Abstract
We used loss-of-function mutants to study three Arabidopsis thaliana sensor histidine kinases, AHK2, AHK3, and CRE1/AHK4, known to be cytokinin receptors. Mutant seeds had more rapid germination, reduced requirement for light, and decreased far-red light sensitivity, unraveling cytokinin functions in seed germination control. Triple mutant seeds were more than twice as large as wild-type seeds. Genetic analysis indicated a cytokinin-dependent endospermal and/or maternal control of embryo size. Unchanged red light sensitivity of mutant hypocotyl elongation suggests that previously reported modulation of red light signaling by A-type response regulators may not depend on cytokinin. Combined loss of AHK2 and AHK3 led to the most prominent changes during vegetative development. Leaves of ahk2 ahk3 mutants formed fewer cells, had reduced chlorophyll content, and lacked the cytokinin-dependent inhibition of dark-induced chlorophyll loss, indicating a prominent role of AHK2 and, particularly, AHK3 in the control of leaf development. ahk2 ahk3 double mutants developed a strongly enhanced root system through faster growth of the primary root and, more importantly, increased branching. This result supports a negative regulatory role for cytokinin in root growth regulation. Increased cytokinin content of receptor mutants indicates a homeostatic control of steady state cytokinin levels through signaling. Together, the analyses reveal partially redundant functions of the cytokinin receptors and prominent roles for the AHK2/AHK3 receptor combination in quantitative control of organ growth in plants, with opposite regulatory functions in roots and shoots.
INTRODUCTION
Cytokinin is a plant hormone that plays positive and negative regulatory roles in many aspects of plant growth and development. It stimulates the formation and activity of shoot meristems, is able to establish sink tissues, retard leaf senescence, inhibit root growth and branching, and plays a role in seed germination and stress responses. Cytokinin also appears to participate in a number of light-regulated processes, such as deetiolation and chloroplast differentiation (Mok, 1994). Some cytokinin functions are executed primarily through the control of cell cycle activity. Analysis of cytokinin-deficient plants has shown that cytokinin plays opposite roles in shoot and root meristems and has suggested that the hormone has an essential function in quantitative control of organ growth (Werner et al., 2001, 2003; Yang et al., 2003).
In Arabidopsis thaliana, the cytokinin signal is perceived by three sensor histidine kinases, AHK2, AHK3, and CRE1/AHK4 (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001). These three receptors show a high degree of sequence identity, but each has distinguishing characteristics. All contain an N-terminal membrane-associated sensor domain. The predicted extracellular ligand binding domain shares the so-called CHASE domain, which is found exclusively in cytokinin receptors of higher plants, as well as the ligand recognition domain of other histidine kinases and guanylyl cyclases of lower eukaryotes and bacteria (Anantharaman and Aravind, 2001; Mougel and Zhulin, 2001). On the predicted cytoplasmic side, all three receptor proteins contain a histidine kinase catalytic domain and a C-terminal response regulator containing a receiver domain. The current model of cytokinin signaling predicts that the receptors feed into the two-component signaling system, which transfers the signal via phosphorelay to the nucleus (reviewed in Hwang et al., 2002; Heyl and Schmülling, 2003; Kakimoto, 2003; Grefen and Harter, 2004; Ferreira and Kieber, 2005).
CRE1/AHK4 and AHK3 were shown to confer cytokinin-sensing ability to yeast (Saccharomyces cerevisiae), Escherichia coli, and tobacco (Nicotiana tabacum) protoplasts (Hwang and Sheen, 2001; Inoue et al., 2001; Suzuki et al., 2001; Yamada et al., 2001). CRE1/AHK4 and AHK3 prefer both the free bases isopentenyladenine and zeatin as a ligand but differ in the recognition of other cytokinin compounds (Yamada et al., 2001; Spíchal et al., 2004). Less is known about AHK2, but a function in cytokinin perception was reported (Hwang and Sheen, 2001; Higuchi et al., 2004; Nishimura et al., 2004). mRNA of all three receptor genes is found in all organs, although with different abundance. CRE1/AHK4 is predominantly expressed in the root, where its mRNA was mainly localized to the vascular cylinder and the pericycle of the root (Mähönen et al., 2000; Higuchi et al., 2004). Gel blot analyses and promoter–β-glucuronidase fusions have detected CRE1/AHK4 expression also in various shoot tissues, although in lower abundance than in the roots (Ueguchi et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004). The AHK2 and in particular the AHK3 gene show greater expression in the aerial parts of Arabidopsis plants (Ueguchi et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004).
Most known mutations of the CRE1/AHK4 gene do not cause a strong morphological plant phenotype (Inoue et al., 2001; Franco-Zorilla et al., 2002) but alter physiological parameters such as the phosphate starvation response and regulation of sulfate acquisition (Franco-Zorilla et al., 2002; Maruyama-Nakashita et al., 2004). Exceptions are the wooden leg alleles, a class of cre1/ahk4 mutations, which show a severe defect in the development of vascular tissue (Scheres et al., 1995; Mähönen et al., 2000; García-Ponce de León et al., 2004). Recently, a lack of morphological phenotypes was also reported for single ahk2 and ahk3 mutant plants. Combination of ahk2 and ahk3 mutations led to reduced shoot growth, which was further enhanced by introgression of a mutated cre1/ahk4 allele (Nishimura et al., 2004). A comprehensive analysis of all single, double, and triple mutants of the cytokinin receptor genes is missing.
The primary aim of this study was to investigate the role of each receptor and their combinations in various cytokinin-regulated processes during plant development. It was anticipated that the analyses of receptor mutants would be informative about processes for which cytokinin is a limiting and therefore possibly regulatory factor. We show that the receptor functions partially overlap but that each receptor contributes to a different extent to the cytokinin response in various assays. For example, the combined activities of the AHK2 and AHK3 receptors are particularly relevant to control organ growth and cytokinin-dependent retardation of senescence. Importantly, we discover an unexpected role for cytokinin in various aspects of seed biology and find further support for a negative regulatory role of cytokinin in controlling root growth under physiological conditions. Furthermore, a feedback control of cytokinin on its own steady state concentration is discovered. The data contribute to a better understanding of the in planta functions of cytokinin, which has long been hindered owing to the lack of loss-of-function mutants.
RESULTS
Isolation of Novel T-DNA Insertion Alleles in the AHK2 and AHK3 Genes
The initial isolation and characterization of cytokinin receptor mutants is illustrated in Supplemental Figures 1 and 2 online. Arabidopsis lines carrying T-DNA insertions in the AHK2 (At5g35750) and AHK3 (At1g27320) genes were identified in different mutant collections. Two independent mutant alleles were isolated for each gene (see Supplemental Figure 1A online). All mutations are in the Arabidopsis Columbia (Col-0) background. The exact T-DNA insertion sites were determined by sequencing (see Supplemental Figure 1B online). One insertion in the AHK2 gene was found to be in the fourth exon. This is a novel allele, which was named ahk2-5, as four other ahk2 insertion mutants were described previously (Higuchi et al., 2004; Nishimura et al., 2004). The second ahk2 mutant allele is identical to ahk2-2 (Nishimura et al., 2004); it carries an insertion 19 bp upstream of the predicted translational start. ahk3-7 is a novel insertion allele of the AHK3 gene and carries an insertion at the 5′ end of intron 1. The second isolated ahk3 mutant allele is identical to ahk3-3 (Higuchi et al., 2004); it has an insertion in the sixth intron (see Supplemental Figures 1A and 1B online). Supplemental Figure 1C online shows that using RT-PCR with primers spanning the insertion site, gene-specific transcripts were detected in the wild type but no gene-specific transcripts were detected in the ahk2-5 and ahk3-7 single mutants or in the ahk2-5 ahk3-7 double mutant. We conclude therefore that both mutations present novel null or strong hypomorphic alleles.
ahk2-5, ahk3-7, and cre1-2, which carries an insertion in the first exon (Inoue et al., 2001), were used for experiments with single knockouts and all reported receptor mutant combinations. In the figures, we use the designations ahk2, ahk3, and cre1 for these alleles. Experiments with the ahk2 ahk3 double mutants were performed using both the ahk2-5 ahk3-7 and ahk2-2 ahk3-3 allele combinations. In all cases, similar results were obtained with both combinations, but only those for ahk2-5 ahk3-7 are shown.
We used several assays to test the cytokinin sensitivity of ahk mutants, in particular that of the novel ahk2-5 and ahk3-7 alleles and their combination. Hypocotyl explants of 10-d-old seedlings were placed on media with different concentrations of auxin and cytokinin. Callus and organ formation was scored after 4 weeks. Supplemental Figure 2A online shows that ahk2-5 mutant explants displayed growth comparable to the wild type. By contrast, explants of ahk3-7 mutants had a significantly reduced ability to respond to cytokinin by callus or shoot formation. Explants of the double mutant ahk2-5 ahk3-7 showed further enhancement of this phenotype. Threefold to tenfold higher cytokinin concentrations than in the wild type were required to induce callus formation and growth, and shoot formation was observed only very rarely in ahk2-5 ahk3-7 explants. A similar result was obtained when the ability of the whole plant to respond to cytokinin was tested in vitro. In the wild type, elevated levels of cytokinin in the medium inhibit shoot growth, reduce the accumulation of leaf chlorophyll, and delay flower induction. ahk2-5 mutants showed a similar inhibition. ahk3-7 mutant seedlings grew better and formed darker green leaves, and ahk2-5 ahk3-7 double mutants showed further enhanced cytokinin resistance in this assay (see Supplemental Figure 2B online).
Cytokinin sensitivity of roots was tested in vitro on medium with increasing cytokinin concentrations. All single mutants and mutant combinations were included in this test to study the relative contribution of all three receptors. Root growth of cre1-2 and combinations of cre1-2 with other ahk mutants showed increased cytokinin resistance (see Supplemental Figure 2C online). This confirms a major role for CRE1/AHK4 in sensing of exogenous cytokinin in the primary root, with some contribution from AHK2 and AHK3 (Inoue et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004). Functioning of AHK2 and AHK3 in the root independent of CRE1/AHK4 became apparent after prolonged exposure to cytokinin. Supplemental Figure 2D online shows that roots of ahk2-5 ahk3-7 double mutants were able to continue to grow slowly on medium containing 0.1 mg/L benzyladenine (BA), while wild-type roots ceased growing.
Shoot Phenotype of Cytokinin Receptor Mutants
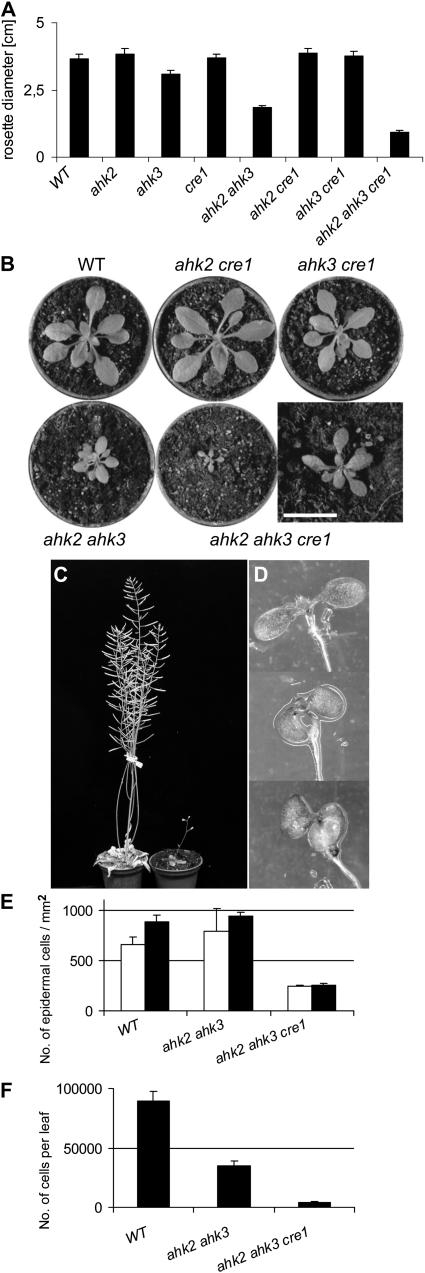
We compared the development of single, double, and triple mutant shoots. Shoot development of ahk2-5 and cre1-2 single mutants was indistinguishable from the wild type (Figure 1; data not shown). By contrast, the rosette diameter of ahk3-7 mutants was reduced ∼15% compared with the wild type (Figure 1A). Introgression of an ahk2-5 mutant allele reduced rosette diameter further to about half the size of the wild-type rosette (Figures 1A and 1B). The rate of leaf formation was not altered in ahk2-5 ahk3-7 double mutants, and flowering was induced at the same time as in the wild type. The final height of these mutant plants was about two-thirds of the wild type.
Figure 1.
Shoot Development of ahk Mutant Plants.
(A) Rosette sizes of wild-type and cytokinin receptor mutants 25 d after germination (DAG). The mutant alleles were ahk2-5, ahk3-7, and cre1-2. Error bars represent se (n = 30).
(B) Rosettes of plants grown on soil 25 DAG. Bar = 10 mm for the close-up of the ahk2-5 ahk3-7 cre1-2 triple mutant shown in the bottom right.
(C) A triple ahk2-5 ahk3-7 cre1-2 mutant plant (right) 70 DAG compared with the wild type.
(D) Downward bending of cotyledons of ahk2-5 ahk3-7 (middle) and ahk2-5 ahk3-7 cre1-2 (bottom) seedlings compared with the wild type (top) (3 DAG).
(E) Number of epidermal cells per mm2 on the adaxial (white bars) and abaxial sides (black bars) of the seventh leaf of the wild type and ahk2-5 ahk3-7 (21 DAG) and of the full expanded seventh leaf of ahk2-5 ahk3-7 cre1-2 (28 DAG). Cell size was measured at three different positions on the seventh leaf in the middle of the leaf blade. Error bars represent se (n = 5).
(F) Number of epidermal cells on the adaxial side on the seventh leaf. Error bars represent se (n = 5).
Triple mutant plants showed an even stronger reduction of shoot development, resulting in miniature plants (Figure 1C) and revealing the CRE1/AHK4 function in the shoot. Cotyledons of ahk2-5 ahk3-7 and triple mutant seedlings bent downwards, indicating differential growth in the adaxial and abaxial sides (Figure 1D). Twenty-five days after germination (DAG), wild-type plants had developed 14 to 16 leaves and started to flower. At 25 DAG, triple mutant plants had only seven to eight leaves, indicating a longer plastochrone. Flower induction was variable; on average, triple mutant plants flowered 2 to 3 weeks later than the wild type. Triple mutant plants were almost completely infertile. However, in contrast with complete sterility reported by others (Higuchi et al., 2004; Nishimura et al., 2004), they self-fertilized and formed a few seeds under favorable temperature and light conditions. This suggests that infertility of triple mutants is a conditional phenotype or is associated with specific mutant alleles.
Leaves of ahk2-5 ahk3-7 double mutants had a reduced length and width, but the overall form and heteroblasty were not altered. Leaves of triple mutants were even smaller but had an unaltered length-to-width ratio as well. Microscopic inspection of epidermal cells at different sites of the fifth leaf at maturity revealed that the average cell size on the upper and lower epidermis of ahk2-5 ahk3-7 mutant leaves was about double the size of wild-type cells. Interestingly, cell size became similar to the wild type in leaves that were formed at later stages (Figure 1E; see also Nishimura et al., 2004). Leaf cell size of triple mutants was increased approximately threefold in young and older leaves as well (Figure 1E), and in total, only ∼5% of cells per leaf were formed compared with the wild type (Figure 1F). The increase in cell size to compensate for the reduced number of cells has been reported for different cell cycle mutants (reviewed in Tsukaya, 2003) and appears to be transient in the ahk2 ahk3 double mutant but persistent in the triple mutant.
Chlorophyll Retention by Cytokinin in the Receptor Mutants
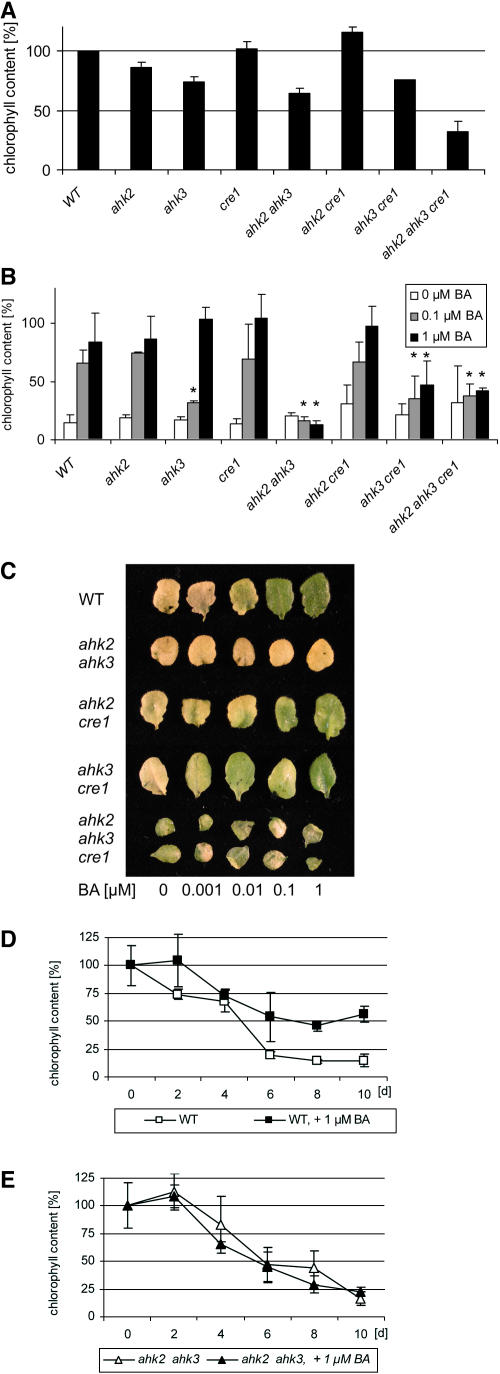
The influence of cytokinin on the chlorophyll content of leaves and their ability to retard leaf senescence was described soon after their discovery (Richmond and Lang, 1957; Mothes and Baudisch, 1958). Figure 2A shows that mutations in specific cytokinin receptors cause a reduction of the leaf chlorophyll content. ahk3-7 reduced the chlorophyll content to ∼75% of the wild type, ahk2-5 had a weak effect, and cre1-2 had no effect. The combination of ahk3-7 with ahk2-5 further reduced the chlorophyll content. A role for CRE1/AHK4 in assuring chlorophyll formation only became apparent in the triple mutant, which has ∼35% of the chlorophyll of the wild type (Figure 2A).
Figure 2.
AHK2 and AHK3 Are Required to Mediate Cytokinin-Dependent Chlorophyll Retention in the Dark.
(A) Chlorophyll content of in vitro–grown plants 24 DAG. Wild type (1.92 ± 0.01 μg/g leaf fresh weight) was set at 100%. For each of five independent samples per clone, five seventh leaves from different plants were pooled and analyzed. Error bars represent se (n = 5). The mutant alleles used were ahk2-5, ahk3-7, and cre1-2.
(B) Dark-induced senescence in a detached leaf assay and its inhibition by cytokinin. The leaf chlorophyll content before the start of dark incubation was set at 100% for each genotype tested. Bars: white, water plus DMSO; gray, 0.1 μM BA; black, 1 μM BA. Chlorophyll content at the beginning of the assay was for the wild type, 1.92 ± 0.01; ahk2-5, 1.64 ± 0.11; ahk3-7, 1.46 ± 0.11; cre1-2, 1.95 ± 0.14; ahk2-5 ahk3-7, 1.23 ± 0.03; ahk2-5 cre1-2, 2.23 ± 0.17; ahk3-7 cre1-2, 1.44 ± 0.09; ahk2-5 ahk3-7 cre1-2, 0.61 ± 0.31 μg/g leaf fresh weight. Asterisks represent significant changes to wild-type control at respective hormone concentrations. Error bars represent se (n = 5).
(C) Leaves of different genotypes at the end of the chlorophyll retention assay described in (B).
(D) Time course of dark-induced leaf senescence in wild-type leaves. The chlorophyll content at the beginning of the experiment was set at 100%. Three independent plates with five leaves per plate were examined at each time point and concentration. The graph shows pooled results from three independent experiments ±se (n = 3).
(E) Time course of dark-induced leaf senescence in leaves of the ahk2-5 ahk3-7 mutant. Conditions are as described in (D).
We investigated the participation of the different receptors in mediating chlorophyll retention by exogenous cytokinin using dark treatment of detached leaves. Dark treatment of leaves causes so-called dark-induced senescence, which mimics partially natural senescence, including chlorophyll degradation (Buchanan-Wollaston et al., 2005). Figure 2B shows that loss of a single receptor did not strongly alter the cytokinin responsiveness. A functional AHK3 receptor alone was sufficient to fully respond to exogenous cytokinin. However, the ahk2-5 ahk3-7 and ahk3-7 cre1-2 double mutants as well as the triple mutant had completely or largely lost the ability to retain chlorophyll in response to cytokinin (Figures 2B and 2C). Analysis of the kinetics of chlorophyll loss revealed that cytokinin slowed the loss of chlorophyll in the wild type, where its content was stabilized after 6 d at ∼60% of the original content (Figure 2D). By contrast, the kinetics of chlorophyll loss was similar in ahk2-5 ahk3-7 mutants in the absence and in the presence of cytokinin (Figure 2E). These data reveal a major contribution of AHK3 in mediating cytokinin-dependent chlorophyll retention in leaves.
Seed Phenotype of Cytokinin Receptor Mutants
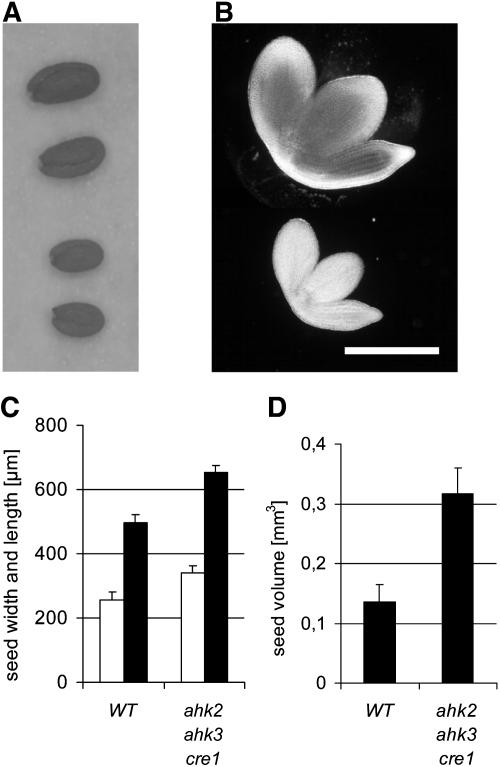
Seed size of single and double mutants was unchanged and microscopic inspection of embryos did not indicate gross developmental differences between mutant and wild-type embryos. However, seeds of the triple mutant were significantly increased in size (Figure 3A). The increase of seed size was mainly due to increased size of the embryos (Figure 3B). Microscopic inspection of embryonic root epidermis revealed that both the cell number and the cell size were increased ∼15 and 30%, respectively.
Figure 3.
Seeds and Embryos of the Triple Mutant Are Increased in Size.
(A) ahk2-5 ahk3-7 cre1-2 triple mutant seeds (top two seeds) compared with the wild type (bottom two seeds).
(B) Embryos of the ahk2-5 ahk3-7 cre1-2 triple mutant (top) and the wild type (bottom). Bar = 3 mm.
(C) Width (white bars) and length (black bars) of seeds from the wild type and the triple mutant. Error bars represent se (n = 60).
(D) Calculated volume of wild-type and ahk2-5 ahk3-7 cre1-2 triple mutant seeds. Error bars represent se (n = 60).
Because of the low number of seeds, we did not measure seed mass but determined seed volume instead. The average seed length and width of triple mutant seeds was ∼30% greater than the wild type (Figure 3C). Calculation of the seed volume by a spheroid formula revealed that the volume of triple mutant seeds was increased up to ∼250% of the wild-type seed volume (Figure 3D).
It could be that the increase in seed size is (1) an autonomous function of the embryo proper, (2) a function linked to the genotype of the triploid endosperm, which results from fertilization of the central cells of the ovule, and/or (3) of the maternal tissue, which includes the seed coat. In order to investigate this further, we analyzed progeny seeds of reciprocal crosses between the wild type and the triple mutant. Seeds formed by cross-pollinated triple mutants were as large as triple mutant seeds or even larger, while seeds formed on cross-pollinated wild-type plants were only slightly larger than wild-type seeds obtained by self-fertilization. This indicated that the genotype of the embryo proper, which is heterozygote for all receptor mutant alleles in both progenies, is not the most important parameter in determining seed size but that the endospermal and/or maternal genotypes have a major influence.
Germination Phenotype of Cytokinin Receptor Mutant Seeds
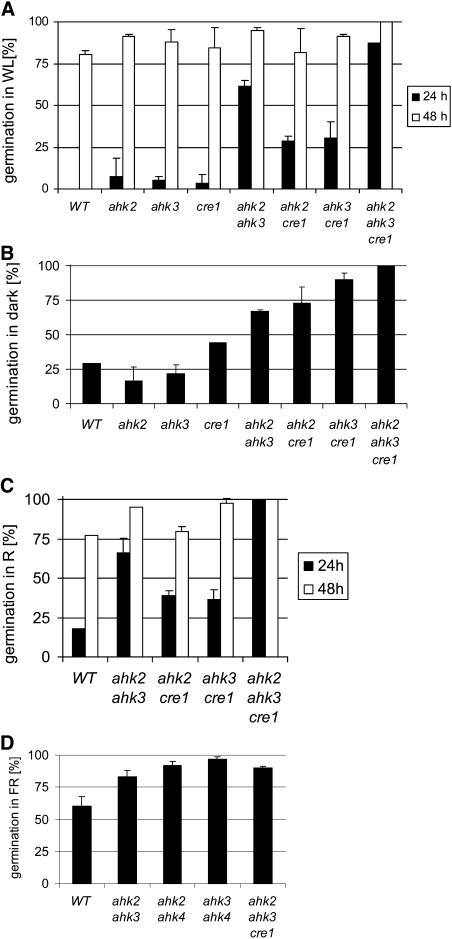
The timing of germination was different in the receptor mutants. After sowing, all seeds were pretreated for 2 d at 4°C in the dark. Wild-type seeds started to germinate ∼24 h after transfer to light and ambient temperature. By contrast, a higher portion of single receptor mutant seeds had started to germinate, a trait which was further enhanced in multiple mutants (Figure 4A). For example, 24 h after transfer to the light, 50% of ahk2-5 ahk3-7 seeds and almost all triple mutant seeds were germinated, indicating the redundant function of all three receptors in mediating cytokinin control of germination (Figure 4A). This notion was further supported by early germination of cytokinin-deficient seeds overexpressing AtCKX2 or AtCKX4 (data not shown).
Figure 4.
Seeds of Cytokinin Receptor Mutants Show Early Germination and Resistance to Far-Red Light.
(A) Percentage of germinated seeds after transfer to white light (WL) and incubation under long-day conditions on sugar-free Murashige and Skoog (MS) medium.
(B) Percentage of seeds germinated after 7 d of incubation in the dark on sugar-free MS medium.
(C) Percentage of seeds germinated under constant red light (R; 660 nm/3.4 μE).
(D) Percentage of germinated seeds after 3 d of incubation under constant far-red light (FR; 724 nm/3.4 μE). Germination rate was determined after an additional 3 d in the dark. Data are means of four to six replicates of two independent seed batches.
The mutant alleles used in all assays were ahk2-5, ahk3-7, and cre1-2.
Wild-type Arabidopsis seeds of most ecotypes require light for efficient germination, and action of phytochrome is considered the primary event in seed germination (Koornneef and Karssen, 1994). Seven days after transfer to dark conditions at room temperature, only ∼25% of wild-type seeds had germinated (Figure 4B). By contrast, 42% of the cre1-2 seeds, >60% of the ahk2-5 ahk3-7 and ahk3-7 cre1-2 double mutant seeds, ∼80% of the ahk2-5 cre1-2 double mutant seeds, and all triple mutant seeds were germinated at this time point (Figure 4B). This indicates a combined activity of all receptors to suppress germination of Arabidopsis seeds in the dark, the largest contribution coming probably from CRE1/AHK4.
Germination is inhibited by far-red light, and the active red light–induced Pfr form of phytochrome is required to trigger the germination pathway (Koornneef and Karssen, 1994). Therefore, we compared the germination under red and far-red light. Figure 4C shows that mutant seeds germinated earlier than the wild type, and a comparable portion of wild-type and mutant seeds had germinated after 48 h in red light, similar to what was seen in white light (Figure 4A). By contrast, far-red light was inhibitory for seed germination in wild-type and single mutants but not for double and triple mutants (t test, P < 0.01) (Figure 4D; data not shown).
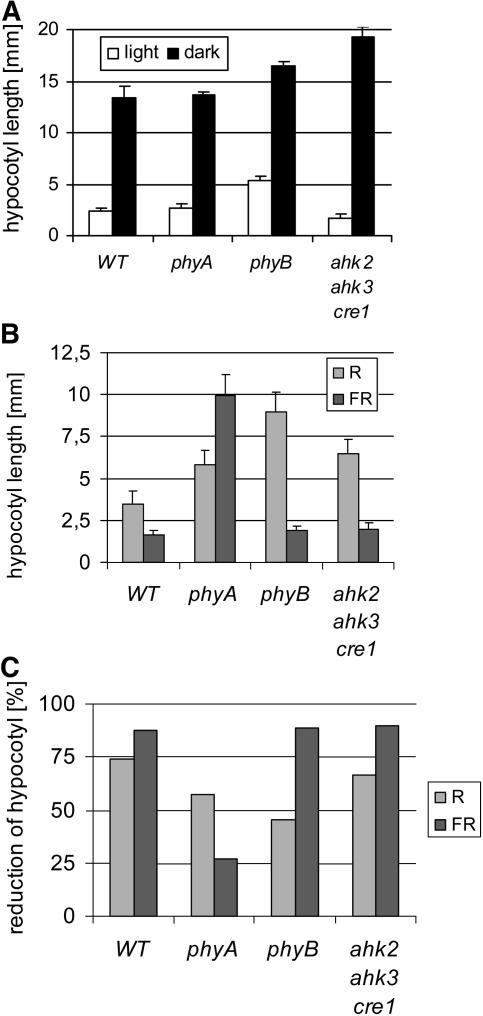
Red Light Sensitivity of Hypocotyl Elongation
Altered red light sensitivity was previously noted for the hypocotyl of A-type arr mutants (Sweere et al., 2001; To et al., 2004), indicating a possible crosstalk between cytokinin and red light signaling in control of hypocotyl elongation. However, a definite role for cytokinin in this process was not shown. Hypocotyl elongation of receptor mutants was analyzed in the dark and under different light regimes. Triple mutants had slightly shorter hypocotyls than the wild type when grown in white light but showed an ∼25% increase in length when grown in darkness (Figure 5A). In red light, hypocotyls grew shorter compared with darkness (Figure 5B), but the relative growth reduction was similar in the wild type and the triple mutant (Figure 5C). Likewise, growth in far-red light led to a shorter hypocotyl, and also under these light conditions, the relative reduction in length was similar in the triple mutant and in the wild type. As a control, phyA and phyB mutants showed resistance to far-red and red light, respectively (Figure 5). Together, these results argue against a role of cytokinin in regulating A-type ARR-dependent modulation of active phytochrome levels in this process.
Figure 5.
Hypocotyl Elongation of Cytokinin Receptor Mutants Shows No Altered Red Light Sensitivity.
(A) Hypocotyl elongation in the light and in the dark 7 DAG. Error bars represent se (n = 15).
(B) Hypocotyl elongation of Arabidopsis plants grown in continuous red light (R; 660 nm/3.4 μE, 4 DAG) or far-red light (FR; 724 nm/3.4 μE, 4 DAG). Error bars represent se (n > 30).
(C) Reduction of hypocotyl elongation in red light or far-red light shown in (B) compared with the elongation in the dark.
The ahk2-5 ahk3-7 cre1-2 triple mutant was used in all assays.
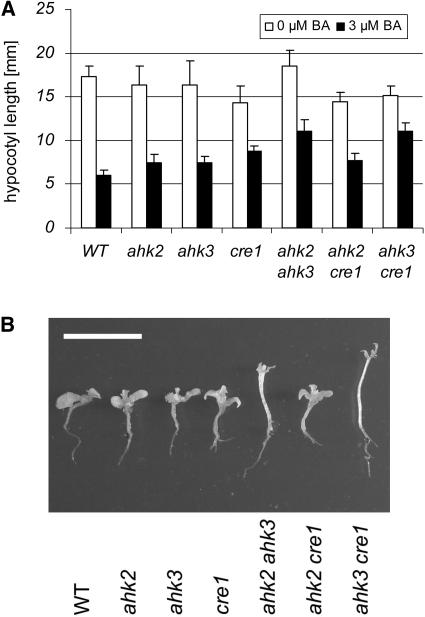
Cytokinin-Induced Photomorphogenesis
High concentrations of cytokinin induce some characteristics of light-grown plants in dark-grown wild-type seedlings, such as inhibition of hypocotyl elongation and development of leaves (Chory et al., 1994). To explore which receptors participate in mediating this response, wild-type and mutant seeds were grown in the dark on media supplemented with different concentrations of cytokinin. Figure 6A shows that hypocotyls of ahk2-5 ahk3-7 and ahk3-7 cre1-2 mutants were most resistant to cytokinin-induced hypocotyl shortening. The same genotypes also showed reduced formation of leaves in the dark, compared with the wild type, single mutants, and the ahk2-5 cre1-2 double mutant (Figure 6B). We conclude that AHK3 in combination with either AHK2 or CRE1/AHK4 is important to mediate cytokinin-dependent deetiolation.
Figure 6.
Cytokinin-Dependent Deetiolation of Dark-Grown Seedlings Is Altered in Receptor Mutants.
(A) Hypocotyl elongation in the dark 7 DAG on MS medium containing no BA or 3 μM BA. Error bars represent se (n = 15).
(B) Deetiolation of wild-type and mutants seedlings 14 DAG on MS medium containing 60 μM BA. Bar = 10 mm.
The mutant alleles used in all assays were ahk2-5, ahk3-7, and cre1-2.
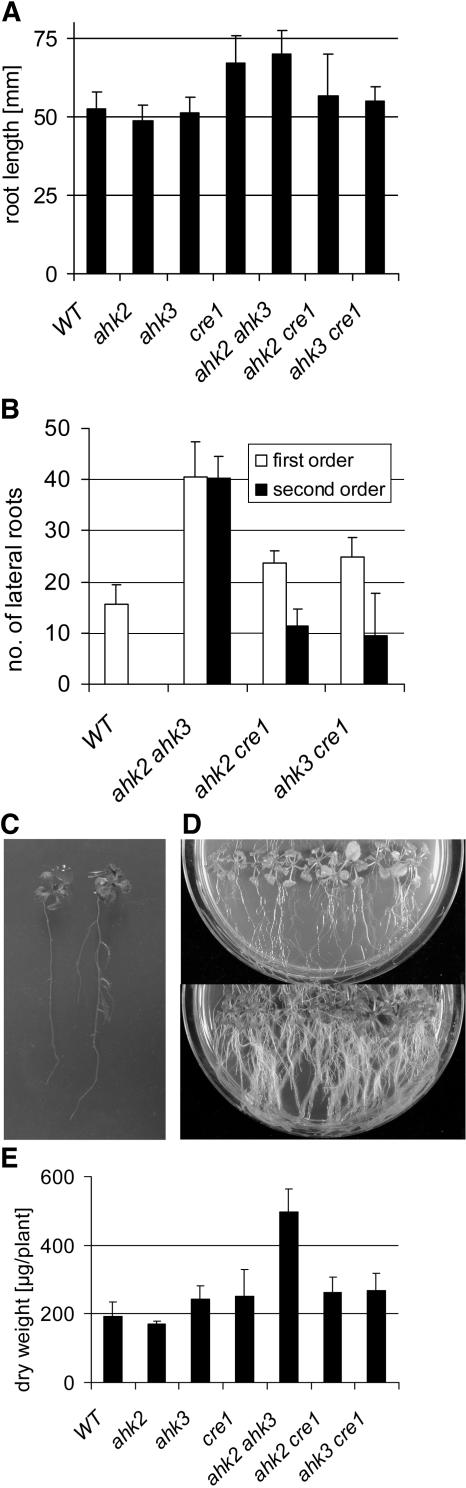
Root Phenotype of Cytokinin Receptor Mutants
At 14 DAG, the primary roots of cre1-2 and the ahk2-5 ahk3-7 double mutant were longer than in the wild type (Figure 7A). A similar difference in root elongation was detected when the growth was measured in the interval between 4 and 7 DAG (see Supplemental Figure 2 online). Two weeks after germination, all three double mutants had formed a larger number of lateral roots, while no significant difference was found for single mutant plants (Figure 7B; data not shown). The strongest increase was found for ahk2-5 ahk3-7 mutants that had more than twice the number of side roots compared with the wild type. Furthermore, all double mutant plants, and in particular ahk2-5 ahk3-7 plants, had formed secondary lateral branches, which were completely absent in the wild type (Figures 7B and 7C). As a consequence, ahk2-5 ahk3-7 mutant plants formed a dense and highly branched root system (Figure 7D). Three weeks after germination, the dry weight of the root system of ahk2-5 ahk3-7 mutants had increased to ∼250% of the wild type, while the enhancement in the other double mutant combinations was in the range of 20% (Figure 7E).
Figure 7.
ahk2 ahk3 Double Mutants Form an Enhanced Root System.
(A) Elongation of primary roots 14 DAG on standard MS medium. Error bars represent se (n ≥ 10).
(B) Number of lateral roots of first and second order 14 DAG on standard MS medium. Error bars represent se (n ≥ 15).
(C) Root system of in vitro–grown wild-type (left) and ahk2-5 ahk3-7 mutant plants 14 DAG.
(D) Enhanced root system of ahk2-5 ahk3-7 mutant plants grown in vitro for 28 d on vertical plates (bottom) compared with the wild type (top).
(E) Dry weight of the root system 21 DAG. The root system was harvested 3 weeks after germination, the roots of eight plants were pooled, and the dry weight of three independent pools per genotype was determined. Error bars represent se (n = 3).
The mutant alleles used in all assays were ahk2-5, ahk3-7, and cre1-2.
Multiple Cytokinin Receptor Mutants Have an Increased Cytokinin Content
To explore whether reduced cytokinin signaling has an influence on the endogenous cytokinin content, we measured the concentrations of different cytokinins in shoots of 22-d-old plants. Table 1 shows that the cytokinin concentrations of single receptor mutant plants were similar to the wild type, with the exception of ahk3-7 mutants, which showed a twofold to threefold increase of the concentrations of all zeatin metabolites. Double mutants containing the ahk3-7 allele (ahk2-5 ahk3-7 and ahk3-7 cre1-2) showed a similar increase, while ahk2-5 cre1-2 mutants showed no increase or only an up to twofold increase for some zeatin compounds (Table 1). Triple mutant plants showed much stronger increases: the concentrations of the biologically most active trans-zeatin and its riboside were increased 16-fold and 9-fold, respectively. The concentrations of N- and O-conjugates were increased as well, up to >50-fold for zeatin O-glucoside. The concentrations of isopentenyladenine-type cytokinins, dihydrozeatin-type cytokinins, and aromatic cytokinins were unchanged in most mutant lines. Only in the triple mutant was a small increase found for these cytokinin compounds (Table 1; data not shown).
Table 1.
Cytokinin Content of Receptor Mutant Arabidopsis Plants
| Line | tZ | tZR | tZRMP | tZ9G | tZOG |
|---|---|---|---|---|---|
| Wild type | 1.0 ± 0.1 | 15.8 ± 4.1 | 36.5 ± 3.8 | 11.9 ± 0.4 | 5.8 ± 0.8 |
| ahk2-5 | 1.0 ± 0.1 | 21.7 ± 6.2 | 48.7 ± 3.6 | 15.3 ± 0.7 | 7.1 ± 0.3 |
| ahk3-7 | 2.0 ± 0.2 | 54.0 ± 16.2 | 108.4 ± 12.4 | 19.6 ± 1.1 | 16.0 ± 1.8 |
| cre1-2 | 0.8 ± 0.0 | 17.9 ± 5.9 | 49.9 ± 14.2 | 9.5 ± 0.8 | 5.1 ± 0.4 |
| ahk2-5 ahk3-7 | 3.7 ± 0.5 | 55.2 ± 14.4 | 115.8 ± 8.2 | 33.9 ± 2.0 | 59.9 ± 4.4 |
| ahk2-5 cre1-2 | 1.2 ± 0.1 | 35.1 ± 10.4 | 66.5 ± 5.1 | 11.8 ± 1.2 | 6.7 ± 0.8 |
| ahk3-7 cre1-2 | 2.1 ± 0.2 | 57.2 ± 20.0 | 114.5 ± 3.3 | 23.9 ± 2.3 | 18.6 ± 0.9 |
| ahk2-5 ahk3-7 cre1-2 | 15.9 ± 4.7 | 145.9 ± 55.8 | 212.7 ± 35.3 | 88.2 ± 6.0 | 394.1 ± 32.6 |
| Line | tZROG | iP | iPR | iPRMP | iP9G |
| Wild type | 5.7 ± 2.1 | 1.8 ± 0.6 | 4.6 ± 1.0 | 11.7 ± 1.1 | 1.1 ± 0.1 |
| ahk2-5 | 5.5 ± 1.4 | 0.8 ± 0.2 | 5.3 ± 1.2 | 14.3 ± 1.5 | 1.0 ± 0.1 |
| ahk3-7 | 18.5 ± 7.7 | 1.2 ± 0.1 | 7.6 ± 1.8 | 17.1 ± 1.9 | 1.2 ± 0.1 |
| cre1-2 | 5.9 ± 2.0 | 0.6 ± 0.1 | 5.3 ± 1.7 | 14.0 ± 1.0 | 0.9 ± 0.1 |
| ahk2-5 ahk3-7 | 21.1 ± 8.4 | 0.9 ± 0.0 | 5.8 ± 1.5 | 15.6 ± 2.0 | 1.8 ± 0.2 |
| ahk2-5 cre1-2 | 13.2 ± 5.6 | 1.2 ± 0.1 | 6.7 ± 1.7 | 16.2 ± 1.1 | 0.9 ± 0.1 |
| ahk3-7 cre1-2 | 15.6 ± 4.9 | 0.9 ± 0.0 | 7.3 ± 2.5 | 18.6 ± 1.2 | 1.4 ± 0.1 |
| ahk2-5 ahk3-7 cre1-2 | 90.9 ± 25.5 | 2.2 ± 0.4 | 13.2 ± 5.1 | 19.1 ± 1.9 | 12.1 ± 1.5 |
One gram of Arabidopsis seedlings (22 DAG) per sample was pooled, and three independent biological samples were taken for each genotype. Data shown are pmol/g fresh weight ± se; n = 3. Increases ≥2.5-fold are in bold. tZ, trans-zeatin; tZR; trans-zeatin riboside; tZRMP, trans-zeatin riboside 5′-monophosphate; tZ9G, trans-zeatin 9-glucoside; tZROG, trans-zeatin riboside O-glucoside; iP, N6-(Δ2isopentenyl)adenine; iPR, N6-(Δ2isopentenyl)adenosine; iPRMP, N6-(Δ2isopentenyl)adenosine 5′-monophospate; iP9G, N6-(Δ2isopentenyl)adenine 9-glucoside.
DISCUSSION
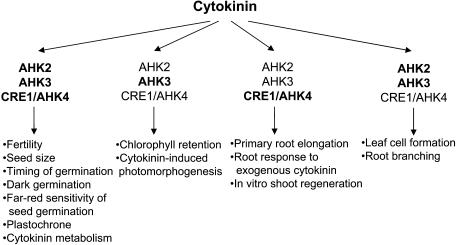
We have undertaken a detailed loss-of-function mutant analysis to study the roles of three cytokinin receptors in development and their participation in a variety of cytokinin-dependent processes. The results of this study are summarized in Figure 8. The general outcome is that the signal perception system is redundant, with all three receptors participating in most of the analyzed reactions. However, the three receptors and their combinations contribute to a different extent to different processes. One example is root branching, which was enhanced in all double mutants but particularly strong in the ahk2 ahk3 combination (Figure 8). This and other examples will be discussed below. The results will be, whenever possible, compared with the consequences of cytokinin deficiency in Arabidopsis and tobacco (Werner et al., 2001, 2003; Yang et al., 2003). Comparisons will also be made with previous reports on cytokinin receptor mutants, which analyzed some aspects of single mutants and specific combinations of multiple mutants (Inoue et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004).
Figure 8.
Contributions of Different Cytokinin Receptors and Receptor Combinations to Cytokinin-Regulated Processes.
Cytokinin receptors that make a major contribution to a given process are printed in bold letters. The figure is based on data from this article. Effects of receptor loss-of-function mutations on fertility, plastochrone, leaf cell formation, primary root elongation, the root response to exogenous cytokinin, and in vitro shoot regeneration were also reported by others (Inoue et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004).
All Three Receptors Participate in Regulating Shoot Development
Mutations in single receptors did not cause strong changes of shoot growth, indicating a high degree of redundancy of receptor functions in shoot growth regulation. The only phenotype we noted was a slight reduction of rosette size in ahk3 mutant plants. Redundancy was not complete as combined loss of AHK2 and AHK3 restricted shoot growth; in particular, chlorophyll and leaf cell formation was reduced. This shows that CRE1/AHK4 alone does not support all cytokinin functions in the shoot, while these functions were maintained by either AHK2 or AHK3 alone. By contrast, the rate of leaf initiation and the timing of flower induction in ahk2 ahk3 mutants were similar to the wild type, indicating that CRE1/AHK4 is sufficient to transmit the cytokinin signal for these processes. Consistently, introgression of the cre1-2 mutation in the ahk2-5 ahk3-7 mutant slowed leaf formation and caused retarded flowering. Interestingly, changed leaf size did not alter overall leaf shape and heteroblasty, indicating that these traits are regulated independent of cytokinin.
These alterations are generally in accordance with the shoot phenotype of cytokinin-deficient Arabidopsis plants (Werner et al., 2003; Yang et al., 2003). However, an important difference is that a strong reduction of the cytokinin content led to complete growth arrest of the apical shoot meristem (Werner et al., 2003), while triple receptor mutants are still able to establish and maintain a functional shoot meristem (Higuchi et al., 2004; Nishimura et al., 2004; this article). It could be that the receptor mutant alleles were not null alleles, but three different allele combinations caused a similar phenotype, which makes this explanation less likely. In order to explain this discrepancy, we propose that two separate cytokinin response systems exist. A second system that operates independently from the receptors should maintain cell cycling, ensuring the formation of a sufficient number of cells for a basic plant body. This system may operate in an autocrine (i.e., cell-autonomous) fashion. Each cell's cytokinin may act on the cell cycle machinery in the producing cell and thus make it independent from an extracellular cytokinin signal. Lack of cytokinin signaling via membrane-located receptors would not affect the basic maintenance function. Consistently, growth arrest of shoots was only obtained when a CKX enzyme localized in the vacuole was overexpressed, while overproduction of CKX enzymes localized in the endoplasmic reticulum and/or extracellularly caused only weak shoot phenotypes and did not lead to growth arrest (Werner et al., 2003). Thus, the proposed receptor-independent maintenance function of cytokinin appears to depend on an intracellular hormone pool. A molecular mechanism to realize cytokinin functions in the cell cycle independent of membrane-located receptors could be a direct regulation of enzymatic activity. In accordance with this proposal is the fact that cytokinin compounds and structural derivatives inhibit directly the activity of cell cycle–regulating kinases (Veselý et al., 1994; Binarová et al., 1998). In our model, the main function of cytokinin perception and signaling via the two-component system would be to mediate information arriving from elsewhere in the plant or from the plant's environment in order to modulate growth and physiological processes in response to these cues.
AHK2 and AHK3 Mediate Cytokinin-Dependent Chlorophyll Retention
The chlorophyll content of several cytokinin receptor mutant combinations was reduced compared with the wild type, similar to cytokinin-deficient plants (Figure 2; T. Werner, personal communication). This clearly shows that the full cytokinin level or the full signaling competence is required to achieve wild-type levels of chlorophyll. However, a basic level of chlorophyll is formed independent of cytokinin, indicating that cytokinin is not necessary per se for chlorophyll formation but may function to modulate the chlorophyll content above a threshold level in response to environmental cues.
Cytokinin delays senescence in detached leaves (Richmond and Lang, 1957) and in planta (Mothes and Baudisch, 1958; Gan and Amasino, 1995). Loss of chlorophyll is one of the most obvious signs of leaf senescence, and it was used here to determine that AHK3 plays the most important role in mediating cytokinin-dependent chlorophyll retention in dark-treated leaves (Figure 2). AHK3 alone was sufficient to mediate the full response, while AHK2 or CRE1/AHK4 alone did not or only weakly transmitted the cytokinin signal. However, the latter two receptors together also provided full cytokinin responsiveness at higher cytokinin concentrations (Figure 2). Interestingly, we have not noted earlier leaf senescence in cytokinin receptor mutants or cytokinin-deficient plants, and the loss of chlorophyll in detached leaves was in the absence of exogenous cytokinin not faster in ahk2-5 ahk3-7 mutants than in the wild type (Figures 2D and 2E). This is consistent with the idea that a low cytokinin content or reduced cytokinin signaling are not triggering factors for the onset of the senescence process but that a low cytokinin status is a license for senescence to occur (Werner et al., 2003).
Cytokinin Controls Seed Size
Despite its agronomic importance, not much is known about the factors regulating seed size and a possible role for cytokinin has only been reported recently (Werner et al., 2003). Control of seed size involves control of growth in the embryo, the surrounding triploid endosperm, and the seed coat. In Arabidopsis, the seed coat and endosperm growth precedes embryo growth, and the seed reaches almost its final size before the enlargement of the embryo, which happens during the later phase of embryogenesis (Mansfield and Bowman, 1993). Genetic studies have shown that maternal and nonmaternal factors are involved in seed size regulation and that crosstalk occurs between maternal and zygotic tissue to coordinate seed size (Alonso-Blanco et al., 1999; Garcia et al., 2005). Genetic analysis of cytokinin receptor mutants has indicated that the increase of triple mutant seed size does not depend on the genotype of the embryo but rather is governed by the maternal and/or endospermal genotypes. Recently, it was shown that mutations in the transcription factor gene APETALA2 act as well maternally and/or in the endosperm to increase seed size (Jofuku et al., 2005; Ohto et al., 2005).
It is an important question whether the increased seed size is causally linked to the reduced number of seeds. It is conceivable that reduced availability of sink organs for fixed carbon may lead to an enhanced deposition in the available sink tissues (i.e., embryos), and it is known that the seed mass is generally negatively correlated with the total seed yield in Arabidopsis (Alonso-Blanco et al., 1999). However, it was shown by hand-pollination of a few flowers of male sterile mutant plants or removal of all flowers on wild-type plants with the exception of three flowers that were left for self-pollination that in Arabidopsis a reduction of seed number causes a mass increase in the individual seed of 22 to 33% (Jofuku et al., 2005; Ohto et al., 2005). This is far lower than the 150% increase in size reported here. We conclude therefore that the increase of seed size in the triple cytokinin receptor mutant is a direct consequence of loss of receptor functions and their role in growth control.
Cytokinin Controls Seed Germination
Arabidopsis seeds develop dormancy during the late stages of maturation, a process that depends on abscisic acid (Koornneef and Karssen, 1994). Breakage of dormancy and seed germination is primarily controlled by a reversible red light–dependent equilibrium of the photoreceptors phyA and phyB (Borthwick et al., 1952; Shinomura et al., 1994; Bentsink and Koornneef, 2002). One additional important factor to overcome abscisic acid–induced dormancy and germinate is gibberellin (GA), which is formed as a consequence of light action (Yamaguchi et al., 1998). Miller et al. (1956) found that cytokinin in the dark could replace red light to induce seed germination in lettuce (Lactuca sativa), but it was believed that cytokinin at physiological levels hardly affects seed germination and probably plays no role (Koornneef and Karssen, 1994). Now the more rapid germination, increased dark germination, and reduced far-red light sensitivity of cytokinin receptor mutant seeds proves the relevance of cytokinin in regulating this process.
Both dark germination and far-red light sensitivity are altered in receptor mutants, which are controlled in different ways. Is it plausible that cytokinin acts on the balance of active phytochromes, but because of the complexity of the phytochrome system, detailed action spectra are required to gain further insight. An alternative possibility to explain the dark germination of receptor mutants is an enhanced GA signaling as a consequence of reduced cytokinin signaling because seed treatment with GA overcomes partially the inhibition of dark germination (Koornneef et al., 1985). Consistent with this idea is the more rapid germination of the receptor mutant seeds and the fact that GA and cytokinin signaling pathways interact (Brenner et al., 2005; Greenboim-Wainberg et al., 2005). Currently, our experiments do not distinguish between a phytochrome- and a GA-dependent effect.
It is noteworthy that different receptors contribute differently to mediate cytokinin control of seed germination. Mutation of CRE1/AHK4 caused the greatest enhancement of germination in the dark (Figure 4B), while mutation of AHK3 alone was sufficient for partial resistance to far-red light (our unpublished results). Together, the data show that cytokinin is, under physiological conditions, a negative regulator of seed germination and that different pathways are controlled through different receptors.
Reduced Cytokinin Signaling Does Not Alter Red Light Sensitivity of Hypocotyl Elongation
Hypocotyl elongation is another red light–controlled process (Borthwick et al., 1952) that was tested in the cytokinin signaling mutants. phyB is more important than phyA in control of hypocotyl elongation in white light and red light, while phyA is required to respond to far-red light (Reed et al., 1994; Neff and Chory, 1998). Previously, it was shown that overexpression of the A-type ARR4 gene (Sweere et al., 2001) and insertional mutation of A-type ARR genes (To et al., 2004) alters red light but not far-red light sensitivity of the hypocotyl. The slightly increased red light sensitivity of ARR4 overexpressers was explained by a direct physical interaction of ARR4 and phyB, stabilizing the active form of the latter (Sweere et al., 2001). Paradoxically and inconsistent with this proposal, arr mutants showed increased red light sensitivity (To et al., 2004). Neither study showed whether A-type ARR proteins play a role in red light signaling independent of their function in cytokinin signaling or whether cytokinin and red light signaling are functionally linked. In the hypocotyl elongation assay, the receptor triple mutant reacted under all light conditions similar to the wild type (Figure 5). Thus, light signaling seemed not to be affected, at least under the experimental conditions used by us, indicating that regulation of phyB by ARR4 may not be cytokinin dependent.
Cytokinin Regulates Root Architecture
The enhanced root system of ahk2 ahk3 mutants is achieved in two ways. The primary root grows faster than in wild-type plants, which is comparable to the consequences of a reduced endogenous cytokinin content (Werner et al., 2003). More importantly, the ahk2 ahk3 mutants form more lateral roots than the wild type (Figure 7), which led to a significantly stronger root enhancement than did the reduction of the cytokinin content. The prominent roles of AHK2 and AHK3 in regulating root growth is a bit surprising as the analysis of cytokinin resistance of mutant roots and elongation of the primary root had shown a major role for CRE1/AHK4 in roots (Inoue et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004; this article). This indicates that the resistance to exogenous cytokinin, the regulation of primary root elongation, and root branching are separate functions and that separate cytokinin receptors are involved in regulating these functions (Figure 8).
Lateral roots in Arabidopsis originate from pericycle cells (Casimiro et al., 2003). Various hormones, mainly auxin (Casimiro et al., 2003), but also ethylene (Lynch and Brown, 1997), brassinosteroids (Bao et al., 2004), abscisic acid (De Smet et al., 2003), as well as different nutrients, such as nitrate, phosphate, sulfate, and iron (reviewed in López-Bucio et al., 2003), regulate lateral root formation. A critical event in lateral root formation is reentry of differentiated pericycle cells into the cell cycle and initiation of the root developmental program. From this result, it appears that cytokinin at physiological levels suppresses induction of cell division in the root pericycle and that this function is redundantly regulated by AHK2 and AHK3. Paradoxically, cytokinin would normally prevent reentry of cells into the cell cycle, although the hormone is usually considered to be a positive regulator of the cell cycle. Consistent with a function of cytokinins in precursor cells of lateral roots is the observation that initiation of lateral roots is associated with a localized repression of a cytokinin-responsive reporter gene, indicating spatial and temporal regulation of the cytokinin status during lateral root formation (Lohar et al., 2004).
Together, these data corroborate the relevance of the cytokinin status in regulating root architecture. We hypothesize that physiological levels of cytokinins are superoptimal for maximal root elongation and initiation of the lateral root formation program. Optimal conditions may be achieved by decreasing the endogenous cytokinin content or by decreasing cytokinin signaling.
Crosstalk between Cytokinin Signaling and Cytokinin Metabolism
An important observation is that reduced cytokinin signaling led to an increase of the cytokinin content, in particular when AHK3 was mutated (Table 1). Our data do not distinguish between increased cytokinin synthesis, decreased cytokinin breakdown, or both as the reason(s) for the increased steady state concentration. Although the increase in cytokinin content is apparently not sufficient to compensate for the loss of receptor activity, it indicates the existence of homeostatic control mechanisms. Previous work has shown that transcript levels of type-A response regulator genes and of the CRE1/AHK4 gene react sensitively to changes in cytokinin concentration, while other cytokinin signaling genes do not respond (D'Agostino et al., 2000; Franco-Zorilla et al., 2002; Rashotte et al., 2003; Werner et al., 2003; Brenner et al., 2005), indicating that the cellular cytokinin level modulates signaling. Thus, apparently mutual control mechanisms exist between metabolism and signaling, which may contribute to fine-tuning of the cytokinin response.
Implications for Cytokinin Biology
The mutant analyses have yielded novel information about the involvement of different receptors and their combinations in various cytokinin-regulated processes. In addition, this loss-of-function analysis has revealed hitherto unknown functions of cytokinin, for example in seed biology. Many of the responses are driven by multiple cytokinin receptors in an additive manner. In other cases, the contribution of a given receptor could only be identified in the absence of others. Noteworthy, mutation of AHK2 alone did not cause a significant change of cytokinin sensitivity in any of the tests. However, in several assays, the ahk2 mutation enhanced the cytokinin resistance of ahk3 (leaf cell formation, senescence retardation, root branching) or cre1/ahk4 (seed germination) mutants. This indicates that AHK2 may function primarily in combination with AHK3 or CRE1/AHK4. Cooperation between AHK3 and CRE1/AHK4 exists as well, as seen in the arrest of chlorophyll loss in the dark (Figure 4B). This raises the question of whether in some instances the formation of receptor heterodimers is relevant for cytokinin signaling. Indeed, recent results of protein–protein interaction studies indicate that interactions between different cytokinin receptors do occur (H. Dortay, A. Heyl, and T. Schmülling, unpublished results).
ahk2 ahk3 receptor mutants phenocopy to a large extent cytokinin-deficient plants, providing substantial support for the concept of a function of cytokinin in positive control of shoot development and negative control of root growth. Further work has to show how the cytokinin receptors are linked downstream to different signaling pathways in order to achieve positive or negative regulatory control on the cell cycle and/or exit of cells from the meristems. It will be interesting to study how far differences between the receptor functions depend on gene dosage, level or specificity of expression, coupling efficiency to downstream signaling elements, and/or availability of cytokinin molecules that act preferentially on a subset of receptors (Spíchal et al., 2004).
Last but not least, it is noteworthy that several of the cytokinin-regulated traits are of agronomic importance. Water and nutrient availability limit yield in most agricultural ecosystems (Lynch, 1995). Improved root systems are therefore of considerable interest in agriculture. Control of senescence is relevant for yield and shelf life. Seed size profoundly influences total harvest. It is therefore relevant to study the underlying regulatory molecular mechanisms in greater detail.
METHODS
Plant Material and Growth Conditions
The Columbia (Col-0) ecotype of Arabidopsis thaliana was used as the wild type (obtained from Lehle Seeds). ahk2-2 and ahk3-3 were from the SALK Institute (SALK_052531 and SALK_069269, respectively; Alonso et al., 2003). ahk2-5 was obtained from the SAIL collection (SAIL_575_E05; Sessions et al., 2002) and ahk3-7 from GABI-KAT (105E02; Rosso et al., 2003). cre1-2 was kindly provided by Tatsuo Kakimoto. Plants were grown in the greenhouse on soil at 22°C under long-day conditions (16 h light/8 h dark). For seedling assays in vitro, seeds were surface-sterilized and cold treated at 4°C for 3 d in the dark and then exposed to white light (∼75 μE). Seedlings were grown at 22°C on horizontal or vertical plates containing MS medium as modified by Kemper et al. (1992), 3% sucrose, and 0.9% agarose (Merck) unless otherwise specified. For germination and red light/darkness experiments, sucrose was omitted from the medium.
Genotyping of Plant Material
Wild-type AHK2 and ahk2-2 and ahk2-5 alleles were identified using the following primers in PCR: AHK2-2-F (5′-GTGTGAAGATTCGGCCTTGT-3′) and AHK2-2-R (5′-TGCGAAGCAGATGGACTATG-3′) or AHK2-5-F (5′-GCAAGAGGCTTTAGCTCCAA-3′) and AHK2-5-R (5′-TTGCCCGTAAGATGTTTTCA-3′), respectively, for detection of the AHK2 wild-type allele. AHK2-2-F in combination with SALK LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) for detection of the ahk2-2 allele and AHK2-5-F and SAIL IT-1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′) were used for detection of the ahk2-5 allele. Wild-type AHK3 and ahk3-3 and ahk3-7 alleles were identified using the following primers in PCR: AHK3-3-F (5′-CACCATGGCCAGTGCTATC-3′) and AHK3-3-R (5′-CTCAAATCAAACCGCACCTC-3′) or AHK3-7-F (5′-CCTTGTTGCCTCTCGAACTC-3′) and AHK3-7-R (5′-CGCAAGCTATGGAGAAGAGG-3′), respectively, for detection of the AHK3 wild-type alleles. AHK3-3-R and SALK LBa1 were used for detection of the ahk3-3 allele and AHK3-7-R and GABI-KAT IG-1 (5′-CCCATTTGGACGTGTAGACAC-3′) for detection of the ahk3-7 allele. Wild-type AHK4/CRE1 and cre1-2 alleles were detected using PCR as described by Inoue et al. (2001).
Analysis of Gene Expression
Total RNA was extracted from seedlings with the TRIzol method. TRIzol reagent (38% phenol, 0.8 M guanidinium thiocyanate, 0.4 M ammonium thiocyanate, 0.1 M sodium acetate, pH 5, and 5% glycerol) was made as described in the GIBCO TRIzol manual of Invitrogen. One microgram of total RNA was treated with RNase-free DnaseI at 37°C for 30 min. One microliter of 25 mM EDTA was added at 65°C for 10 min. RNA (0.5 μg) was used for an RT-PCR reaction. The primers AHK2-5-RT-F (5′-TGAACCATGTTCATGCCTTG-3′) and AHK2-5-RT-R (5′-TTGCCCGTAAGATGTTTTCA-3′) were used to identify AHK2 transcripts and AHK3-7-RT-F (5′-ATCAAAGCCTCCCCATTCTT-3′) and AHK3-7-RT-R (5′-AACCATTGAGGGCGAGTATG-3′) to identify AHK3 transcripts. Both primer pairs span the respective T-DNA insertion site. In all RT-PCR reactions, the Actin2 primers ACTIN2-F (5′-TACAACGAGCTTCGTGTTGC-3′) and ACTIN2-R (5′-GATTGATCCTCCGATCCAGA-3′) were used as controls. RT-PCR was performed with the One-Step RT-PCR kit (Qiagen) according to the manufacturer's instructions. The PCR comprised 30 cycles of 30 s at 94°C, 1 min at 58°C, and 2 min at 72°C.
Shoot Induction Assay
Seedlings were grown under low-light conditions (∼5 μE) for 3 d. Ten hypocotyls of ∼10 mm length were excised and placed on plates containing MS medium supplemented with 36 different combinations of isopentenyladenine and naphthylacetic acid. Each of the hormones was combined with the other hormone at concentrations of 0, 30, 100, 300, 1000, and 3000 ng/mL. Hypocotyls were cultured for 28 d on these media and scored for organ formation. Typical representatives were arranged together for an overview picture of each genotype. Experiments were done in triplicate with 20 seedlings for each experiment.
Seed Germination Assay
Only seed batches that had been harvested and stored at the same time and under the same conditions were used, except for the triple mutant, due to its delayed shoot growth. Seeds were sown on MS medium, vernalized at 4°C and then transferred to either white light (∼75 μE), red light (660 nm, 3.4 μE), dark red light (724 nm, 3.4 μE), or darkness at 22°C. The germination rate was counted at different time points. Experiments were done in triplicate with 50 seeds for each experiment and genotype.
Hypocotyl Elongation Assay
Seedlings were grown on vertical plates under different light conditions. After 4 d of growth in either white light (∼75 μE), red light (660 nm, ∼3.5 μE), dark red light (724 nm, ∼3.5 μE), or darkness, seedlings were photographed with a digital camera (Nikon Coolpix 8800). Hypocotyl length was measured using the Scion Image program version beta 4.02 (National Institute of Health; www.scioncorp.com). Experiments were done in triplicate with 20 seeds for each experiment and genotype.
Photomorphogenesis
Seedlings were grown in vitro on MS medium with addition of 3 to 60 μM BA. After sterilization, vernalization, and 8 h of light, Petri dishes were transferred to darkness for 1 to 2 weeks and then pictures were taken for phenotypic evaluation. The experiment was repeated twice.
Chlorophyll Retention Assay
Seedlings were grown in vitro on horizontal plates for 24 d. Approximately the seventh leaf was detached and floated on distilled water supplemented with 0, 0.001, 0.01, 0.1, or 1 μM BA in small Petri dishes for 10 d in the dark. Three samples were measured for each genotype, each sample consisting of five leaves. Chlorophyll was extracted with methanol for 24 h in the dark. Light absorption at 647 and 664 nm was determined with a spectrophotometer, normalized to fresh weight, and the chlorophyll content was calculated as described by Porra et al. (1989). The chlorophyll content at the start of the experiments was taken as a reference and set at 100%.
Root Growth Assays
Seedlings were grown on vertical plates, and the length of the primary root was marked on the Petri dish at 14 DAG. Photographs were taken and the root length determined with the Scion Image program. In independent experiments, the root length was determined 7 or 10 DAG, yielding similar relative differences between the genotypes. Emerging lateral roots that had been grown through the root exodermis were counted using a microscope 14 DAG on 10 different roots for each genotype. Twenty-one-day-old in vitro–grown plants were harvested, and the root system was detached, dried for 24 h at 60°C, and then weighed with a fine balance LE244S (Sartorius).
Determination of Seed Size and Rosette Diameter
Seed size of wild-type and ahk mutant lines was determined measuring length and width of 60 seeds of two independent wild-type and ahk2 ahk3 cre1 mutant lines. The volume was estimated by calculating with the formula for a spheroid (volume = 4/3 · π · length · width · depth). For this calculation, the measured width was also taken as depth. Rosette diameter of plants grown on soil in the greenhouse was determined 25 DAG using a ruler, taking the mean value of two measurements on the same rosette.
Microscopic Analysis
Seeds were fixed with ethanol:acetic acid (6:1, v/v) for 4 h and cleared with chloral hydrate solution (100 g chloral hydrate, 2.5 g gum arabicum, and 30 mL water). Epidermal cells of embryos were visualized by imprints in 3% Gelrite. Leaves were cleared following the protocol of Mähönen et al. (2000). All microscopic observations were accomplished with a Zeiss Axioskop microscope.
Identification and Quantification of Endogenous Cytokinins
Plants were grown on soil until 22 DAG. At this stage, all plants had formed approximately eight leaves. Triple mutants were harvested between 8 and 14 d later at a similar developmental stage. Each sample was made up of ∼1 g of pooled shoots. Three independent biological samples were taken for each genotype. The procedure used for cytokinin analysis was a modification of the method described by Novák et al. (2003). Freeze-dried samples were extracted in ice-cold 70% ethanol (v/v) and deuterium-labeled cytokinin standards added (Olchemim), each at 5 pmol per sample to check recovery during purification and to validate determination. After 3 h extraction, the homogenate was centrifuged (15,000g, 4°C) and the pellets reextracted the same way. The combined supernatants were concentrated to ∼1.0 mL under vacuum at 35°C, diluted to 20 mL with ammonium acetate buffer (40 mM, pH 6.5), and purified using a combined DEAE-Sephadex (1.0 × 5.0 cm) octadecylsilica (0.5 × 1.5 cm) column and immunoaffinity chromatography based on generic cytokinin monoclonal antibody (Faiss et al., 1997). This resulted in three fractions containing (1) the free bases, ribosides, and N-glucosides, (2) a nucleotide fraction, and (3) an O-glucoside fraction. Fraction 2 containing ribotides was obtained after alkaline phosphatase treatment and the fraction 3 after hydrolysis by β-glucosidase; both fractions were also immunopurified. The samples were subjected to HPLC (Waters Alliance 2690) linked to a Micromass ZMD 2000 single quadrupole mass spectrometer equipped with an electrospray interface [LC(+)ES-MS] and photodiode array detector (Waters PDA 996). Using a post-column split of 1:1, the effluent was introduced into an electrospray source (source block temperature 100°C, desolvation temperature 250°C, capillary voltage +3.0 V, and cone voltage 20 V) and photodiode array detector (scanning range 210 to 300 nm, with 1.2-nm resolution), and quantitative analysis of the different cytokinins was performed in selective ion recording mode using a standard isotope dilution method (Novák et al., 2003).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (see http://www.arabidopsis.org) with Arabidopsis Genome Initiative locus identifiers At5g35750 (AHK2), At1g27320 (AHK3), and At2g01830 (AHK4/CRE1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification and Molecular Characterization of ahk2 and ahk3 Mutant Alleles.
Supplemental Figure 2. The Cytokinin Sensitivity of Cytokinin Receptor Mutants Is Decreased in Shoots and Roots.
Supplementary Material
Acknowledgments
We thank Ulrike Deppe and Hana Martinková for excellent technical assistance, Monebandith Yinnavong for her contributions during a practical course, Monika Losensky for taking care of the plants, and Tomáš Werner and Alexander Heyl for critical reading of the manuscript. We are indebted to Tatsuo Kakimoto for providing the cre1-2 mutant seeds. The support of Elmar Hartmann and Tilman Lamparter for the light experiments is acknowledged. We also thank the SAIL, SALK, and GABI mutant seed collections as well as the Nottingham Arabidopsis Stock Centre for providing seeds. This work was supported by grants of the Deutsche Forschungsgemeinschaft (Sfb 449 and Arabidopsis Functional Genomics Network program) to T.S. and Grant MSM 6198959216 to M.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thomas Schmülling (tschmue@zedat.fu-berlin.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037796.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Koornneef, M. (1999). Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 4710–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V., and Aravind, L. (2001). The CHASE domain: A predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem. Sci. 10 579–582. [DOI] [PubMed] [Google Scholar]
- Bao, F., Shen, J., Brady, S.R., Muday, G.K., Asami, T., and Yang, Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134 1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., and Koornneef, M. (2002). Seed dormancy and germination. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0050, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Binarová, P., Dolezel, J., Draber, P., Heberle-Bors, E., Strnad, M., and Bogre, L. (1998). Treatment of Vicia faba root tip cells with specific inhibitors to cyclin-dependent kinases leads to abnormal spindle formation. Plant J. 16 697–707. [DOI] [PubMed] [Google Scholar]
- Borthwick, H.A., Hendricks, S.B., Parker, M.W., Toole, E.H., and Toole, V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, W., Romanov, G., Bürkle, L., and Schmülling, T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44 314–333. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P.O., Nam, H.G., Lin, J.-F., Wu, S.-H., Swidzinski, J., Ishizaki, K., and Leaver, C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42 567–585. [DOI] [PubMed] [Google Scholar]
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. [DOI] [PubMed] [Google Scholar]
- Chory, J., Reinecke, D., Sim, S., Washburn, T., and Brenner, M. (1994). A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 104 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, I., Deruere, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family of cytokinin. Plant Physiol. 124 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C.H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33 543–555. [DOI] [PubMed] [Google Scholar]
- Faiss, M., Zalubilova, J., Strnad, M., and Schmülling, T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12 401–415. [DOI] [PubMed] [Google Scholar]
- Ferreira, F.J., and Kieber, J.J. (2005). Cytokinin signalling. Curr. Opin. Plant Biol. 8 518–525. [DOI] [PubMed] [Google Scholar]
- Franco-Zorilla, J.M., Martin, A.C., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation response in Arabidopsis. Plant J. 32 353–360. [DOI] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270 1986–1988. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Fitz Gerald, J.N., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ponce de León, B., Zorrilla, J.M.F., Rubio, V., Dahiya, P., Paz-Ares, J., and Leyva, A. (2004). Interallelic complementation at the Arabidopsis CRE1 locus uncovers independent pathways for the proliferation of vascular initials and canonical cytokinin signalling. Plant J. 38 70–79. [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg, Y., Maymon, I., Borochov, R., Alvarez, J., Olszewski, N., Ori, N., Eshed, Y., and Weiss, D. (2005). Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen, C., and Harter, K. (2004). Plant two-component systems: Principles, functions, complexity and cross talk. Planta 219 733–742. [DOI] [PubMed] [Google Scholar]
- Heyl, A., and Schmülling, T. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6 480–488. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE 1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., Omidyar, P.K., Gee, Z., and Okamuro, J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54 605–627. [DOI] [PubMed] [Google Scholar]
- Kemper, E., Grevelding, C., Schell, J., and Masterson, R. (1992). Improved method for the transformation of Arabidopsis thaliana with chimeric dihydrofolate reductase constructs which confer methotrexate resistance. Plant Cell Rep. 11 118–121. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen-Martinet, E., van Rijn, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65 33–39. [Google Scholar]
- Koornneef, M., and Karssen, C.M. (1994). Seed dormancy and germination. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 313–334.
- Lohar, D.P., Schaff, J.E., Laskey, J.G., Kieber, J.J., Bilyeu, K.D., and Bird, D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214. [DOI] [PubMed] [Google Scholar]
- López-Bucio, J., Cruz-Ramirez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6 280–287. [DOI] [PubMed] [Google Scholar]
- Lynch, J. (1995). Root architecture and plant productivity. Plant Physiol. 109 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J., and Brown, K.M. (1997). Ethylene and plant responses to nutritional stress. Physiol. Plant. 100 613–619. [Google Scholar]
- Mähönen, A.P., Bonke, M., Kaupinnen, L., Riikonen, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Bowman, J. (1993). Embryogenesis. In Arabidopsis: An Atlas of Morphology and Development, J. Bowman, ed (Berlin: Springer-Verlag), pp. 349–362.
- Maruyama-Nakashita, A., Nakamura, Y., Yamaya, T., and Takahashi, H. (2004). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: Implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38 779–789. [DOI] [PubMed] [Google Scholar]
- Miller, C.O., Skoog, F., Okomura, F.S., Von Saltza, M.H., and Strong, F.M. (1956). Isolation, structure and synthesis of kinetin, a substance promoting cell division. J. Am. Chem. Soc. 78 1345–1350. [Google Scholar]
- Mok, M.C. (1994). Cytokinins and plant development—An Overview. In Cytokinins: Chemistry, Activity and Function, D.W.S. Mok and M.C. Mok, eds (Boca Raton, FL: CRC Press), pp. 129–137.
- Mothes, K., and Baudisch, W. (1958). Untersuchungen über die reversibilität der ausbleichung grüner blätter. Flora 146 521–531. [Google Scholar]
- Mougel, C., and Zhulin, I.B. (2001). CHASE: An extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26 582–584. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004). Genetic analysis of Arabidopsis histidine kinase genes encoding cytokinin receptors reveals their overlapping biological functions in the regulation of shoot and root growth in Arabidopsis thaliana. Plant Cell 16 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák, O., Tarkowski, P., Tarkowska, D., Dolezal, K., Lenobel, R., and Strnad, M. (2003). Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass-spectrometry. Anal. Chim. Acta 480 207–218. [Google Scholar]
- Ohto, M., Fischer, R.L., Goldberg, R.G., Nakamura, K., and Harada, J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Rashotte, A.M., Carson, S., To, J.P.C., and Kieber, J.J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, A.E., and Lang, A. (1957). Effect of kinetin on protein content and survival of detached Xanthium leaves. Science 125 650–651.13421662 [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-KAT) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Scheres, B., Di Laurenzio, L., Willemsen, V., Hauser, M.T., Janmaat, K., Weisbeek, P., and Benfey, P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121 53–62. [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Chory, J., and Furuya, M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Cell Physiol. 2 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíchal, L., Rakova, N.Y., Riefler, M., Mizuno, T., Romanov, G.A., Strnad, M., and Schmülling, T. (2004). Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 45 1299–1305. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizumo, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42 107–113. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Bäurle, I., Kudla, J., Nagy, F., Schäfer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with the photoreceptor phytochrome B in modulating red light signalling. Science 294 1108–1111. [DOI] [PubMed] [Google Scholar]
- To, J.P.C., Haberer, G., Ferreira, F.J., Deruère, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signalling. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2003). Organ shape and size: A lesson from studies of leaf morphogenesis. Curr. Opin. Plant Biol. 6 57–62. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001). The AHK4 gene involved in the cytokinin-signalling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42 751–755. [DOI] [PubMed] [Google Scholar]
- Veselý, J., Havlicek, L., Strnad, M., Blow, J.J., Donella-Deana, A., Pinna, L., Letham, D.S., Kato, J., Detivaud, L., and Leclerc, S. (1994). Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem. 224 771–786. [DOI] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42 1017–1023. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G., Kamiya, Y., and Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.H., Yu, H., and Goh, C.J. (2003). Functional characterisation of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol. Biol. 51 237–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.