagging mutagenesis, we identified a mutation, sob2-D (for suppressor of phytochromeB-4 [phyB-4]#2 dominant), which suppresses the long-hypocotyl phenotype of a phyB missense allele, phyB-4. This mutant phenotype is caused by the overexpression of an APETALA2 transcription factor, SOB2, also called DRN-like. SOB2/DRN-like transcript is not detectable in wild-type seedling or adult tissues via RT-PCR analysis, suggesting that SOB2/DRN-like may not be involved in seedling development under normal conditions. Adult sob2-D phyB-4 plants have curled leaves and club-like siliques, resembling plants that overexpress a closely related gene, LEAFY PETIOLE (LEP). Hypocotyls of a LEP-null allele, lep-1, are shorter in the light and dark, suggesting LEP involvement in seedling development. This aberrant hypocotyl phenotype is due at least in part to a delay in germination. In addition, lep-1 is less responsive to GA and more sensitive to the GA biosynthesis inhibitor paclobutrazol, indicating that LEP is a positive regulator of GA-induced germination. RT-PCR shows that LEP transcript accumulates in wild-type seeds during imbibition and germination, and the transcript levels of REPRESSOR OF ga1-3-LIKE2 (RGL2), a negative regulator of GA signaling during germination, is unaffected in lep-1. These results suggest LEP is a positive regulator of GA-induced germination acting independently of RGL2. An alternative model places LEP downstream of RGL2 in the GA-signaling cascade.
INTRODUCTION
Germination, one of the key steps in seedling development, occurs when a dormant seed begins to imbibe water and is complete when the embryonic axis, or radicle, elongates (Bewley, 1997). In order for the radicle to begin elongating, a number of external signals, such as proper temperature and light quality are required. In many species, including Arabidopsis thaliana, germination is promoted by exposure to lower temperatures during imbibition (Cone and Spruit, 1983). Light quality is also critical as red light promotes germination and can often be photoreversed by far-red light (Borthwick et al., 1952). Light also affects the synthesis of internal signals required for germination (Kamiya and Garcia-Martinez, 1999).
The key internal signals required for germination include plant hormones, the most influential being abscisic acid (ABA) and gibberellins (GA). ABA inhibits germination as is evident by the loss of dormancy in ABA-insensitive mutants (Koornneef et al., 1984; Finkelstein, 1994). GA plays an essential role in promoting germination, since the Arabidopsis GA biosynthetic mutant ga1-3 does not germinate without application of exogenous GA (Koornneef et al., 1983; Sun et al., 1992). In addition, chemical inhibitors of GA biosynthesis, such as paclobutrazol (PAC), inhibit germination (Jacobsen and Olszewski, 1993), indicating that following imbibition, de novo synthesis of GA is required for germination in Arabidopsis.
Loss-of-function mutations in REPRESSOR OF ga1-3-LIKE2 (RGL2) can germinate in the presence of PAC, suggesting it is a negative regulator of GA signaling during germination (Lee et al., 2002). RGL2 encodes a member of the GRAS family of putative transcriptional regulators and contains a DELLA domain, which is important for protein stability (Sun and Gubler, 2004). RGL2 expression is induced by imbibition and is downregulated both transcriptionally and posttranslationally by GA during germination (Lee et al., 2002; Tyler et al., 2004). Like RGL2, another DELLA-containing protein, RGL1 may be a negative regulator of GA-induced germination, since a RGL1-silenced line is resistant to the affects of PAC (Wen and Chang, 2002). However, this phenotype could not be confirmed with a T-DNA insertional mutation in RGL1, suggesting RGL1 may not be regulating germination (Tyler et al., 2004).
Other than RGL2 and possibly RGL1, few GA signaling components involved in germination have been identified. Using activation-tagging mutagenesis, we have indirectly identified a new GA signaling component affecting germination. The activation-tagged sob2-D phyB-4 (for suppressor of phytochromeB-4 [phyB-4]#2 dominant) mutant phenotypes are caused by the misexpression of a putative APETALA2 (AP2) transcription factor, SOB2, which has also been called DRN-like (Kirch et al., 2003). Though SOB2/DRN-like is likely not involved in seedling development, the sob2-D adult phenotype prompted us to genetically examine a similar AP2 transcription factor, LEAFY PETIOLE (LEP), which when overexpressed has a similar adult phenotype (van der Graaff et al., 2000).
LEP was also identified in an activation-tagging screen, where it was shown that LEP overexpression confers curled leaves lacking petioles and misshaped siliques in adult plants (van der Graaff et al., 2000). Interestingly, LEP is not expressed in adult tissues, and the loss-of-function mutant (lep-1) has no aberrant leaf phenotype (van der Graaff et al., 2000, 2002). However, LEP is expressed in seedling tissue (van der Graaff et al., 2000), suggesting that it may be involved in seedling development. Using the lep-1 mutant, we investigated LEP's possible role in seedling development, demonstrating that it is a positive regulator of GA-induced germination.
RESULTS
The sob2-D Mutation, Caused by the Overexpression of the AP2 Transcription Factor SOB2/DRN-like, Suppresses the Long-Hypocotyl Phenotype of phyB-4
Activation-tagging mutagenesis of a missense mutation in Arabidopsis phyB (phyB-4) was performed as by Ward et al. (2005). In this screen of ∼7000 primary transformants, we have identified and cloned six gain-of-function dominant mutations, which suppress the long-hypocotyl phenotype of phyB-4 (Turk et al., 2005; Ward et al., 2005; data not shown). This article will focus on one of these mutations, sob2-D, and a homologous gene LEP.
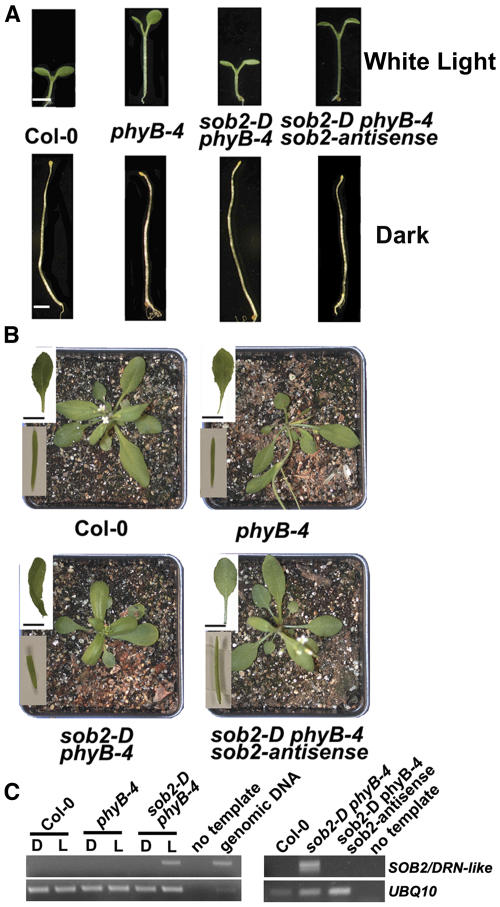
When grown in white light, the sob2-D phyB-4 double mutant had a dramatically shorter hypocotyl compared with the phyB-4 mutant (Figure 1A). However, the sob2-D phyB-4 hypocotyl elongated normally in the dark (Figure 1A), suggesting that the gene responsible for this mutant phenotype is involved in light signaling. As an adult, the sob2-D phyB-4 mutant had curled leaves lacking petioles and irregularly shaped siliques (Figure 1B). Segregation analysis suggested that the sob2-D phyB-4 mutant contained a single locus insertion, and DNA gel blot analysis suggested that there were multiple T-DNA insertions in a head-to-tail pattern at this locus (data not shown).
Figure 1.
Phenotypic Analysis of sob2-D phyB-4 and Cloning of the SOB2 Gene.
(A) Seedlings were grown in continuous white light (∼35 μM/m2/s) or in the dark for 5 d. Bars = 2 mm.
(B) Three-week-old plants were grown in long-day (16 h light/8 h dark) growth conditions. The top inset shows rosette leaf. Bars = 10 mm. The bottom inset shows mature silique.
(C) Total RNA was isolated from 5-d-old seedlings grown in continuous light (L) or in the dark (D). PCR was performed on cDNA using SOB2/DRN-like–specific primers for 24 cycles. The UBQ10 cDNA, amplified for 24 cycles, was used to normalize the amount of cDNA in each of the samples.
Genomic DNA flanking the T-DNA insertion was cloned via plasmid rescue, and the resulting plasmid was sequenced. BLAST analysis of this sequence showed that the T-DNA was inserted into chromosome I. The nearest open reading frame (SOB2; At1g24590) encodes a 306–amino acid protein containing one AP2 DNA binding domain. The SOB2 gene has been previously termed DRN-like, and when overexpressed in a wild-type background, is a dwarf plant with alterations in silique shape (Kirch et al., 2003), very similar to the sob2-D phyB-4 mutant phenotype.
RT-PCR analysis showed that this AP2 transcription factor, SOB2/DRN-like, was overexpressed and light regulated in sob2-D phyB-4 seedlings (Figure 1C). Though the activation-tagging enhancer elements should enhance the endogenous expression levels of the tagged gene, no transcript was detected in the wild-type or phyB-4 mutant seedlings after one round of PCR amplified for 24 cycles (Figure 1C). A second round of PCR was performed on dilute template from the first round, and still no transcript was detected (data not shown), suggesting that SOB2/DRN-like is not expressed in 5-d-old seedlings. Thus, it is likely that the light regulation of SOB2/DRN-like transcript in the sob2-D phyB-4 mutant is an artifact of activation tagging.
To confirm that SOB2/DRN-like overexpression is responsible for the mutant phenotype, sob2-D phyB-4 plants were transformed with a T-DNA harboring a SOB2-antisense cDNA. Resulting transgenic plants, which had reduced SOB2/DRN-like transcript, reverted back to the seedling and adult phyB-4 mutant phenotypes (Figure 1), demonstrating that SOB2/DRN-like overexpression is responsible for the sob2-D phyB-4 mutant phenotype.
Since the sob2-D phyB-4 mutant had noticeable adult phenotypes (Figure 1B), RT-PCR was performed on wild-type adult tissues. SOB2/DRN-like transcript could not be detected from RNA isolated from rosette and cauline leaves, stems, flowers, or roots (see Supplemental Figure 1 online). It has been previously reported that SOB2/DRN-like transcript accumulates in petals and stamen, and there is one EST that has been reported for SOB2/DRN-like (Gong et al., 2004); however, we have been unable to confirm the presence of this transcript by RT-PCR. In addition, the Massively Parallel Signature Sequencing database has been unable to detect any of the signatures specific to this putative gene (Meyers et al., 2004). Because we were unable to detect SOB2/DRN-like transcript during normal seedling or adult development, we focused our attention on related AP2 transcription factors that may be involved in seedling development.
LEP-Overexpressing and sob2-D phyB-4 Lines Have Similar Adult and Seedling Phenotypes
The family of AP2 transcription factors contains 144 members, all of which have at least one AP2 DNA binding domain (Riechmann and Meyerowitz, 1998; Sakuma et al., 2002). These proteins have been grouped into subfamilies based on the number of AP2 domains and the presence or absence of other domains (Sakuma et al., 2002). The dehydration-responsive element binding (DREB or A-subfamily) and ethylene response factor1-like (ERF1-like or B-subfamily) subfamilies are a group of ∼120 proteins containing just one AP2 domain and a conserved WLG domain. Protein alignments of the AP2 DNA binding domains of the B-subfamily allow further organization into distinct subgroups, B-1 through B-6 (Gutterson and Reuber, 2004).
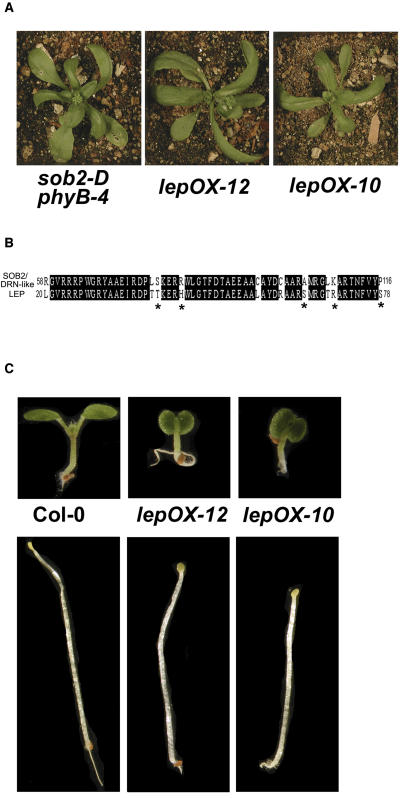
A protein alignment using the AP2 domains of the Arabidopsis B-1 subgroup showed that SOB2/DRN-like is most similar to At1g12980 (see Supplemental Figure 2 online), which has been characterized and previously named by two independent labs. At1g12980 has been named ENHANCER OF SHOOT REGENERATION1 (ESR1) (Banno et al., 2001) and DORNRÖSCHEN (DRN) (Kirch et al., 2003). Although the alignment suggests these are homologs (see Supplemental Figure 2 online), overexpression of ESR1/DRN in Arabidopsis results in premature arrest of shoot meristem activity, a phenotype, which does not resemble the sob2-D phyB-4 mutant or the SOB2/DRN-like overexpression phenotype in a wild-type background (Kirch et al., 2003). The only other member from the B-1 subfamily that has been characterized is LEP (At5g13910) (van der Graaff et al., 2000). Interestingly, overexpression of LEP results in plants that look very similar to the sob2-D phyB-4 mutant (Figure 2A; van der Graaff et al., 2000).
Figure 2.
LEP-OX Adult and Seedling Phenotypes.
(A) Four-week-old plants were grown in long-day growth conditions.
(B) Protein sequence of the SOB2/DRN-like and LEP AP2 domains. Identical residues are highlighted in black, and similar residues are marked with an asterisk.
(C) Seedlings were grown in continuous white light (∼35 μM/m2/s) or in the dark for 5 d.
LEP was identified in an activation-tagging screen targeting genes involved in leaf development (van der Graaff et al., 2000). Like the sob2-D phyB-4 mutant, LEP-overexpressing (LEP-OX) plants have curled leaves that lack petioles (Figure 2A) as well as irregularly shaped siliques (van der Graaff et al., 2000). SOB2/DRN-like and LEP have 82% amino acid identity and 90% similarity within the AP2 domain (Figure 2B). Like most of the AP2 transcription factors, these two proteins have lower amino acid identity (36%) and similarity (64%) outside of the DNA binding domain (see Supplemental Figure 3 online). In addition to the similar protein sequences and overexpression phenotypes, LEP, unlike SOB2/DRN-like, is expressed in wild-type seedling tissues (van der Graaff et al., 2000).
To determine if LEP may be involved in seedling development, we grew LEP-OX lines in the light and dark. These lines had short-hypocotyl phenotypes in both conditions (Figure 2C). Taken together, these data suggest that LEP may be involved in seedling development. To complement our gene overexpression studies, which can cause neomorphic phenotypes, we focused the rest of this study on a previously identified loss-of-function allele, lep-1 (van der Graaff et al., 2002), to determine if LEP plays an additional role in seedling development.
lep-1 Has a Short Hypocotyl in the Light and Dark, Which Is Caused at Least in Part by a Delay in Germination
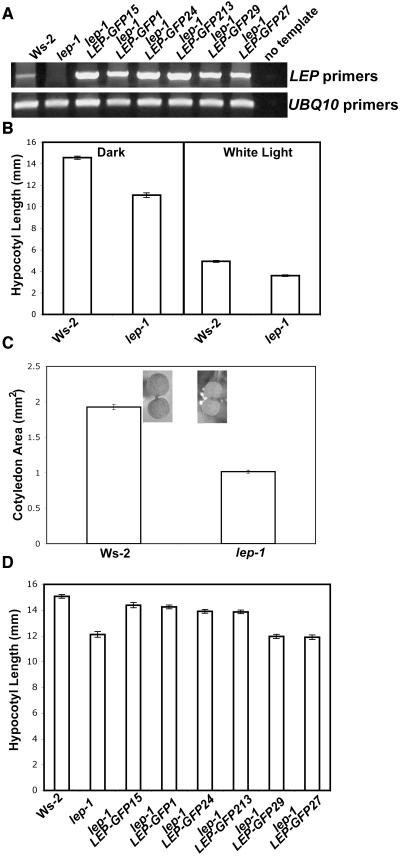
The lep-1 mutant contains a T-DNA insertion 424 bp 3′ from the LEP translational start site (see Supplemental Figure 4A online; van der Graaff et al., 2002). LEP transcript could not be detected in the lep-1 mutant with primers that amplified the whole gene (Figure 3A; van der Graaff et al., 2002). Primers that amplified a portion of the LEP gene 5′ of the T-DNA insertion detected a transcript, which contained the T-DNA (see Supplemental Figure 4B online). The portion of the LEP transcript 3′ of the T-DNA insertion could not be detected via RT-PCR (see Supplemental Figure 4B online). Taken together, these data suggest that the LEP transcript present in the lep-1 mutant is likely not functional.
Figure 3.
lep-1 Light- and Dark-Grown Seedling Phenotypes.
(A) Total RNA was isolated from 5-d-old seedlings grown in continuous white light. PCR was performed on cDNA using LEP-specific primers for 40 cycles. The UBQ10 cDNA, amplified for 26 cycles, was used to normalize the amount of cDNA in each of the samples.
(B) Seedlings were grown in the dark or in continuous white light (∼35 μM/m2/s) for 5 d.
(C) Seedlings were grown in continuous white light (∼35 μM/m2/s) for 5 d.
(D) Seedlings were grown for 5 d in the dark.
Bars = ±1 se.
The lep-1 mutant has no aberrant adult phenotypes (see Supplemental Figure 4C online; van der Graaff et al., 2002). However, it did have a shorter hypocotyl in the light and in the dark (Figure 3B), and the light-grown cotyledons were smaller (Figure 3C), suggesting that LEP is involved in seedling development. To confirm that lack of LEP transcript causes the aberrant hypocotyl phenotype, the lep-1 mutant was transformed with a T-DNA harboring a constitutively expressed LEP-green fluorescent protein (GFP) translational fusion. In the T2 generation, we identified independent lines, segregating this LEP-GFP transgene, which no longer conferred the short-hypocotyl phenotype in the dark (Figure 3D). These lines, which rescued the lep-1 phenotype, had increased LEP transcript accumulation (Figure 3A), suggesting that the lep-1 mutant seedling phenotype is caused by a loss of functional LEP transcript.
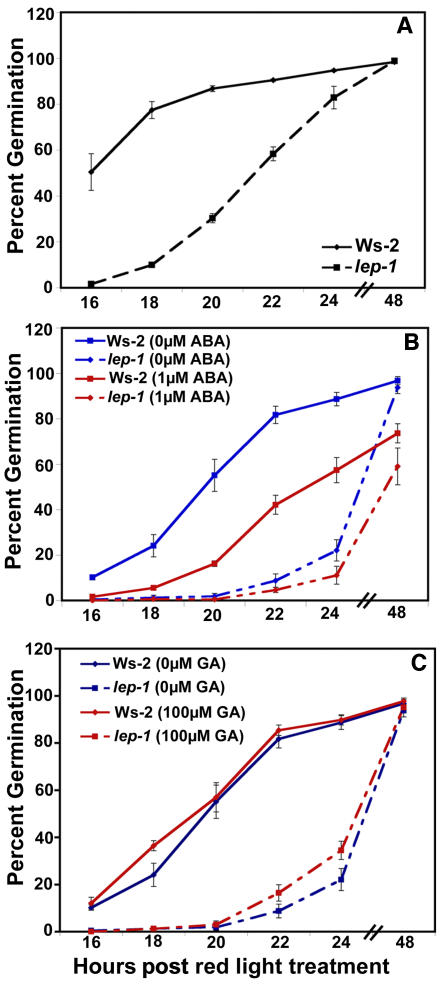
The lep-1 mutation conferred short hypocotyls and small cotyledons when compared with the wild type, suggesting that this mutant is either developmentally delayed or defective in cell elongation. This short-hypocotyl phenotype was caused at least in part by a delay in germination of 6 to 8 h for lep-1 compared with the wild type (Figure 4A). The lep-1 mutant responded like the wild type to ABA (Figure 4B), and neither the mutant nor the wild type were affected after the red light treatment when GA was included in the media (Figure 4C). Together, these results suggest that LEP is a positive regulator of germination and may be affecting germination via a novel mechanism.
Figure 4.
Phenotypic Analysis of the lep-1 Mutant in Response to ABA and GA during Germination.
(A) Seeds were incubated for 4 d at 4°C on plates with no hormones and then treated with 1 h of red light to induce germination.
(B) Seeds were germinated as in (A) but on plates containing 1 μM ABA.
(C) Seeds were germinated as in (A) but on plates containing 100 μM GA.
Germination was measured by radicle emergence. Bars = ±1 se.
LEP Is Expressed during Germination
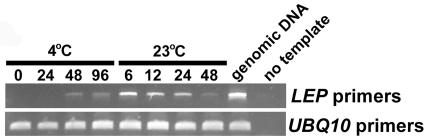
To determine if LEP is expressed during germination, RNA was isolated from seeds that were incubated in water at 4°C for 4 d followed by 2 d at 23°C. RT-PCR analysis showed that LEP transcript began to accumulate after incubation for 48 h at 4°C (Figure 5). After the transition from 4 to 23°C, there was a large increase in LEP transcript accumulation (Figure 5). LEP transcript accumulation remained high during germination, which in the wild type was complete after 24 h following the transition to a higher temperature (Figure 4). This pattern of LEP transcript accumulation is similar to RGL2, a gene involved in germination (Lee et al., 2002), further supporting a role for LEP in this developmental process.
Figure 5.
LEP Transcript Accumulation during Imbibition and Germination.
Seeds were incubated in sterile water for the time (hours) indicated at each temperature. The seeds were transitioned to 23°C after being incubated for 96 h at 4°C. Total RNA was isolated from seeds, and PCR was performed on the cDNA using the LEP primers for 35 cycles. The UBQ10 cDNA amplified for 28 cycles was used to normalize the cDNA templates.
lep-1 Is Less Responsive to GA and More Responsive to PAC during Germination
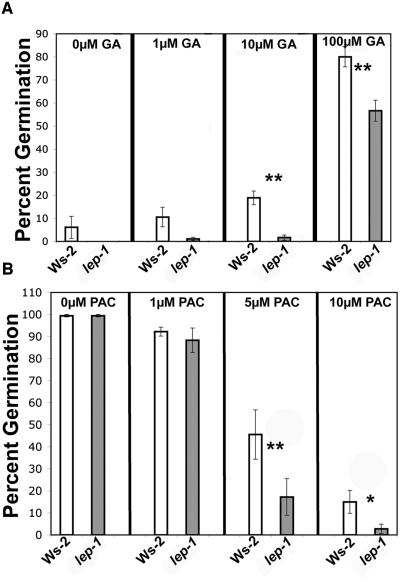
Additional GA seemed to have no effect on the timing of wild-type or lep-1 germination after the red light treatment (Figure 4C). However, if the red light stimulus was removed, both the wild type and lep-1 responded to GA (Figure 6A). Interestingly, when germinated in the dark, the lep-1 mutant was less responsive to multiple concentrations of GA compared with the wild type (Figure 6A). In addition, the lep-1 mutant was more responsive to the GA-biosynthesis inhibitor PAC when seeds were germinated in the light (Figure 6B). These aberrant responses to GA and PAC by the lep-1 mutant suggest that LEP is a positive regulator of GA-induced germination.
Figure 6.
Phenotypic Analysis of the lep-1 Mutant in Response to GA and PAC during Germination.
(A) Seeds were incubated for 4 d at 4°C on plates containing varying concentrations of GA and then placed in the dark without red light treatment for 5 d at 25°C.
(B) Seeds were incubated as in (A) but on plates containing varying concentrations of PAC then placed in white light for 5 d at 25°C.
Germination was measured by radicle emergence. Bars = ±1 se. One asterisk indicates P < 0.05, and two asterisks indicate P < 0.005 from a Student's paired two-tail t test comparing the mutant and its control at each hormone concentration.
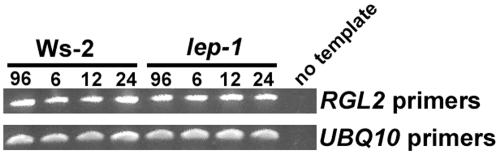
A small family of negative regulators of GA signaling, which contain a DELLA domain, have been identified in Arabidopsis (Sun and Gubler, 2004). One of these proteins, RGL2, has been shown to be involved in GA-induced germination (Lee et al., 2002). Since RGL2 is regulated transcriptionally during germination and appears to have an overlapping expression pattern with LEP (Figure 5; Lee et al., 2002), RT-PCR analysis was performed on seeds to determine if RGL2 transcript levels were altered in the lep-1 mutant. In the wild type and lep-1 mutant, there was no difference in transcript accumulation of RGL2 (Figure 7). We also tested the transcript accumulation of RGL1, which may be involved in germination, and it too was unaffected by the genetic state of LEP (see Supplemental Figure 5 online). Taken together, these data suggest models where LEP is functioning in the GA signaling pathway either downstream or independently of the DELLA proteins (Figure 8).
Figure 7.
RGL2 Expression in Wild-Type and lep-1 Seeds during Germination.
Seeds were incubated as described in Figure 4 for 96 h at 4°C followed by either 6, 12, or 24 h at 23°C. cDNA was synthesized from total RNA, and RGL2 cDNA was amplified for 30 cycles. The UBQ10 cDNA amplified for 28 cycles was used to normalize the cDNA templates.
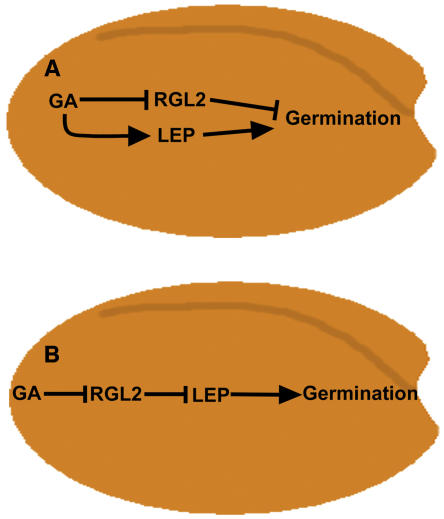
Figure 8.
Possible Models Describing the Role of LEP in GA Signaling during Germination.
(A) LEP is a positive regulator of GA-induced germination. In a parallel signaling cascade, GA promotes germination by affecting the transcription and protein stability of RGL2, a negative regulator of germination.
(B) GA negatively affects RGL2, allowing a downstream signaling component, LEP, to positively regulate germination.
DISCUSSION
SOB2/DRN-Like Misexpression Leads to the Identification of a New Role for LEP in Germination
Using activation-tagging mutagenesis, we have identified the AP2 transcription factor SOB2/DRN-like, which when overexpressed suppressed the long-hypocotyl phenotype of a weak allele of phyB (Figure 1). We were unable to detect SOB2/DRN-like expression in wild-type seedling and adult tissues, suggesting that this gene does not play a prominent role in normal development; however, a sob2-null allele is needed to test this hypothesis. SOB2/DRN-like has high protein sequence similarity to another AP2 transcription factor (see Supplemental Figure 2 online), ESR1/DRN, which has been identified in two independent overexpression screens (Banno et al., 2001; Kirch et al., 2003).
ESR1 was identified by overexpressing cDNAs to identify genes involved in cytokinin-induced shoot formation (Banno et al., 2001). ESR1 expression is induced by cytokinin and when overexpressed increases the efficiency of shoot regeneration from callus (Banno et al., 2001). The activation-tagged drn-D mutant, which is caused by the overexpression of DRN, aborts shoot apical meristem growth prematurely (Kirch et al., 2003). DRN is expressed in meristem stem cells and when overexpressed affects the expression of CLAVATA3 and WUSCHEL, which are involved in meristem stem cell fate (Kirch et al., 2003).
Despite the high protein sequence similarity between SOB2/DRN-like and ESR/DRN, ESR/DRN-overexpressing plants do not resemble the sob2-D phyB-4 mutant (Kirch et al., 2003), suggesting that sequence similarity may not be the best indicator of function. However, another closely related AP2 transcription factor, LEP, when overexpressed has very similar adult phenotypes as sob2-D phyB-4 (Figure 2; van der Graaff et al., 2000). LEP and SOB2/DRN-like also have high protein sequence similarity within the AP2 DNA binding domain (Figure 2).
LEP was identified in an activation-tagging screen for mutations that alter leaf development (van der Graaff et al., 2000). LEP-OX lines, like the sob2-D phyB-4 mutant, have curled leaves, which lack petioles, and misshaped siliques (Figure 2; van der Graaff et al., 2000). Interestingly, LEP is not expressed in mature leaves but is expressed in young shoots and leaf primordia (van der Graaff et al., 2000). A T-DNA insertional mutant, lep-1, has no aberrant adult phenotypes, suggesting that another protein may be functionally redundant to LEP with regards to modulating leaf development (van der Graaff et al., 2002). Through analysis of the lep-1 mutant, we have identified a new role for this AP2 transcription factor in germination.
Although a number of AP2 transcription factors are involved in seed dehydration or dormancy, including DROUGHT-RESPONSIVE DRE/CRT binding PROTEIN2, maize (Zea mays) DRE binding FACTOR1, ABA-INSENSITIVE4, Hordeum vulgare DEHYDRATION-RESPONSE FACTOR1, and Triticum aestivum DRE binding PROTEIN1, very few AP2 transcription factors have been identified that are directly involved either in the promotion of germination or GA biosynthesis/signaling (Finkelstein et al., 1998; Liu et al., 1998; Kizis and Pages, 2002; Shen et al., 2003; Xue and Loveridge, 2004). DWARF AND DELAYED-FLOWERING1 was identified in activation-tagging mutagenesis and when overexpressed has decreased levels of bioactive GAs (Magome et al., 2004). Herein, we have identified a new role for another AP2 transcription factor, LEP, in the promotion of germination by positively regulating GA biosynthesis or signaling.
New Role for LEP: Positive Regulator of GA-Induced Germination
To determine if LEP is involved in seedling development, the lep-1 mutant was grown in the light and dark (Figure 3). The lep-1 mutant had a short hypocotyl in the light and in the dark, and this phenotype was due at least in part to a delay in germination (Figure 4A). Together with LEP mRNA accumulation during imbibition and germination (Figure 5), this mutant phenotype suggests that LEP is a positive regulator of germination. The lep-1 mutant responded normally to ABA (Figure 4B); however, it was less responsive to GA when germinated in the dark (Figure 6A). In addition, lep-1 was more responsive to the GA biosynthesis inhibitor PAC (Figure 6B). Together, these data suggest LEP is not involved in the interaction between the ABA and GA signaling pathways but is a positive regulator of GA signaling during germination. Interestingly, LEP may be affecting signaling by altering GA transport since LEP-OX lines have an increased number of xylem cells (van der Graaff et al., 2002).
To date, no other mutations have been identified that show a delay in germination; however, other positive regulators of GA signaling have been identified in Arabidopsis. GA-INSENSITIVE DWARF2 and SLEEPY1 encode F-box proteins, which modulate GA responses by regulating the protein stability of at least some of the DELLA domain–containing negative regulators of GA signaling (McGinnis et al., 2003; Sasaki et al., 2003). In addition, mutations in SLEEPY1 result in a reduced ability to germinate (Steber et al., 1998; Steber and McCourt, 2001). PICKLE encodes a CHD3 chromatin remodeling factor, which when mutated resembles other GA-response mutants (Ogas et al., 1997). GTP BINDING (G) PROTEINα-SUBUNIT1 (GPA1) and G PROTEIN-COUPLED RECEPTOR1 (GCR1) encode the α-subunit of a heterotrimeric G protein and a seven-transmembrane cell-surface receptor, respectively (Ma et al., 1990; Colucci et al., 2002). gpa1 and gcr1 mutants are less sensitive to GA during germination, though double mutant analysis indicates they act independently of each other (Ullah et al., 2002; Chen et al., 2004). Thus far, LEP is the only transcriptional activator identified that is a positive regulator of GA-induced germination.
Current models of GA signaling during germination include the negative regulators RGL2 and possibly RGL1 (Sun and Gubler, 2004). RGL2 is downregulated transcriptionally and posttranslationally in response to GA during germination (Lee et al., 2002; Tyler et al., 2004). Since the genetic state of LEP does not affect the transcript accumulation of RGL1 or RGL2 (Figure 7; see Supplemental Figure 5 online), LEP may regulate GA signaling through a novel signaling cascade independent of these proteins. In our experimental conditions, RGL2 transcript accumulation was unaltered in wild-type seeds (Wassilewskija [Ws-2]) after a 24 h shift to 23°C (Figure 7), though the same treatment with a different ecotype (Landsberg erecta) conferred a reduction in RGL2 transcript levels (Lee et al., 2002). This difference could be attributed to the wild-type genetic background used in these experiments.
Based on the data presented here, we propose two possible models to explain the role that LEP plays during germination. The first model suggests that there are two GA-signaling cascades acting independently of each other (Figure 8A). The primary cascade includes RGL2, which acts as a negative regulator. GA promotes germination by altering RGL2 mRNA transcript and/or protein levels. In the second independent cascade, LEP positively modulates GA-induced germination.
In an alternative model, LEP is acting in conjunction with RGL2 via direct or indirect interactions to promote germination (Figure 8B). GA-induced reduction of RGL2 enables LEP and other transcription factors to promote germination possibly through activation of GA-responsive genes. A number of both transcriptional activators and repressors must play an important role in GA-induced germination, as microarray analysis of the GA-biosynthesis mutant, ga1-3, show that a number of genes are upregulated or downregulated in response to GA at different time points during germination (Ogawa et al., 2003). Transcription factors, such as LEP, that are positive regulators of GA signaling must be involved in promoting germination in order to cause the upregulation of genes in response to GA.
Lines overexpressing LEP suggest that its role in GA signaling may be more complex than the above models indicate (Figure 8). Strong LEP-OX lines are severe dwarfs and sterile, resembling mutants that are GA deficient or that constitutively express negative regulators of GA signaling. In addition, LEP-OX seedlings, like lep-1, have short hypocotyls in the light and in the dark (Figures 2 and 3), suggesting there is a deficiency in hormone production or response. Taken together, LEP may be negatively regulating the biosynthesis of GA through a feedback mechanism. There is precedent for GA signaling components affecting GA levels in Arabidopsis, since the constitutive GA-response mutant, repressor of ga1-3 #24, has lower transcript accumulation of a GA-biosynthetic gene (Dill and Sun, 2001).
In summary, we have identified a new role for the AP2 transcription factor LEP in GA-induced germination. A number of factors are important for germination to occur, two of the most critical being GA and light (Borthwick et al., 1952). The interconnection between light and GA during germination is not well understood, and LEP may represent a point of interaction between light and hormone signaling. The lep-1 aberrant germination response is unaffected by GA when a red light treatment is provided (Figure 4C); however, lep-1 is less responsive to GA when no light treatment is given (Figure 6), suggesting that both light and GA may play a critical role in LEP-induced germination. Future experiments will further explore the role of light on LEP-induced germination as well as the genetic and/or physical interactions between LEP and RGL2.
METHODS
Plant Materials and Growth Conditions
The phyB-4, sob2-D phyB-4, and LEP-OX mutants are in the Columbia (Col-0) ecotype. The LEP-OX plants are described in detail by van der Graaff et al. (2000). The lep-1 mutant, which contains a T-DNA in the LEP gene, is in the Wassilewskija (Ws-2) ecotype. The lep-1 mutant is from the Wisconsin knockout collection, and its isolation is described by van der Graaff et al. (2002).
Seeds were sterilized as by Ward et al. (2005) and were sown on media containing either 0.8% phytagar (w/v) (Gibco BRL), half-strength LS salts (PhytoTechnology Laboratories), and 1.5% sucrose with antibiotic or 1.0% phytagel (Sigma-Aldrich) and half-strength LS salts without antibiotic. After incubation for 4 d at 4°C, germination was induced by treating seeds with 1 h of red light (90 μM/m2/s) followed by 23 h in the dark. Seeds were then put in the appropriate light condition.
All chambers were at 25°C. White light was supplied as described by Ward et al. (2005). Red light was supplied by red light emitting diodes from an E-30-LED incubator (Percival Scientific).
Activation-Tagging Mutagenesis and Cloning of the SOB2 Gene
Arabidopsis thaliana phyB-4 mutant plants were transformed with the activation-tagging vector pSKI074 (GenBank accession number AF218466) (Weigel et al., 2000) as by Neff et al. (1999). Plants were transformed via the floral dip method using Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). Analysis of T1, T2, and T3 plants was performed as by Ward et al. (2005). Segregation ratios of T2 plants suggested that there were two linked T-DNAs in the sob2-D phyB-4 mutant. The mutant phenotype segregated in a 3:1 ratio, suggesting that this phenotype is caused by one of the T-DNAs, so the two T-DNAs were separated in the T2 and T3 generations. The presence of the phyB-4 mutation in the sob2-D phyB-4 mutant was confirmed as by Neff et al. (1999).
DNA gel blot analysis and plasmid rescue were performed essentially the same as by Neff et al. (1999). The DNA gel blot indicated that the sob2-D phyB-4 mutant contained multiple T-DNA insertions, although segregation analysis indicates that they were inserted in a single locus. The location of this T-DNA was determined by cloning flanking genomic DNA by plasmid rescue. Genomic DNA was digested with SpeI and religated with T4 DNA ligase (New England Biolabs), resulting in a ∼13.5-kb plasmid containing ∼8 kb of genomic DNA. The genomic DNA flanking the T-DNA was sequenced using the T7 primer (5′-GTAATACGACTCACTATAGGG-3′). BLASTn analysis indicated that the genomic DNA was from chromosome I, and the closest open reading frame was a putative AP2 transcription factor (At1g24590).
RT-PCR Analysis
Seedlings were grown for 5 d in continuous light (Figures 1 and 3) or in the dark (Figure 1). Total RNA was isolated from seedlings using the RNeasy Plant Mini kit (Invitrogen). Total RNA was isolated from 50 mg of seeds (Vicient and Delseny, 1999) and DNaseI treated using DNA-free (Ambion). cDNA synthesis and PCR was performed as by Ward et al. (2005). The SOB2/DRN-like gene was amplified using the following primers: AMLBG1F, 5′-GAAGCAATCTCTAGACTCGAAGGTGCCG-3′; AMLBG1R, 5′-GAAGAGCTCCCATTCTCATGATCAGCCC-3′.
The LEP gene was amplified using the following primers: SpeLEP-F, 5′-GGACTAGTATGAACACAACATCATC-3′; SpeLEP-R, 5′-GGACTAGTGGAGCCAAAGTAGTTG-3′.
The RGL2 gene was amplified using the following primers: RGL2-F, 5′-CCGAAATGTTCGAAACCCGACCC-3′; RGL2-R, 5′-TCAGGCGAGTTTCCACGCCGAGG-3′.
The following primers were used to characterize LEP transcript in the lep-1 mutant: LEP 5′ primers: SpeLEP-F; midLEP-R, 5′-GTCCACTTGATCACAATGAGGC-3′; 5′ T-DNA primers: SpeLEP-F; WILB, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′; 3′ T-DNA primers: GUS-F, 5′-GATTCACCACTTGCAAAGTCCC-3′; SpeLEP-R.
The RGL1 gene was amplified using the following primers: RGL1-F, 5′-CGGTCTTCGAGCTTCATCGCC-3′; RGL1-R, 5′-TTCCACACGATTGATTCGCCACGC-3′.
The ubiquitin10 (UBQ10) gene was used as an internal control to normalize each of the templates. UBQ10 was amplified using the following primers: RPED1, 5′-GGTATTCCTCCGGACCAGCAGC-3′; RPED2, 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGGAACGG-3′.
The linear range of accuracy for the detection of each transcript was determined by comparing samples run at different number of cycles. All RT-PCR reactions shown were completed at least in duplicate.
Generation of the SOB2-Antisense Construct
The SOB2/DRN-like gene was amplified from genomic DNA using the AMLBG1F and AMLBG1R primers. The PCR product, cut with SacI and KpnI, was ligated into pCHF1 cut with these same enzymes. The resulting plasmid was transformed into the sob2-D phyB-4 mutant via the floral dip method using Agrobacterium strain GV3101 (Clough and Bent, 1998). Transgenic seedlings were selected by sowing seeds on plates containing gentamycin (60 mg/L). T2 lines that segregated 3:1 resistant:sensitive were selected from self-fertilized T1 plants. The photograph in Figure 1 is from a T2 seedling, and gentamycin-resistant seedlings from this line were used in the RT-PCR analysis.
Generation of the LEP-GFP Construct
The LEP cDNA was amplified using the SpeLEP-F and SpeLEP-R primers and cut with the SpeI restriction endonuclease. The LEP cDNA was ligated into pCAMBIA1302 vector cut with SpeI. Colony PCR was performed with the SpeLEP-F and SpeLEP-R primers to determine if the LEP insert was present in the vector. DNA from colonies that contained the insert was isolated and cut with BsaI to determine the orientation of the insert. Plasmids containing the LEP insert in the correct orientation were sequenced to determine if the LEP cDNA sequence was correct. The resulting plasmid was transformed into the lep-1 mutant via the floral dip method using Agrobacterium strain GV3101 (Clough and Bent, 1998).
Transgenic seedlings were selected by sowing seeds on plates containing hygromycin (20 mg/L). T2 lines that segregated ∼3:1 resistant:sensitive were selected from the self-fertilized T1 plants, and these lines were grown in the dark on plates containing no selection. Many of the T1 plants exhibited severe LEP overexpression phenotypes and were sterile (data not shown). However, a subset of antibiotic resistant plants had wild-type or mild LEP-OX phenotypes, and these are the lines for which we examined hypocotyl length. The hypocotyls of these T2 plants were measured as by Ward et al. (2005). These T2 dark-grown seedlings were also used for the RT-PCR analysis in Figure 3.
Hypocotyl and Cotyledon Measurements and Germination Assays
Hypocotyls and cotyledons were measured as by Ward et al. (2005). For the germination assays, seeds were incubated in the dark at 4°C for 4 d on plates containing no hormone, ABA, GA (GA3), or PAC (PhytoTechnology Laboratories). Following this incubation, seeds were treated with red light (90 μM/m2/s) for 1 h to induce germination and finally placed in the appropriate light condition or in the dark. Germination was measured by radicle emergence. Radicle emergence was observed using the Nikon SMZ800 dissecting microscope.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under accession numbers At1g24590 (SOB2/DRN-like) and At5g13910 (LEP) and in GenBank under the accession number AF218466 (pSK1074).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SOB2 Expression in Wild-Type and sob2-D phyB-4 Adult Tissues.
Supplemental Figure 2. Phylogenic Analysis of the AP2 Domains of the B-1 Subfamily of AP2 Transcription Factors.
Supplemental Figure 3. Alignment of the SOB2/DRN-Llike and LEP Proteins.
Supplemental Figure 4. Identification of the lep-1 Mutation.
Supplemental Figure 5. RGL1 Expression in Wild-Type and lep-1 Seeds during Germination.
Supplementary Material
Acknowledgments
We thank Miguel A. Blazquez, John Chandler, Ian Street, and Jingyu Zhang for their critical review of this manuscript. We would also like to thank the anonymous reviewers and coeditor for their careful review and helpful criticisms. In addition, we thank them for their appreciation that scientific discovery often takes a circuitous path with several hard left turns. This work was directly supported by the Department of Energy (grant number DE-FG02-02ER15340 to M.M.N.). We are also grateful for support from the National Science Foundation (Grant 0114726 to M.M.N.) and from Monsanto Corporation (Grant 46011J to M.M.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael M. Neff (mneff@biology2.wustl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036707.
References
- Banno, H., Ikeda, Y., Niu, Q.W., and Chua, N.H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. (1997). Seed germination and dormancy. Plant Cell 9 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick, H.A., Hendricks, S.B., Parker, M.W., Toole, E.H., and Toole, V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.G., Pandey, S., Huang, J., Alonso, J.M., Ecker, J.R., Assmann, S.M., and Jones, A.M. (2004). GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 135 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colucci, G., Apone, F., Alyeshmerni, N., Chalmers, D., and Chrispeels, M.J. (2002). GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, J.W., and Spruit, C.J.P. (1983). Imbibition conditions and seed dormancy of Arabidopsis thaliana. Plant Physiol. 59 416–420. [Google Scholar]
- Dill, A., and Sun, T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5 765–771. [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson, N., and Reuber, T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 465–471. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, Y., and Garcia-Martinez, J.L. (1999). Regulation of gibberellin biosynthesis by light. Curr. Opin. Plant Biol. 2 398–403. [DOI] [PubMed] [Google Scholar]
- Kirch, T., Simon, R., Grunewald, M., and Werr, W. (2003). The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis, D., and Pages, M. (2002). Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30 679–689. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Plant Physiol. 61 377–383. [Google Scholar]
- Koornneef, M., van Eden, J., Hanhart, C., and de Jongh, A. (1983). Genetic fine-structure of the GA-1 locus in the higher plant Arabidopsis thaliana (L.) Henynh. Genet. Res. Camb. 41 57–68. [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1990). Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 87 3821–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37 720–729. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Lee, D.K., Vu, T.H., Tej, S.S., Edberg, S.B., Matvienko, M., and Tindell, L.D. (2004). Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol. 135 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Nguyen, S.M., Malancharuvil, E.J., Fujioka, S., Noguchi, T., Seto, H., Tsubuki, M., Honda, T., Takatsuto, S., Yoshida, S., and Chory, J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290 998–1009. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Shen, Y.G., Zhang, W.K., He, S.J., Zhang, J.S., Liu, Q., and Chen, S.Y. (2003). An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 106 923–930. [DOI] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 1998 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T., Goodman, H.M., and Ausubel, F.M. (1992). Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Turk, E.M., et al. (2005). BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 42 23–34. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Wang, S., and Jones, A.M. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff, E., Dulk-Ras, A.D., Hooykaas, P.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127 4971–4980. [DOI] [PubMed] [Google Scholar]
- van der Graaff, E., Hooykaas, P.J., and Keller, B. (2002). Activation tagging of the two closely linked genes LEP and VAS independently affects vascular cell number. Plant J. 32 819–830. [DOI] [PubMed] [Google Scholar]
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268 412–413. [DOI] [PubMed] [Google Scholar]
- Ward, J.M., Cufr, C.A., Denzel, M.A., and Neff, M.M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, G.P., and Loveridge, C.W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37 326–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.