Abstract
We screened for suppressors of repressor of silencing1 (ros1) using the silenced 35S promoter-neomycin phosphotransferase II (Pro35S:NPTII) gene as a marker and identified two allelic mutants, ror1-1 and ror1-2 (for suppressor of ros1). Map-based cloning revealed that ROR1 encodes a 31-kD protein similar to DNA replication protein A2 (RPA2A). Mutations in ROR1 reactivate the silenced Pro35S:NPTII gene but not RD29A promoter-luciferase in the ros1 mutant. DNA methylation in rDNA, centromeric DNA, and RD29A promoter regions is not affected by ror1. However, chromatin immunoprecipitation data suggest that histone H3 acetylation is increased and histone H3K9 dimethylation is decreased in the 35S promoter in the ror1 ros1 mutant compared with ros1. These results indicate that release of silenced Pro35S:NPTII by ror1 mutations is independent of DNA methylation. ROR1/RPA2A is strongly expressed in shoot and root meristems. Mutations in ROR1/RPA2A affect cell division in meristems but not final cell sizes. Our work suggests important roles of ROR1/RPA2A in epigenetic gene silencing and in the regulation of plant development.
INTRODUCTION
Foreign genes integrated into plant genomes and some endogenous genes often become silenced at the transcriptional level. Transcriptional gene silencing (TGS) involves the establishment of a condensed chromatin structure that is often closely related to DNA hypermethylation (Bender, 2004; Tariq and Paszkowski, 2004; Chan et al., 2005). Genes that affect TGS by altering DNA methylation can be divided into two major groups. The first group, including DECREASE IN DNA METHYLATION1 (DDM1) (Vongs et al., 1993; Jeddeloh et al., 1999; Morel et al., 2000; Scheid et al., 2002), DNA METHYLTRANSFERASE1 (Vongs et al., 1993; Morel et al., 2000), CHROMOMETHYLASE3 (CMT3) (Bartee et al., 2001; Lindroth et al., 2001; Tompa et al., 2002), KRYPTONITE1/SU(VAR)3-9 HOMOLOG4 (KYP1/SUVH4) (Jackson et al., 2002; Malagnac et al., 2002), SUVH2 (Naumann et al., 2005), and HOMOLOGY-DEPENDENT GENE SILENCING1 (HOG1) (Rocha et al., 2005) (our unpublished results), affects DNA methylation at the whole genome level. The second group of genes, such as DOMAINS-REARRANGED METHYLASE1 (DRM1) and DRM2 (Cao and Jacobsen, 2002), HISTONE DEACETYLASE6 (Aufsatz et al., 2002a; Probst et al., 2004), DICER-LIKE3 (Xie et al., 2004), ARGONAUTE4 (Zilberman et al., 2003, 2004), DNA-DEPENDENT RNA POLYMERASE IV (DRD1) (Kanno et al., 2004), and NUCLEAR RNA POLYMERASE IV 1B (NRPD1B)/DRD3 and NRPD2A/DRD2 (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005), affect DNA methylation only in some specific regions of the genome. Recent studies also identified several other genes that regulate TGS without changing DNA methylation. Among them, MORPHEUS' MOLECULE1 (MOM1), which encodes a protein with limited similarity to the SWI2/SNF2 family of proteins, affects TGS probably through chromatin remodeling (Amedeo et al., 2000; Scheid et al., 2002; Tariq et al., 2002). BRUSHY1 (BRU1) (a DNA repair-related protein) (Takeda et al., 2004), FASCIATA1 (FAS1) and FAS2 (subunits of chromatin assembly factor [CAF-1], the condensin complex) (Kaya et al., 2001), and MEIOTIC RECOMBINATION11 (MRE11) regulate epigenetic gene silencing during DNA/chromatin replication, repair, or recombination (Takeda et al., 2004).
The analysis of Arabidopsis thaliana mutants has revealed several other chromatin-related genes that regulate flowering time, suggesting that a chromatin-mediated gene regulation system functions during plant development (Fransz and de Jong, 2002; He and Amasino, 2005). For example, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), encoding an ISWI family chromatin remodeling protein, is an activator of the flower repressor FLOWER LOCUS C (FLC), a MADS box transcriptional regulator, and a mutation in PIE1 leads to early flowering (Noh and Amasino, 2003). VERNALIZATION2, a Polycomb-group protein, suppresses the expression of FLC in response to vernalization (Gendall et al., 2001). EARLY BOLTING IN SHORT DAYS, encoding a chromatin remodeling factor, is required to repress the expression of FLOWER LOCUS T (FT), a key gene in the floral promotion pathways in Arabidopsis (Pineiro et al., 2003). TERMINAL FLOWER2 (TFL2), encoding a homolog of HETEROCHROMATIN PROTEIN1 (HP1), negatively regulates FT expression (Kotake et al., 2003; Takada and Goto, 2003). Mutations in TFL2 lead to early flowering and a terminal floral structure (Larsson et al., 1998). HP1 in mammals is thought to bind methylated lysine 9 of histone H3 (H3K9), which results from the activity of histone methyltransferase SUV39, to form a repressive complex (Bannister et al., 2001; Lachner et al., 2001). In plants, TFL2 binds in vitro to CMT3, which is required for maintenance of CNG methylation (Bartee et al., 2001; Lindroth et al., 2001; Tompa et al., 2002; Lindroth et al., 2004). CMT3, in turn, interacts with the histone 3 tail, which is methylated at H3K9, controlled by histone methyltransferase KYP, and at H3K27, regulated by an unknown protein (Jackson et al., 2002; Lindroth et al., 2004). DNA methylation and H3K9 methylation are marks of heterochromatin in plants and mammals, whereas DNA hypomethylation, H3K4 methylation, and histone H3 hyperacetylation are characteristics of euchromatin (Johnson et al., 2002; Mathieu and Bender, 2004; Tariq and Paszkowski, 2004).
Previously, Gong et al. (2002) described the isolation of a repressor of TGS, Repressor of Silencing1 (ROS1), which, when mutated, causes TGS of two originally active genes in a T-DNA region, RD29A promoter-luciferase (ProRD29A:LUC) and 35S promoter-neomycin phosphotransferase II (Pro35S:NPTII). ROS1 encodes a DNA repair protein, and recombinant ROS1 protein can cut methylated but not unmethylated DNA in an in vitro assay, which suggests that ROS1 probably functions as a DNA demethylation enzyme to keep the RD29A promoter active (Gong et al., 2002).
In this study, we isolated mutants that show a release of TGS using the silenced Pro35S:NPTII gene as a selection marker. We isolated two allelic mutants in the ros1 background, ror1-1 and ror1-2 (for suppressor of ros1), that show a kanamycin-resistant phenotype. The mutated gene was cloned and shown to encode a protein similar to DNA replication protein A2 (RPA2A). RPA is a three-subunit protein complex that has a critical function during DNA metabolism, including DNA replication, repair, and recombination (Wold, 1997; Binz et al., 2004). In the yeast Saccharomyces cerevisiae, each subunit of RPA is encoded by a single essential gene. Cell growth is arrested when expression of any of the three genes is disrupted (Brill and Stillman, 1991). In Arabidopsis, there are four RPA1 (including At2g06510/RPA1A/RPA70a, At5g08020/RPA1B/RPA70b, At5g45400/RPA1C, and At5g61000/RPA1D) (Ishibashi et al., 2005), two RPA2 (including At2g24490/RPA2A/ROR1 and At3g02920/RPA2B), and two RPA3 homologues (including At3g52630/RPA3A and At4g18590/RPA3B). Our results suggest that ROR1/RPA2A plays a critical role in maintaining epigenetic gene silencing and in the regulation of plant development.
RESULTS
Identification of ros1 Suppressor Mutations That Release TGS of Pro35S:NPTII
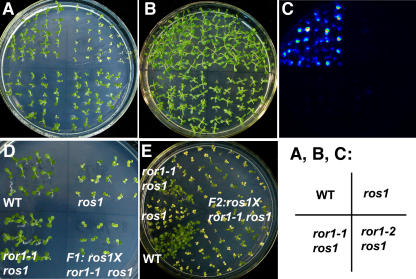
The ros1-1 plants (ros1, C24 accession) (Gong et al., 2002) were mutagenized with ethyl methanesulfonate, and M2 generation seeds were screened by germination on Murashige and Skoog (MS) medium containing 50 mg/L kanamycin, a condition under which ros1 mutants are very sensitive and are not able to grow. Putative mutants able to grow on the kanamycin-containing medium were selected from the M2 population of mutagenized ros1 plants. We identified two allelic mutants, ror1-1 and ror1-2, in the ros1 background. ror1-1 ros1 and ror1-2 ros1 mutants but not ros1 plants grew well on medium containing kanamycin (Figure 1A). We compared the kanamycin-resistant phenotype of ror1-1 ros1 and C24 wild type at different concentrations of kanamycin ranging from 50 to 200 mg/L and found that the suppressor mutants have the wild-type level of kanamycin resistance. RT-PCR analysis indicated that the transcripts of NPTII could be detected in the wild type and ror1-1 ros1 but not in ros1 mutant plants (Figure 2B). The ror1 ros1 mutants were each backcrossed with the original ros1 mutant, and all F1 plants were kanamycin sensitive (Figure 1D, only ror1-1 ros1 is shown). The progeny obtained by selfing of F1 plants showed a ratio of ∼3:1 kanamycin-sensitive:kanamycin-resistant seedlings (a total of 1240 seedlings tested, 918 sensitive, 322 insensitive) (Figure 1E), which indicates that the suppressor mutant phenotype is caused by a recessive mutation in a single nuclear gene (ror1-2 ros1 shows similar results).
Figure 1.
ror1 Mutations Release the TGS of the Pro35S:NPTII Transgene in the ros1 Mutant.
(A) ror1-1 ros1 and ror1-2 ros1 mutants containing the Pro35S:NPTII reporter transgene exhibit similar kanamycin-resistant phenotypes as C24 wild-type plants, whereas the ros1 mutant is sensitive to kanamycin.
(B) and (C) Comparison of luminescence after ABA treatment for 5 h in ror1-1 ros1, ror1-2 ros1, ros1, and C24 seedlings.
(B) C24 wild type (top left), ros1 (top right), ror1-1 ros1 (bottom left), and ror1-2 ros1 (bottom right) seedlings grown on MS medium.
(C) Luminescence comparison of different plants in (B).
(D) F1 seedlings of ror1 ros1 crossed with ros1 are sensitive to kanamycin. C24 wild type (top left), ros1 (top right), ror1-1 ros1 (bottom left), and F1 (crossed between ror1-1 ros1 and ros1, bottom right) seedlings grown on MS medium supplemented with 50 mg/L kanamycin.
(E) Seedlings of selfed F1 progeny segregate at a ratio of 3:1 kanamycin-sensitive:kanamycin-resistant seedlings. Plants were grown on MS medium containing 50 mg/L kanamycin.
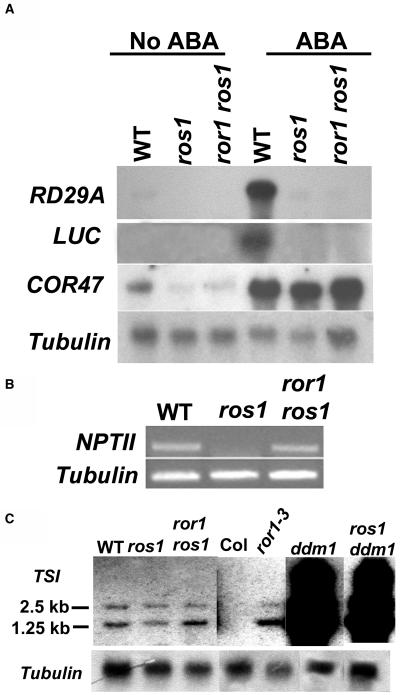
Figure 2.
ror1 Mutations Have No Effect on Expression of Either RD29A or LUC but Increase the Expression of NPTII and TSI Transcripts.
(A) RNA gel blot analysis with RD29A and LUC as probes reveals that RD29A and LUC remain silenced in ror1-1 ros1 and ros1 mutants after ABA treatment. By contrast, RD29A and LUC expression was induced to a high level in C24 wild-type plants. COR47 was used as a positive control for ABA treatment. Tubulin was used as a loading control.
(B) RT-PCR analysis of NTPII expression. NPTII transcripts were detected in both ror1 ros1 and the wild type but not in ros1.
(C) RNA gel blot analysis with the TSI fragment as a probe shows that ror1 mutations increase the level of TSI transcripts. The expression of TSI in ror1-3, a T-DNA insertion mutant, was compared with that in the wild type. ddm1 and ddm1 ros1 mutants were used as positive controls for TSI expression. Tubulin was used as a loading control. Col, Columbia accession.
The Silencing of ProRD29A:LUC Transgene and Endogenous RD29A Gene Is Not Released in ror1 ros1 Mutants
Because Pro35S:NPTII is adjacent to ProRD29A:LUC, which is also silenced in ros1 mutants (Gong et al., 2002), we next analyzed the LUC gene expression in ror1-1 ros1 and ror1-2 ros1 mutants by determining the bioluminescence in response to abscisic acid (ABA) treatment. Without ABA treatment, no bioluminescence could be detected in any of the plants (data not shown). Figure 1C shows the bioluminescence images of the wild type, ros1, and the suppressor mutants after ABA treatment. Compared with the wild type, which emitted strong luminescence, ros1, ror1-1 ros1, and ror1-2 ros1 emitted virtually no luminescence. RNA gel blot analysis indicated that both ProRD29A:LUC and the endogenous RD29A gene remained silenced in ror1-1 ros1 mutants just as in ros1 plants (Figure 2A). As a control, RD29A or LUC was induced to a high level in wild-type plants (Figure 2A). Because ror1-1 ros1 and ror1-2 ros1 show similar phenotypes, we used ror1-1 ros1 (indicated as ror1 ros1 herein unless specifically noted), which was backcrossed to ros1 twice, in all subsequent experiments.
The Transcripts of Transcriptionally Silent Information Are Expressed at Higher Levels in Mutant Plants Compared with Wild-Type Plants for the ror1 Gene
The transcript of a retrotransposon-related Athila sequence called Transcriptionally Silent Information (TSI) is generated from the repeats of heterochromatic pericentromeric regions of chromosomes (Steimer et al., 2000). TSI transcript levels in some TGS mutants, such as mom1, hog1, ddm1, ddm2, som1-8, sil, cmt3, kyp, bru1, fas1, and fas2, are increased (Steimer et al., 2000; Lindroth et al., 2001; Jackson et al., 2002; Takeda et al., 2004). We checked the TSI transcripts in ror1 ros1 mutants. Total RNAs extracted from 18-d-old seedlings were used for RNA gel blot analysis. As shown in Figure 2C, the TSI transcript is expressed at a higher level in ror1 ros1 compared with ros1 or the wild type. We also compared the levels of TSI transcripts in a ror1 T-DNA insertion mutant (ror1-3, SALK_129173). TSI transcripts in ror1-3 were expressed at a higher level compared with those in the Columbia wild type, in which TSI transcripts were not detected under our experimental conditions. As a positive control, the expression of TSI was dramatically activated in ddm1 or ros1 ddm1 (Figure 2C). These results suggest that the ror1 mutation reactivates the expression of TSI, albeit to a much lesser degree compared with the effect of ddm1.
ror1 Mutations Do Not Affect DNA Methylation
Transgene repeats can lead to the production of small interference RNAs (siRNAs) and can trigger cytosine methylation of homologous DNA from the transgene and endogeneous gene (Finnegan and Matzke, 2003; Mathieu and Bender, 2004; Matzke and Birchler, 2005). To determine the structure of the inserted T-DNA, we digested genomic DNA with three different enzymes, PstI, HindIII, and BamHI, each of which has only one site in the original T-DNA construct. DNA gel blot analysis with LUC as a probe shows that there are two main bands in HindIII and BamHI digested products (Figure 3E). In PstI-digested product, there are two bands with stronger signal, one with a weaker signal. However, after a longer exposure, a weaker, smaller band was found in both HindIII- and BamHI-digested products. The largest and strongest bands have the same size in all three digested products, and the other strong bands have different sizes. The weaker, smaller band might have come from a partial LUC gene insertion. According to these and other DNA gel blot results (data not shown), we deduced that the T-DNA insertion contains two T-DNA repeats in the same orientation and a partial T-DNA fragment that may be located on either side of the two head-end T-DNA repeats (Figure 3F). The precise structure of the T-DNA insertion needs to be confirmed by sequencing of the genomic region containing the inserted DNA.
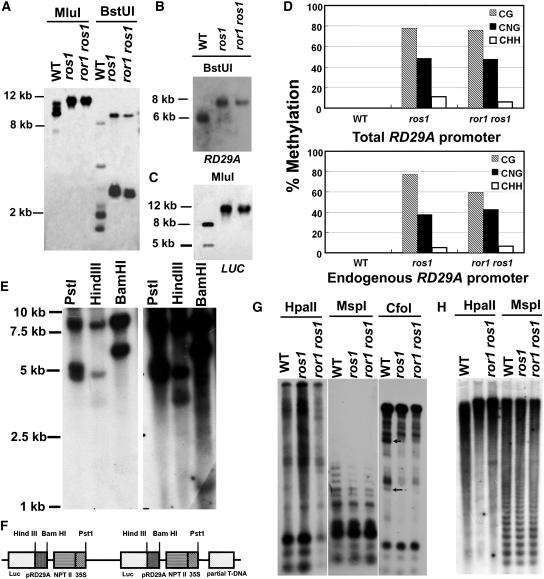
Figure 3.
ror1 Mutations Have No Effect on DNA Methylation of the RD29A Promoter, rDNA, and Centromeric DNA.
Genomic DNA samples isolated from C24 wild-type, ros1, and ror1 ros1 plants were digested with different methylation-sensitive restriction enzymes and subjected to DNA gel blot analysis or assayed by bisulfite sequencing.
(A) MluI and BstUI, with the RD29A promoter used as a probe to detect the total methylation of the RD29A promoter.
(B) BstUI, with RD29A used as a probe to detect the methylation of the endogenous RD29A promoter.
(C) MluI, with LUC used as a probe to detect the methylation of the transgene RD29A promoter.
(D) Bisulfite sequencing analysis of cytosine methylation in the upper strand of RD29A. Top: overall methylation of both the transgene RD29A promoter and the endogenous RD29A gene promoter. Bottom: methylation in endogenous RD29A promoter (Yamaguchi-Shinozaki and Shinozaki, 1994).
(E) Analysis of T-DNA copy number in ProRD29A:LUC transgenic plants. The genomic DNA was digested with restriction enzymes with unique sites in the T-DNA region, including PstI, HindIII, and BamHI, and hybridized with luciferase as probe.
(F) The deduced T-DNA insertion structure in ProRD29A:LUC transgenic plants.
(G) HpaII, MspI, and CfoI, with rDNA used as a probe. Arrows point to two extra bands in C24 wild-type plants not found in ror1 ros1 and ros1 mutant plants.
(H) HpaII and MspI, with 180-bp centromeric DNA used as a probe.
In order to test whether ROR1 plays a role in DNA methylation, we assayed for DNA methylation at the RD29A promoter using two methylation-sensitive enzymes, BstUI (CGCG) and MluI (ACGCGT), as described previously (Gong et al., 2002). No apparent difference was found between ror1 ros1 and ros1 in DNA methylation at the RD29A promoter regions when either the RD29A promoter (to test methylation of the RD29A promoter in both the transgene and endogenous RD29A) (Figure 3A), endogenous RD29A (to check the endogenous RD29A promoter) (Figure 3B), or LUC (to check the transgene RD29A promoter) (Figure 3C) was used as a probe. Bisulfite sequencing for overall methylation of the RD29A promoter (including both the transgene and endogenous RD29A promoter) indicates that little DNA methylation exists in the wild type. By contrast, heavy DNA methylation in CG, CNG (N is A, T, G, or C), and CHH (H is A, C, or T) sites is detected in both ros1 and ror1 ros1, but there was no apparent difference in DNA methylation patterns between them (Figure 3D, top panel). We further determined the methylation of the endogenous RD29A promoter using bisulfite sequencing. Although the methylation level at the endogenous RD29A promoter in CG sites is slightly lower in ror1 ros1 than that in ros1, the overall DNA methylation pattern is similar between ros1 and ror1 ros1 (Figure 3D, bottom panel). The slight methylation difference in CG sites might be difficult to detect by DNA gel blot analysis. This result is consistent with the previous observation that neither ror1 ros1 nor ros1 releases the silencing of the ProRD29A:LUC transgene or endogenous RD29A.
We also tested whether the ror1 mutations would influence DNA methylation in centromeric and rDNA regions. No DNA methylation difference was detected at rDNA and centromeric DNA between ror1 ros1 and ros1 plants (Figures 3G and 3H). However, digestion with the restriction enzyme CfoI (GCGC) revealed that methylation was increased in the rDNA regions in ros1 and ror1 ros1 plants compared with wild-type plants (Figure 3G). This result indicates that the ros1 mutation might cause increased DNA methylation in some genomic regions (Gong et al., 2002), but the ror1 mutation does not affect such DNA methylation.
Since the transgene of Pro35S:NPTII was reactivated in the ror1 ros1 double mutant but not in ros1 mutants, we wondered if there were any differences in DNA methylation level at the 35S promoter between the two mutants. Using DNA methylation sensitive restriction enzymes (DdeI for CTNAG and XmiI for GTMKAC) (Aufsatz et al., 2002b), we did not detect any DNA methylation difference in the 35S promoter among ror1 ros1, ros1, and the wild type (Figure 4A). In addition, we used bisulfite sequencing to compare the methylation status of the 35S promoter and found little accumulation of DNA methylation in the 35S promoter in the wild type, ros1, and ror1 ros1 (Figure 4B). The fact that there is no apparent difference in the methylation level between ros1 and the wild type suggests that the silencing of the 35S promoter is different from that of the RD29A promoter and is not caused by DNA hypermethylation.
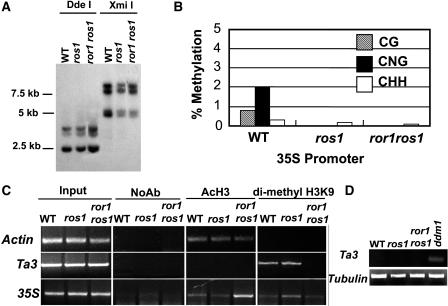
Figure 4.
Reactivation of the Silenced Pro35S:NPTII Gene Is Independent of DNA Methylation but Related to Histone Modification.
(A) DNA gel blot analysis of genomic DNA digested with methylation-sensitive enzymes DdeI (CTNAG) and XmiI (GTMKAC) hybridized with the 35S promoter as probe.
(B) Bisulfite sequencing analysis of cytosine methylation in the upper strand of the 35S promoter (from 61 to 400, GenBank accession number AJ007625).
(C) ChIP analysis of histone H3 acetylation and H3K9 dimethylation in 35S promoter region. Chromatin was precipitated using specific antibodies against acetylated histone H3 and methylated H3 (anti-H3K9). Equal input used for ChIP was confirmed by equal PCR amplification. Actin was used as a control for anti-AcH3 and anti-H3K9 precipitation. NoAb, precipitation without antibody as a negative control.
(D) The expression of Ta3 in ror1 ros1, ros1, ddm1, and wild type by RT-PCR. Tubulin was used as a control.
Reactivation of the 35S Promoter by the ror1 Mutation Is Related to Histone Modification
Lines of evidence suggest that histone modification plays important roles in TGS (Aufsatz et al., 2002a; Jackson et al., 2002). Therefore, we investigated whether histone modification is involved in reactivation of the 35S promoter. We performed chromatin immunoprecipitation (ChIP) experiments using antibodies specific for acetyl H3 known to be associated with transcriptionally active genes or dimethylated H3K9 known to be associated with TGS to examine the status of histone H3 modification of the 35S promoter in wild-type, ros1, and ror1 ros1 plants (Jenuwein and Allis, 2001; Noma et al., 2001; Gendrel et al., 2002). As a control (Johnson et al., 2002), the Actin promoter representing an active gene showed similar PCR amplification for acetyl H3 antibody, and no amplification was observed for the antibody against dimethylated H3K9 that in immunostaining experiments on Arabidopsis chromosomes predominantly binds to heterochromatic regions (Figure 4C, top panel). On the other hand, Ta3 representing a silenced gene is associated with dimethylated H3K9 antibody but not acetyl H3 antibody in wild-type plants (Figure 4C, middle panel). As shown in Figure 4C, it appears that in both ror1 ros1 and wild-type plants, the 35S promoter was associated more with acetyl H3 but more weakly with H3K9 dimethylation compared with the ros1 mutant. This result suggests that reactivation of the 35S promoter in ror1 ros1 mutants might be partially due to changes in chromatin modification. We also noticed that in ror1 ros1mutants, H3K9 dimethylation was greatly reduced at Ta3. In order to further examine whether this loss of H3K9 dimethylation could release TGS, we performed RT-PCR at Ta3 using ddm1 as a positive control. Intriguingly, as in ros1 and the wild type, no transcript could be detected in ror1 ros1, but the transcript could be detected in ddm1 by RT-PCR (Figure 4D). This suggests that although the ror1 mutation might affect H3K9 methylation at Ta3, the mutation has no apparent effect on Ta3 expression. It is conceivable that alterations in histone modification at the Ta3 region measured in this study might not have affected Ta3 expression, or the expression of Ta3 was too low to be detected under our experimental conditions. Our ChIP data suggest that histone alterations occur at certain sequences in the mutant, but more comprehensive follow-up studies are needed to fully understand these changes.
ror1 Mutations Influence Cell Division but Not Final Cell Size
ror1 ros1 plants are smaller than wild-type plants (Figures 5A to 5C), and the leaves and flowers are all smaller (Figures 5A to 5C and 5H). There are fewer trichomes on the leaves of the wild-type plant (accession C24) used in this study than that of other accessions, such as Columbia. However, virtually no trichomes could be found on the leaves of ror1 mutants (Figure 5H). To investigate whether the smaller leaf phenotype is caused by defects in cell division or cell expansion, we compared the epidermal cells of the abaxial side of the mature leaf in ror1 ros1, ros1, and wild-type plants and found no difference among the genotypes (Figures 6A to 6C). We further compared the cell morphology in a cross section of the middle part of mature leaves (the fourth leaf of 3-week-old plants) and found no difference in cell shape among C24, ros1, and ror1 ros1 plants (Figures 6D to 6F). Because the wild-type or ros1 leaf is bigger than that of ror1 ros1 (Figures 5A to 5C and 5H), the ROR1 mutation might reduce cell division but not the final leaf cell size during leaf development.
Figure 5.
Growth Phenotypes of ror1 ros1 Mutants.
(A) Seedlings grown in soil for 3 weeks under long-day conditions (16 h light/8 h dark). (a) C24 wild-type; (b) ros1 mutant; (c) ror1-1 ros1 mutant; (d) ror1-2 ros1 mutant. The seedlings of ror1-1 ros1 and ror1 -2 ros1 have begun to flower.
(B) Seedlings grown on MS plates for 18 d.
(C) Seedlings grown in soil under long-day conditions for 32 d. Note that the ror1 ros1 mutant has many branches without an apparent main shoot.
(D) Comparison of flower sizes among ror1 ros1 (left), ros1 (middle), and C24 wild type (right) is shown in (a). (b) to (f) The different floral organs observed in ror1 ros1 plants. These abnormal phenotypes are seldom observed in normal ros1 or C24 plants. (b) and (c) Several siliques developing from the terminal flower. (d) Terminal floral structure in a main shoot. (e) Terminal floral structure in an axillary shoot. (f) Carpels bearing ovules.
(E) The comparison of growth phenotypes between the ror1 single mutant and ror1 ros1 double mutant.
(F) and (G) Young inflorescence structures and silique size comparison among ror1 ros1 (left), ros1 (middle), and C24 wild type (right).
(H) Mature leaf size comparison between ros1 and ror1 ros1. Note trichomes on leaf surface of ros1 but not on ror1 ros1.
Figure 6.
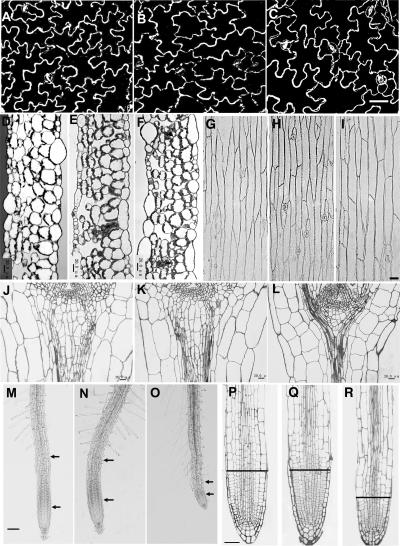
Cell Shape Comparison among ror1 ros1, ros1, and C24 Wild Type.
(A) to (C) Epidermal cells of adaxial side mature leaf. (A) C24 wild type; (B) ros1; (C) ror1 ros1. Bar = 60 μm.
(D) to (F) Cross section of the middle parts of mature leaves. (D) C24 wild type; (E) ros1; (F) ror1 ros1. Bars = 20 μm.
(G) to (I) Epidermal cells from the first node of mature inflorescence stem. (G) C24 wild type; (H) ros1; (I) ror1 ros1. Bar = 40 μm.
(J) to (L) The structures of median longitudinal sections of shoot apices. (J) C24 wild type; (K) ros1; (L) ror1 ros1. Bars = 20 μm.
(M) to (O) Primary root of 12-d-old seedlings. (M) C24 wild type; (N) ros1; (O) ror1 ros1. The elongation zone is the region between the two arrows. Bar = 50 μm.
(P) to (R) The structures of median longitudinal sections of root tips. (P) C24 wild type; (Q) ros1; (R) ror1 ros1. The region below the line is the root meristem zone. Bar = 20 μm.
ror1 ros1 plants flower earlier than wild-type or ros1 plants under long-day conditions. The number of rosette leaves 1 d before the onset of flowering is ∼12 ± 1 in wild-type and ros1 plants but only ∼8 ± 1 in ror1 ros1 plants (120 plants for counting, two independent experiments). The flowering time for ror1 ros1 is ∼20 ± 1 d compared with 26 ± 1 d for wild-type or ros1 plants. The flowers and siliques of ror1 ros1 are smaller than those of ros1 or C24 (Figures 5Da, 5F, and 5G). During the later developmental stages, ∼31% (117 plants examined) of ror1 ros1 shoots showed defective inflorescence development, which results in an early formation of terminal floral structures (Figures 5Db to 5Df). The degree of terminal floral defects varies between individual ror1 ros1 plants or the different branches of the same plant. The length of internodes in ror1 ros1 plants is shorter than that of wild-type plants when the plants are mature (Figure 5C). Compared with ros1 or wild-type plants, ror1 ros1 plants have more branch shoots (Figure 5C). The earlier flowering and terminal flower structure phenotypes in ror1 ros1 are very similar to those described for the tfl2 mutant (Larsson et al., 1998). We crossed ror1 ros1 with C24 wild type and obtained the ror1 single mutant without the ros1 mutation. The ror1 single mutant shows the same developmental phenotypes as the ror1 ros1 double mutant (Figure 5E).
To investigate whether the shoot phenotype is caused by an altered shoot apical meristem (SAM), we analyzed the structure of median longitudinal sections of SAM under a light microscope, and the representative samples are shown in Figures 6J to 6L. SAM consists of three classical zones in dicots grown under long-day conditions: the central zone at the SAM apex; the peripheral zone, which encircles the central zone; and the rib zone below the central and peripheral zones. The cells in rib zones divide rapidly, and their daughter cells differentiate into subapical pith in which the cells are elongated and contain more vacuoles (Nakajima and Benfey, 2002; Jacqmard et al., 2003). Seedlings grown for 8 d under long-day conditions were used for the SAM cell morphology comparison. It is difficult to tell any difference in cell shapes in the central and peripheral zones among ror1 ros1, ros1, and C24 plants. However, the growth of cells in the rib zone of ror1 ros1 appears to be greatly inhibited, resulting in round, smaller and shorter cells in the subapical pith as compared with those in ros1 or the wild type. The cells in each layer between the stele and epidermis are shorter, which is likely caused by delayed cell division or slower growth (Figures 6J to 6L).
We found no differences in node epidermal cell shapes among ror1 ros1, ros1, and C24 mature plants (Figures 6G to 6I). Because the nodes are much shorter in ror1 ros1 than ros1 or C24 mature plants (Figure 5C), ror1 mutations must cause a reduction in cell division. Taken together, the above results suggest that ror1 mutations affect cell division but not final cell sizes.
Since the ror1 ros1 roots are much shorter than those of ros1 or C24 (Figure 5B), we examined the organization of root tips. The Arabidopsis root tip consists of the meristem and elongation and differentiation zones (Dolan et al., 1993). The elongation zone in ror1 ros1 plants is much shorter than that of ros1 or C24 plants (Figures 6M to 6O). Examination of median longitudinal sections indicated that the meristematic zone in ror1 ros1 is only about half that of ros1 or C24 (Figures 6P to 6R). These results suggest that the lesions in ROR1 disrupt regular cell division in the root.
ROR1 Encodes a Putative DNA RPA2
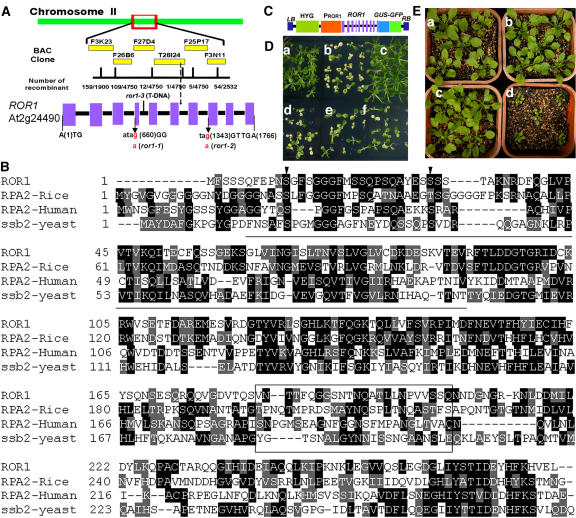
We identified the ROR1 gene using a map-based cloning strategy. Plants with ror1 phenotypes were selected from an F2 population originating from the progeny of a cross between ror1 (C24 accession) and gl1 (Columbia accession). The ROR1 mutation was finely mapped to a region in BAC clone T28I24 (Figure 7A). We compared the sequences of all open reading frames on BAC clone T28I24 amplified from ros1 and ror1-1 ros1 mutants and found a G-to-A point mutation in the At2g24490 gene (G660 to A660, counting from the first putative ATG) in ror1-1 (Figure 7A). We also sequenced the At2g24490 gene from ror1-2 plants and found another G-to-A mutation (G1343 to A1343, Figure 7A). Both mutations change the 3′ splicing sites (from AG to AA), which are predicted to disrupt pre-RNA splicing, and would result in open reading frames of 69 (for the ror1-1 mutation) and 218 (for the ror1-2 mutation) amino acids, respectively. A T-DNA insertion line (SALK_129173, ror1-3) was obtained from the SALK T-DNA collection, and the T-DNA was found to be inserted in intron 4 of At2g24490 (between 1225 and 1226, counting from the first putative ATG) (Figure 7A). Compared with the Columbia wild type, ror1-3 shows phenotypic characters similar to ror1 ros1 mutants, such as earlier flowering and smaller size. A DNA fragment of 2433 bp containing the At2g24490 gene (1763 bp counted from the first putative ATG, but without the last putative TGA) fused in frame to the N terminus of the β-glucuronidase-green fluorescent protein (GUS-GFP) chimeric gene (Figure 7C) complemented the kanamycin (Figure 7D) and growth phenotypes of the ror1 ros1 mutant (Figure 7E) when introduced into ror1 ros1, thus confirming that At2g24490 is indeed ROR1.
Figure 7.
Positional Cloning of the ROR1 Gene and Mutant Complementation.
(A) Map-based isolation and gene structure of ROR1 (At2g24490), which consists of 10 exons. Blocks, exons; lines, introns. Mutations in ror1-1, ror1-2, or ror1-3 are indicated in the bottom panel.
(B) ROR1/RPA2A encodes a DNA RPA2 protein. The deduced primary amino acid sequence of ROR1 is aligned with three other RPA2s (RPA2-rice, BAD25304; RPA2-human, CAI21775; RPA2-yeast, CAA22775). Two conserved Ser residues in the N terminus are indicated with arrowheads. The conserved central domain for DNA binding activity is underlined. The less conserved domain in the C terminus is boxed.
(C) T-DNA construct used for complementation. HYG, hygromycin-resistant gene.
(D) ror1 ros1 mutant is complemented by the ROR1/RPA2A gene (At2g24490). Seedlings were grown on MS medium supplemented with 50 mg/L kanamycin. (a) C24 wild type; (b) ros1; (c) ror1 ros1; (d) to (f) three independent complemented lines (T3). Some seedlings in (e) show the kanamycin-resistant phenotype, which suggests that the segregation happened in progeny of this line.
(E) Growth phenotype of complemented plants. (a) C24 wild type; (b) ros1; (c) complemented plants; (d) ror1 ros1.
ROR1 cDNA obtained through RT-PCR was compared with the ROR1 genomic DNA. The ROR1 open reading frame contains 10 exons (Figure 7A) and is predicted to encode a 31-kD polypeptide of 279 amino acids with an isoelectric point of 5.2. The ROR1 gene product is most similar to the single-stranded DNA binding RPA2 from other species (Figure 7B). Two conserved Ser residues in the N-terminal part, which are modified by phosphorylation after DNA damage, and the conserved central domain for DNA binding activity, which exists in all single-stranded DNA binding proteins, are all found in ROR1. The C-terminal domain is less conserved in all RPA2 homologues and may function in protein–protein interaction (Binz et al., 2004). ROR1 shows 37% identity with another RPA2 homolog in Arabidopsis, which is encoded by At3g02920 and consists of 279 amino acids with an estimated molecular mass of 31 kD. Searching the EST databases identified one EST of At3g02920 and four ESTs of At2g24490/ROR1, suggesting that both genes are transcribed, and the expression of At3g02920 might be lower than that of ROR1. RT-PCR confirmed the expression of At3g02920 in Arabidopsis (see Supplemental Figure 1 online). We renamed At2g24490/ROR1 as RPA2A and At3g02920 as RPA2B. The two homologous genes might share some redundant functions in Arabidopsis.
ROR1/RPA2A Is Highly Expressed in Meristems and Young Tissues
RNA gel blot analysis with total RNAs from different tissues showed that ROR1 mRNA is abundant in roots, rosette leaves, cauline leaves, and flowers but less so in stems (Figure 8F). It is interesting that ROR1/RPA2A has two transcript sizes: the small one corresponds to the size of the mature mRNA, and the larger one probably corresponds to the pre-RNA. In roots, the main RNA transcript is the smaller one. However, in other tissues, the ratio of the larger to smaller ones differs. These results suggest that ROR1/RPA2A might be regulated at the posttranscriptional level during the splicing of pre-RNA to mRNA.
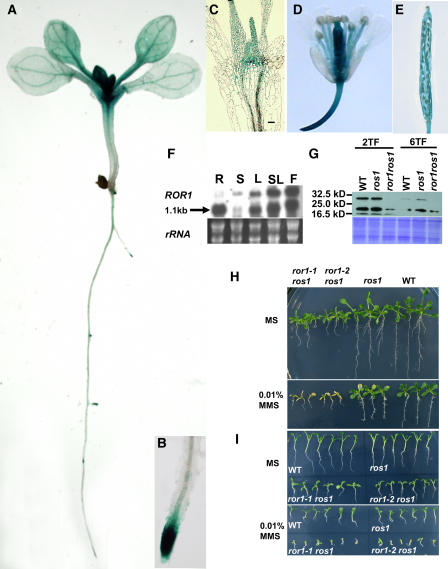
Figure 8.
The Expression Pattern of ROR1/RPA2A and Sensitivity of ror1 ros1 to MMS.
(A) to (E) GUS analysis in Arabidopsis transgenic seedlings. The complemented seedlings obtained from Figure 7 were used for the GUS activity assay.
(A) The overall expression within a young seedling.
(B) Expression within a primary root apex.
(C) Expression within shoot apex tissues in median longitudinal sections.
(D) Expression within a flower.
(E) Expression within a silique.
(F) RNA gel blot analysis of ROR1/RPA2A expression in different tissues. R, root; S, stem; L, leaf; SL, stem leaf; F, flower. rRNA was used as a loading control.
(G) Protein gel blot analysis of ROR1/RPA2A expression in seedlings with different true leaves. Forty micrograms of total proteins was loaded in each lane. 2TF, seedlings with two true leaves; 6TF, seedlings with six true leaves. WT, C24 wild type.
(H) The seedlings of ror1-1 ros1 and ror1-2 ros1 are more sensitive to MMS than ros1. Seven-day-old seedlings were transferred to liquid MS (top panel) or liquid MS medium containing 0.01% MMS (bottom panel). The picture was taken after transferring for 5 d. Note that the growth of ror1-1 ros1and ror1-2 ros1 was seriously inhibited compared with ros1 or the wild type. WT, C24 wild type.
(I) Germination of ror1-1 ros1 and ror1-2 ros1 seeds is more sensitive to MMS. Seeds were germinated on MS (top panel) or MS containing 0.01% MMS (bottom panel) for 12 d. WT, C24 wild type.
To visualize the pattern of ROR1/RPA2A gene expression, we analyzed the plants obtained from the above complementation experiment. The GUS expression pattern is expected to mimic that of the endogenous ROR1/RPA2A gene. Multiple, independent transgenic lines displayed the same expression pattern of the GUS reporter gene. GUS staining was detected in primary roots, SAMs, cotyledons, and vascular tissues (Figure 8A). At later stages of shoot development, GUS staining was detected more in SAM and young leaves but less in older leaves (Figure 8A). Median longitudinal sections of shoot tips of 8-d-old seedlings further confirmed GUS expression to be highest in SAMs, lower in young leaves, and lowest in old leaves (Figure 8C). In later stages of root development, GUS activity was detected in all lateral root primordia, including all stages during primordium formation (Figure 8A), and then localized around the root tips (Figures 8A and 8B). Strong GUS staining was also detected in flowers and weak staining in young siliques (Figures 8D and 8E). Transgenic plants expressing ProROR1:GUS showed similar GUS staining patterns (see Supplemental Figure 2 online) to plants with the whole ROR1/RPA2A gene fused with GUS-GFP. These results suggest that ROR1/RPA2A is expressed more in meristematic and young tissues, where cell division is most active.
We next examined ROR1/RPA2A protein levels in seedlings with two and six true leaves using ROR1/RPA2A protein antibody. Total proteins were extracted from different stages of seedlings and used for protein gel blot analysis. As shown in Figure 8G, the amount of ROR1/RPA2A protein detected in seedlings with two true leaves is higher than that in seedlings with six true leaves. The level of ROR1/RPA2A protein was similar to that in ros1 or C24 plants at the same growing stages. However, no intact ROR1/RPA2A protein could be detected in ror1-1 ros1 (Figure 8G) or ror1-2 ros1 plants (see Supplemental Figure 3 online), which suggests that the ror1 mutations might disrupt the function of ROR1/RPA2A completely. In addition to the 31-kD band of ROR1/RPA2A, the analysis also detected two smaller bands, one at ∼20 kD and another at ∼16.5 kD. These smaller proteins could be degradation products of ROR1/RPA2A protein or a result of mis-spliced ROR1/RPA2A or nonspecific reaction to the antisera. Since these two bands could be weakly identified even in the ror1 ros1 mutant, it is possible that they are translated from truncated ROR1/RPA2A mRNA. However, a similarly sized transcript was detected in the wild type and the two suppressor mutants but not in ror1-3 by RNA gel blot analysis (see Supplemental Figure 4 online), even though both of the point mutations are predicted to disrupt the correct splicing sites of pre-mRNA. It is likely that the smaller proteins represent nonspecific reactions and the transcripts in the mutants are mis-spliced forms that do not give rise to ROR1/RPA2A proteins.
ror1 Mutations Increase the Sensitivity to DNA Base Damage
RPA consists of 70-, 32-, and 14-kD subunits (RPA1, RPA2, and RPA3) and is involved in DNA replication, repair, recombination, and transcription (Binz et al., 2004). RPA is highly conserved in all eukaryotic organisms examined (Wold, 1997; Ishibashi et al., 2001). Disruption of Arabidopsis AtRPA70b causes the mutants to be more sensitive to methyl methanesulfonate (MMS) or UV-B, suggesting that RPA in plants functions in DNA repair (Ishibashi et al., 2005). MMS is a genotoxic agent that damages DNA bases. We used MMS to test whether the ror1 mutations affect DNA repair. Seedlings grown on solid MS medium for 7 d were transferred to new liquid MS medium containing 0.01% MMS. After growing for another 5 d, we noticed that the growth of ror1-1 ros1 and ror1-2 ros1 seedlings was completely inhibited and cotyledons became white, whereas ros1 and the wild type were less affected (Figure 8H). We also tested the germination sensitivity of ror1 ros1 mutants to MMS. The seeds of ror1-1 ros1, ror1-2 ros1, ros1, and wild-type plants were plated on MS medium containing 0.01% MMS or only MS medium as a control. Both ror1-1 ros1 and ror1-2 ros1 were more sensitive to MMS, compared with ros1 or the wild type (Figure 8I). In both seedling growth and germination tests, ros1 is more sensitive to MMS than the wild type, which is consistent with a previous report (Gong et al., 2002), but more tolerant than ror1 ros1. These results indicate that ror1 ros1 is more sensitive to MMS than ros1 and suggest that mutations in ROR1/RPA2A impair its DNA repair function in Arabidopsis.
ROR1/RPA2A Is Localized in the Nucleus and Interacts with ROS1 in Yeast Two-Hybrid Assays
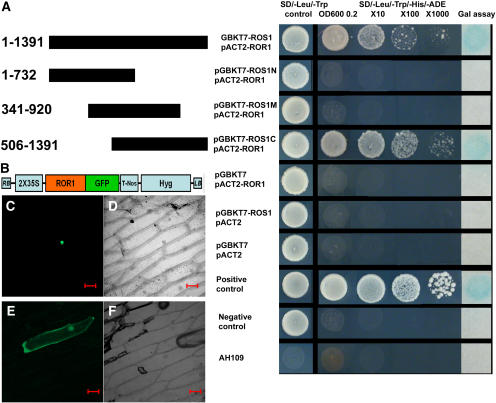
To study the subcellular localization of ROR1/RPA2A protein, we expressed the ROR1/RPA2A-GFP fusion gene under the constitutive 35S promoter. The ROR1/RPA2A-GFP fusion protein was localized in the nucleus by transient expression in onion epidermal cells (Figure 9C). GFP used as a control was shown to be localized more in the cytoplasm (Figure 9E).
Figure 9.
Interaction of ROR1/RPA2A and ROS1 in a Yeast Two-Hybrid System and ROR1/RPA2A-GFP Cellular Localization.
(A) ROR1/RPA2A-ROS1 interaction in a yeast two-hybrid assay. Full-length ROR1/RPA2A cDNA (840 bp) was fused to the GAL4 activation domain in the pACT2 vector. Different parts of ROS1 cDNA were fused with the GAL4 DNA binding domain in pGBKT7 vector. Interaction was observed when ROR1/RPA2A was cotransformed with full-length ROS1 or the C terminus (506 to 1391) of ROS1. ROR1/RPA2A or ROS1 did not show any activity when cotransformed with empty vector.
(B) T-DNA structure used for transient ROR1/RPA2A-GFP expression. Hyg, hygromycin-resistant gene.
(C) Transient expression of the ROR1/RPA2A-GFP fusion protein in onion epidermal cells under confocal microscopy. ROR1/RPA2A-GFP was localized in the nucleus.
(D) Bright-field image of (C).
(E) Transient expression of the GFP protein in onion epidermal cells. Compared with localization of ROR1/RPA2-GFPA fused protein, GFP is more distributed in the cytoplasm.
(F) Bright-field image of (E). Bars = 100 μm.
A previous study indicated that ROS1, a DNA repair protein, is also localized in the nucleus (Gong et al., 2002). We used a yeast two-hybrid system to examine whether ROS1 and ROR1/RPA2A might interact physically. ROS1 interacted with ROR1/RPA2A in the yeast two-hybrid assay, and further experiments indicated that ROR1/RPA2A interacted with the C-terminal but not the N-terminal region of ROS1 (Figure 9A).
DISCUSSION
The molecular basis for the formation and maintenance of TGS is still poorly understood. In this study, we provide evidence that ROR1/RPA2A, a putative RPA2, serves crucial functions in maintaining epigenetic gene silencing and regulating meristem development in Arabidopsis.
The T-DNA locus used in this study is very stable in the original transgenic C24 plants (Gong et al., 2002). Two actively expressed genes, ProRD29A:LUC and Pro35S:NPTII, in the T-DNA region become silenced when ROS1, which serves as a TGS repressor, is mutated (Gong et al., 2002). siRNAs are produced from the transgene RD29A promoter and probably act as a signal for triggering DNA methylation and gene silencing at both the transgene RD29A promoter region and endogenous RD29A promoter (Gong et al., 2002). By contrast, the 35S promoter that is also silenced in the ros1 mutant accumulates hardly any methylation. Using the silenced Pro35S:NPTII gene as a selection marker, we have identified several mutants that reverse the kanamycin-sensitive phenotype of the ros1 mutant. In this study, we show that mutations in a putative DNA replication A2 subunit reactivated the silenced Pro35S:NPTII gene but did not change the silenced status of ProRD29A:LUC and endogenous RD29A gene. By contrast, mutations in DDM1 (Gong et al., 2002) or HOG1 (our unpublished results) released TGS of both ProRD29A:LUC and Pro35S:NPTII in the ros1 background. These findings suggest that maintenance of TGS at the different loci may be regulated by different genes with different mechanisms.
Both DNA methylation and/or histone modification can regulate TGS (Richards and Elgin, 2002). Previous studies in plants suggest that siRNAs could precisely direct DNA hypermethylation in the RNA-DNA matching regions and cause TGS in these regions (Finnegan and Matzke, 2003; Mathieu and Bender, 2004; Matzke and Birchler, 2005). However, our results indicate that there is no clear difference in DNA methylation at the 35S promoter between ros1 and the wild type or between ros1 and ror1 ros1. ChIP analysis suggests that ror1 mutations might disturb the established H3K9 dimethylation in ros1 mutant and increase histone H3 acetylation. These results suggest that TGS of the Pro35S:NPTII gene might not be directly related to DNA methylation in this region but to histone modification. Recent studies on the epigenetic regulation of FLC transcription in Arabidopsis indicate its expression is mediated by histone methylation in response to vernalization, rather than by DNA methylation (Bastow et al., 2004; Sung and Amasino, 2004; He and Amasino, 2005). Mutations in ROR1/RPA2A release Pro35S:NPTII gene silencing, without any effect on ProRD29A:LUC or endogenous RD29A expression or DNA methylation at the RD29A promoter. The results suggest that ROR1/RPA2A functions in the maintenance of Pro35S:NPTII gene silencing independently of DNA methylation. Consistent with this notion, ror1 mutations also increase the expression of TSI to even a higher level than that in the wild type but have no apparent effect on DNA methylation in rDNA and centromeric DNA. TSI repeats are located in pericentromeric regions, which are hypermethylated. It is interesting to note that the expression of TSI is only reactivated to a low level by ror1 mutations when compared with that by ddm1 mutation. However, the silenced Pro35S:NPTII gene in ros1 mutant is reactivated by ror1 mutations to a similar level seen in the wild type. These results suggest that reactivation of the silenced Pro35S:NPTII gene and TSI by ror1 mutations might be controlled by different mechanisms.
It is conceivable that the heterochromatin formed around siRNA-directed DNA methylation at RD29A promoter might spread outward to the Pro35S:NPTII gene without changing the DNA methylation of the latter. The phenomena of heterochromatin spreading are well studied in fission yeast, which has no DNA methylation in the genome, but the exact mechanism is unknown (Huang, 2002). ROR1/RPA2A may play a critical role in maintaining the spreading and/or inheritance of the heterochromatin. Although it is theoretically possible that the expression of the NPTII gene might be driven by a promoter in the genomic DNA in which the transgene integrated rather than by 35S promoter, we think that this is unlikely considering the inserted T-DNA copy number and the nice correlations observed between NPTII expression and increased acetyl H3 and reduced H3K9 dimethylation levels. In bru1 and fas mutants, TGS of both heavily methylated GUS transgene and TSI is released in a DNA methylation independent manner (Takeda et al., 2004). By contrast, the observation that ror1 mutations release the silencing of TSI but not RD29A implies that the silencing at these loci might be mediated by different mechanisms. Taken together, we suggest that ROR1/RPA2A functions in maintaining TGS at specific loci without affecting DNA methylation.
T-DNA inserted in RPA1A/RPA70a in Arabidopsis is lethal, but disruption of RPA1B/RPA70b is viable and the mutant shows no morphological phenotype (Ishibashi et al., 2005). Further experiments will determine whether other RPA genes besides ROR1/RPA2A are involved in TGS. Protein gel blot analysis did not detect any ROR1/RPA2A protein in ror1-1 ros1 or ror1-2 ros1 mutants, which indicates that the mutant phenotypes might be caused by null mutations and that ROR1/RPA2A is not an essential gene in Arabidopsis. Some of the functions of the two RPA2 proteins in Arabidopsis might be redundant, but the dramatic mutant phenotypes of ror1 indicate that they clearly also have nonredundant functions.
Silenced chromatin is faithfully maintained during cell division, and DNA replication machinery has been implicated in TGS (Nakayama et al., 2001). RPA binds single-stranded DNA to promote an initial opening of DNA and helps to form a replication fork with the original replication complex (ORC) and other proteins, such as DNA polymerase α, during the early stage of DNA replication (Huang, 2002; Binz et al., 2004). After the replication forks are established, RPA remains associated with the fork during the DNA elongation phase (Wold, 1997). ORC is a protein complex with six subunits that binds initiator sequences at replication origins. Like RPA, ORC remains bound to the origins throughout the cell cycle in yeast (Gerbi et al., 2002). In budding yeast, ORC recruits the Sir1 protein to HML and HMR loci and functions in transcriptional silencing (Dillin and Rine, 1997). Although ORC is closely linked with TGS at the HMR locus in yeast, its role in replication is independent of its function in silencing (Fox et al., 1997). After DNA replication, the transiently disrupted chromatin structure must be rebuilt precisely to ensure proper inheritance of heterochromatin. Establishment of new heterochromatin after each cell cycle involves the participation of many other DNA replication machinery-related proteins, such as PCN1 (for DNA replication and repair), RFC1 (for DNA replication and repair checkpoints), Polɛ (for DNA replication and checkpoints), and Polα (for DNA replication) in fission yeast (Huang, 2002). Polα interacts directly with the HP1 homolog, Swi6, which functions in TGS (Nakayama et al., 2001). Swi6 also interacts with other components of the replication machinery (Bailis et al., 2003). In Drosophila, HP1 interacts with ORC2, and a mutation in ORC2 perturbs HP1 localization (Pak et al., 1997; Huang et al., 1998). These results indicate that HP1 is an integral component of the epigenetic cellular memory mechanism, and it might serve as a molecular bookmark to propagate heterochromatin imprint during cell division (Nakayama et al., 2000; Huang, 2002). ROR1 does not appear to contribute directly to establishing a repressed chromatin state since its mutations do not release TGS in the RD29A promoter, which might be the nucleation site for setting up the repressed chromatin in the T-DNA locus. Nevertheless, mutations in ROR1/RPA2A result in terminal and early flowering phenotypes, which are very similar to those observed in LHP1/TFL2 mutants (Larsson et al., 1998; Kotake et al., 2003). It is possible that RPA2 may interact directly or indirectly with the Arabidopsis HP1 homolog, LHP1 (Kotake et al., 2003), to control TGS at the 35S promoter and other DNA regions, such as TSI. It is also possible that ROR1/RPA2A mutations may affect the chromosomal localization of LHP1/TFL2. It has been shown that mutations in DNA polymerase α affect its binding to Swi6 at the mating-type region and thus promote transition from the silenced epigenetic state to the expressed state (Nakayama et al., 2001).
Human RPA interacts specifically with some essential DNA repair proteins, such as Uracil-DNA glycosylase, and is involved in multiple DNA repair pathways (Binz et al., 2004). ror1 ros1 mutants are more sensitive to DNA base damage compared with ros1, suggesting that ROR1/RPA2A is important for DNA repair. It is possible that ror1 mutations cause instability of the replication fork, which is reflected in defects in cell division, as suggested by the small sizes of organs but not cells. Interestingly, our yeast two-hybrid data suggest that ROR1/RPA2A specifically interacts with the C-terminal region of ROS1, a DNA glycosylase/lyase (Gong et al., 2002). Mutations in ROS1 also increase the sensitivity to DNA base damage. These results suggest that ROR1/RPA2A may function together with ROS1 in the DNA repair pathway. However, because ROS1 acts as a repressor of TGS, whereas ROR1/RPA2A is required for the maintenance of TGS, the physical interaction may be restricted to their function in DNA repair, and ROR1/RPA2A and ROS1 may function independently of one another in the epigenetic silencing pathway.
The expression of ROR1/RPA2A varies in different tissues. Promoter analysis indicates that ROR1/RPA2A is expressed more highly in root tips, shoot apices, and young leaves but is also detected in other differentiated tissues. This expression pattern is similar to that of rice RPA2 (Marwedel et al., 2003). Eukaryotic DNA replication is restricted to the S phase of the cell cycle. Mutations in RPA2 result in defects of S phase progression in yeast (Santocanale et al., 1995). Although ROR1/RPA2A is not an essential protein in Arabidopsis, its mutations greatly reduce cell division in both shoot and root meristems but have no effect on final cell sizes. Previous work showed that most ros1 plants grow normally and show no clear developmental phenotypes, but a few show abnormal phenotypes after inbreeding probably due to epigenetic effects caused by the ros1 mutation (Gong et al., 2002). In this study, we observed that the ror1 single mutant plants showed similar morphological phenotypes to ror1 ros1 double mutants, suggesting that the ror1 mutation alone is responsible for the developmental phenotypes.
Although shoot and root meristems share a fundamentally similar radial structure, most gene mutations required for the formation or maintenance of apical meristems specifically affect either SAMs or root apical meristems in Arabidopsis (Carles and Fletcher, 2003; Doerner, 2003). Only a few genes, such as FAS1 and FAS2 (CAF-1 complex) (Kaya et al., 2001), BRU1/TSK/MGO3 (a DNA repair-related protein) (Guyomarc'h et al., 2004; Suzuki et al., 2004; Takeda et al., 2004), and ROR1/RPA2A (this study), are required for the organization of both SAMs and root apical meristems. BRU1 and CAF-1, together with the condensing complex, or MRE11 were proposed to have cooperative roles for DNA/chromatin replication and stable reconstitution of epigenetic information (Kaya et al., 2001; Bundock and Hooykaas, 2002; Siddiqui et al., 2003; Takeda et al., 2004). Defective expression of WUSCHEL (WUS) mRNA in SAM is found in each mutant of fas1, fas2, and bru1. WUS is required for the maintenance of stem cell fate in Arabidopsis SAM (Mayer et al., 1998). Accumulating evidence suggests that defects in genome integrity affect epigenetic states of genes and meristem function in plants. Despite the similarities between ror1 and bru1 and fas1 and fas2 mutants in DNA repair and developmental phenotypes, ror1 phenotypes seem to be less severe than others in terms of meristem disorganization or MMS sensitivity. By contrast, ror1 appears to affect the cell cycle more drastically than the other fasciation mutants. This suggests that although ROR1/RPA2A and the other fasciation genes may function in some common pathways for DNA repair and gene silencing, ROR1/RPA2A may function through interactions with different partner proteins and may even have its own novel specific functions.
METHODS
Plant Growth and Mutant Isolation
Seedlings were germinated and grown on MS nutrient medium with 3% (w/v) sucrose and 0.7% agar in disk glass plates. After 1 week, the seedlings were transferred to a 340-mL pot filled with a mixture of peat/forest soil and vermiculite (3:1) in a greenhouse at 22°C, with light intensity of 50 μmol m−2 s−1 and 70% RH under long-day conditions (16-h-light/8-h-dark cycle).
Approximately 20,000 seeds of the ros1-1 mutant (accession C24) (Gong et al., 2002) (referred to as ros1 in this study) were mutagenized with use of ethyl methanesulfonate. Mutants that were able to grow on MS medium containing 50 mg/L kanamycin were isolated from ∼3500 single M1 plants in the M2 population. We identified two allelic mutants that show similar kanamycin-resistant and developmental phenotypes, named ror1-1 ros1 and ror1-2 ros1. For genetics analysis, the two mutants were backcrossed to ros1, and the resulting F1 seedlings exhibited the ros1 phenotypes. F2 progeny from self-fertilized F1 plants showed an ∼3:1 segregation of kanamycin sensitivity to resistant phenotypes. The resistant ones also showed the similar developmental phenotypes. We also crossed ror1-1 ros1 or ror1-2 ros1 to C24, and F2 progeny exhibited ∼3:1 of normal plants to development defect ones, which is consistent with the developmental phenotypes in ror1-1 ros1 or ror1-2 ros1 mutants being independent of ros1 mutations. The combined results indicate that ror1 mutants are caused by single nuclear recessive mutations. A T-DNA insertion line (ror1-3), SALK_129173 was obtained from the Arabidopsis Stock Center, and the insertion site were confirmed by PCR and sequencing. In this study, we mainly used ror1-1 ros1 as experimental materials; if not specially stated in the text, ror1-1 ros1 is ror1 ros1.
Seedlings grown on MS medium for 2 weeks were subjected to luciferase imaging analysis after being treated or not with 100 μM ABA for 5 h as described (Gong et al., 2002).
RNA Gel Blot Analysis
RNAs isolated from 18-d-old seedlings treated with 100 μM ABA for 5 h were transferred to membrane and hybridized with α-32P–labeled RD29A probe to detect the expression of RD29A. The expression of other genes was detected by each α-32P–labeled probe in RNAs extracted from 18-d-old seedlings. The TSI fragment was amplified with the forward primer 5′-CACTCTTGTTAATCCAAGTAGCTGACTCTCC-3′ and reverse primer 5′-GGGCTTTTGCCCATCTTCAATAGCT-3′ and was used as a probe (Steimer et al., 2000). Total RNAs isolated from the root, stem, leaf, and flower of Columbia accession were used for RNA gel blot analysis of ROR1 expression. Full-length ROR1 cDNA (840 bp) was α-32P labeled and used as a probe. Tubulin was used as a loading control. All labeling involved use of the Random Primer-labeled kit (TaKaRa).
DNA Gel Blot Analysis
DNA methylation in the RD29A promoter, 35S promoter, rDNA, and centromere DNA regions was determined by DNA gel blot analysis with methylation-sensitive restriction enzymes as previously described (Gong et al., 2002).
Genomic Bisulfite Sequencing
Two micrograms of genomic DNA from C24, ros1, and ror1 ros1 was digested with restriction enzymes BamHI, EcoRI, and HindIII. Sodium bisulfite treatment was performed as described (Jacobsen et al., 2000). The top strand of 35S promoter corresponds to positions 228 to 406 or 61 to 400 (GenBank accession number AJ007625). Primers for 35S promoter region of 228 to 406 are as follows: forward, 5′-GATAGTGGTTTTAAAGATGGAC-3′; reverse, 5′-TGAGATTTTTCAATAAAGGGTAATATT-3′. Primers for 35S promoter region of 61 to 400 are as follows: forward, 5′-AATAAAATAAACTTCCTTATATAAAAAAAA-3′; reverse, 5′-AAGATATAGTTTTAGAAGATTAAAGGG-3′. The top strand of RD29A promoter region corresponds to position 12,300 to 12,590 of TAC clone K24M7 (GenBank accession number AB019226), and primers are CX980 5′-GTAAAATGATTATATGATGGGTTAATAGATATGGATT-3′ and CX982 5′-CTTTCCAATAAAAATAATCAAACCCTTTATTCCTAATAATTA-3′. The top strand of endogenous RD29A promoter region corresponds to position 12,300 to 12,684 (GenBank accession number AB019226), and primers are CX980 and CX983 5′-TTTCTAAAATTAAAATCTACCTAAATACTACTAA-3′. PCR products were cloned using T-easy cloning vector (Promega), and 18 to 22 individual clones were sequenced for each PCR-amplified product at 35S and RD29A promoter regions, respectively. DNA methylation status in both long and short fragment amplification of 35S promoter are similar. The data shown in Figure 4B are only from the longer PCR products of the 35S promoter.
ChIP
ChIP assay was performed as previously described by Johnson et al. (2002). Briefly, formaldehyde was added to the leaves from 3- to 4-week-old plants at a final concentration of 1% and incubated for 10 min under vacuum at room temperature. The reaction was stopped by the addition of glycine to a final concentration of 0.125 M. Cross-linked leaves were harvested and lysed. Cross-linked chromatin was sonicated to reduce DNA length to achieve the chromatin solution. The chromatin solution was precleared with Protein A/agarose beads that had been preabsorbed with salmon sperm DNA and BSA (Upstate; 16-157). Immunoprecipitations were performed with anti-acetyl-Histone H3 (Upstate; 06-599) and anti-dimethyl-Histone H3 (Lys9) (Upstate; 07-441) antibodies. Cross-linking was reversed in the immunoprecipitate complexes by the addition of NaCl to a final concentration of 200 mM and incubation at 65°C for 4 to 6 h. The DNA was purified by proteinase K treatment (150 μg/mL) for 1 h, followed by phenol-chloroform extraction and precipitation by ethanol. The PCR analysis was performed using specific primers as follows: Actin primers were 5′-CGTTTCGCTTTCCTTAGTGTTAGCT-3′ and 5′-AGCGAACGGATCTAGAGACTCACCTTG-3′, Ta3 primers were 5′-GCTAAAACGGATTTCTCATGTAGAGGAT-3′ and 5′-TACTTATCCCACTCTAATCTCTTCCATAAACAC-3′, and 35S promoter primers were 5′-GGACTAGTGGCGCGCCACTGTCGGCAGAGGCATCTTGAACGATAG-3′ and 5′-CGGGATCCATTTAAATGGTGGAGCACGACACTCTCGTCTACTCC-3′.
Positional Cloning of the ROR1 Gene
We used the growth defect phenotypes of ror1 mutants to isolate the corresponding gene. The ror1-1 ros1 mutant was crossed to the wild-type gl1 (Columbia accession). The F2 population was screened for ror1 mutants on the basis of ror1 growth phenotypes. We used simple sequence length polymorphism markers to map ROR1 first to chromosome 2 between the markers F3K23 (using the primer pair 5′-GGGCCGATACAGAGAGACCCTAAC-3′ and 5′-ACTCTCCGCCATGTCTCCGATC-3′) and F3N11 (using the primer pair 5′-ATGCACGCACCCTTCTACTC-3′ and 5′-TTCGCACATGTGAGATTATGGG-3′). Then, markers F26B6 (using the primer pair 5′-TAGGGCTTTGAGAACCCACGTG-3′ and 5′-AAAGTACCGTCGCGCACAG-3′), F27D4 (using the primer pair 5′-CGAGTCACATTCGCGCAATGATG-3′ and 5′-GGTAGCAAAGAAGAGAAGACCTCTC-3′), and F25P17 (using the primer pair 5′-CGACACCAAATGTCGGCTCTGAG-3′ and 5′-TAGTGACTTGATGAATGATTGCCGC-3′) were used to narrow down the ror1 mutation to within the BAC clone T28I24. To identify the ror1 mutation, candidate genes from wild-type and ror1 ros1 mutant plants were sequenced. The sequences were compared to find the mutation.
Mutant Complementation and Assay of GUS Activity
A 2433-bp fragment of the ROR1 gene (corresponding to the region 62,628 to 65,060 of T28I24, without the last putative stop codon TGA) was PCR amplified from genomic DNA isolated from gl1 plants (Columbia accession) and inserted into the pCAMBIA 1304 binary vector (http://www.cambia.org/daisy/cambia/585.html) by digestion with restriction enzymes KpnI and SpeI. Thus, the fragment was fused in frame with the GUS-GFP fused gene. The recombinant plasmid was introduced into ror1-1 ros1 plants by Agrobacterium tumefaciens strain GV3101. Approximately 50 independent transgenic plants were selected, and most (∼42 lines) were complemented on the basis of kanamycin and growth phenotypes.
Histochemical detection of GUS activity involved in use of the above complemented plants is as previously described (Gong et al., 2002).
Histological Analysis
Shoot apices or root tip meristems from 6-d-old seedlings of C24, ros1, and ror1 ros1 were fixed in formaldehyde/glutaraldehyde fixative (4% paraformaldehyde and 1% glutaraldehyde in 0.05 mol/L phosphate buffer, pH 7.2). After fixation, tissues were dehydrated in ethanol and, finally, embedded in LR white acrylic resin (Sigma-Aldrich; L9774-100G). Thin sections were prepared and observed by light microscopy. The GUS-stained tissues were treated in the same way as above. Leaf tissues were finally embedded in Spurr's resin (SPI-CHEM; 4221).
ROR1/RPA2A-GFP Expression Construct
RT-PCR was performed on total RNA extracted from 3-week-old C24 (ProRD29A:LUC) seedlings with use of TRIzol (Invitrogen). The first-strand cDNA was synthesized with an 18-mer oligo(dT) primer. Subsequent PCR amplification of ROR1/RPA2A cDNA involved the following primers: ROR1/RPA2A-GFP-F, 5′-CACCATGTTCTCCAGCAGCCAATTCGA-3′ (underline indicates the first putative ATG), and ROR1/RPA2A-GFP-R, 5′-GACTAGTAAGCTCCACGTGCTTGAAGTGATACTC-3′ (underline indicates the codon before the last putative stop codon). The PCR product was cloned into the binary vector pMDC85 (Curtis and Grossniklaus, 2003) downstream from the cauliflower mosaic virus 35S promoter with use of Gateway Technology (Invitrogen). The recombinant ROR1/RPA2A-GFP fused plasmid was introduced into onion epidermal cells by particle bombardment. GFP analysis was performed as described (Gong et al., 2002).
Protein Gel Blot Analysis
For prokaryotic expression, the open reading frame of ROR1/RPA2A cDNA (840 bp) was subcloned into the prokaryotic expression vector pGEX-2T by use of restriction enzymes SacII and EcoRI to obtain a ROR1/RPA2A-GST fusion protein. Expression of the ROR1/RPA2A-GST fusion protein in Escherichia coli strain BL21 (DE3) cells was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside. The fusion protein was purified by use of Glutathione Sepharose 4B (Amersham Pharmacia Biotech) as described (GST gene fusion system; Amersham Pharmacia Biotech). The GST tag was removed and the purified ROR1/RPA2A protein injected into rabbits to obtain the polyclonal antibody of ROR1/RPA2A.
For protein gel blot analysis, total protein was extracted from seedlings with two or six true leaves of C24, ros1, and ror1 ros1 as described (Martinez-Garcia et al., 1999). The protein concentration was determined with the Bio-Rad protein assay kit. Equal amounts of total protein (40 μg) were separated on a 15% (w/v) SDS polyacrylamide gel and transferred onto Hybond-P membrane (A.P.RPN303F). Nonspecific binding was blocked with 5% (w/v) milk powder in TBST (10 mM Tris, 150 mM NaCl, and 0.2% [w/v] Tween-20, pH 8.0) for 1 h at room temperature. The primary antibody was diluted to 1:1000 in TBST and incubated with the blotted membrane for 60 min at room temperature. A peroxidase-coupled secondary antibody (anti-rabbit IgG; Amersham Pharmacia Biotech) at a dilution of 1:5000 in TBST was incubated with the membrane for 45 min at room temperature, and then the ECL Plus Western Blotting detection reagents were added to detect the bands (Applygen Technologies; RPN2132).
Interaction between ROR1/RPA2A and ROS1
Yeast two-hybrid assay was used to detect the interaction between ROR1/RPA2A and ROS1 in vitro. Different parts of ROS1 cDNA fused with the GAL4 DNA binding domain in pGBKT7 were constructed as follows: full-length ROS1 cDNA (1 to 4179) released from pMAL-ROS1 digested by restriction enzymes EcoRI and ScaI was fused to the GAL4 DNA binding domain in pGBKT7 digested by restriction enzymes EcoRI and SmaI. ROS1 cDNA fragment (1 to 2196) was amplified using the following primers: ROS1-F1, 5′-CGGAATTCATGGAGAAACAGAGGAGAGAAGAAAGCAGC-3′, and ROS1- R1, 5′-AACTGCAGCAGGACACGACACTTCTCATCCGTC-3′. A ROS1 cDNA fragment (1024 to 2760) was amplified using the following primers: ROS1- F2, 5′-CCGGAATTCAGTTCCACATCTCAGCTCAGTGCTAATAGA -3′, and ROS1- R2, 5′-CTGCAGCCAGTCGATCAAGGAAGCCCTGTATACGTTC-3′. The ROS1 cDNA fragment (1518 to 4173) was amplified using the following primers: ROS1-F3, 5′-CGGAATTCTTGGTGGTGGCGCTGGAGCAATTGTGCCTG-3′, and ROS1-R3, 5′-AACTGCAGGTTAGCTTGTTGTCCCTTCAGTTTGCTC-3′. These PCR products digested by restriction enzymes EcoRI and PstI were fused to the GAL4 DNA binding domain in pGBKT7 digested by the same enzymes.
Full-length ROR1/RPA2A cDNA (840 bp) was fused to the GAL4 activation domain in pACT2 digested with restriction enzymes BamHI and EcoRI.
pGBKT7-(Δ)ROS1and pACT2-ROR1/RPA2A were cotransformed to the yeast strain AH109, which has three reporter genes, ADE2, HIS3, and MEL1 (lacZ), under the control of GAL4 upstream activating sequences and TATA boxes as described (Matchmaker GAL4 Two-Hybrid System 3 and Libraries User Manual PT3247-1; Clontech). The positive clones grown on Synthetic Dropout medium (Ade–, His–, Leu–, and Trp–) were checked by colony-lift-galactosidase activity filter assay to confirm the lacZ expression.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ284987.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Analysis of At3g02920 Expression in C24 Wild Type, ros1, and ror1 ros1.
Supplemental Figure 2. ProROR1:GUS Analysis in Arabidopsis Transgenic Seedlings.
Supplemental Figure 3. Protein Gel Blot Analysis of ROR1/RPA2A Expression in Seedlings of ror1-1 ros1 and ror1-2 ros1 with Different True Leaves.
Supplemental Figure 4. RNA Gel Blot Analysis of ROR1 Expression in C24 Wild Type, ror1-1 ros1, ror1-2 ros1, ror1-3, and Columbia Wild Type.
Supplementary Material
Acknowledgments
We thank Laura Heraty for critical reading and English correction of the manuscript. We also thank Yan Guo and his colleagues at the National Institute of Biological Sciences (Beijing, China) for their assistance with luminescence analysis, Roldan-Arjona Teresa (Universidad de Cordoba, Spain) for providing the full-length ROS1 cDNA, U. Grossniklaus (University of Zurich, Switzerland) for providing a set of gateway vectors, and the Arabidopsis Stock Center for providing SALK T-DNA insertion lines. This work was supported by the National Natural Science Foundation of China to Z.G. (30225004 and 30421002) and X.C. (30325015), by the Ministry of Education of China to Z.G. (104017), by the National Basic Research Program of China to X.C. (2005CB522400), and by the BaiRen Program to X.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhizhong Gong (gongzz@cau.edu.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037507.
References
- Amedeo, P., Habu, Y., Afsar, K., Scheid, O.M., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405 203–206. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J. (2002. a). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, A.J., and Matzke, M. (2002. b). RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 (suppl. 4), 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis, J.M., Bernard, P., Antonelli, R., Allshire, R.C., and Forsburg, S.L. (2003). Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 5 1111–1116. [DOI] [PubMed] [Google Scholar]
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. [DOI] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. [DOI] [PubMed] [Google Scholar]
- Bender, J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55 41–68. [DOI] [PubMed] [Google Scholar]
- Binz, S.K., Sheehan, A.M., and Wold, M.S. (2004). Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst.) 3 1015–1024. [DOI] [PubMed] [Google Scholar]
- Brill, S.J., and Stillman, B. (1991). Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5 1589–1600. [DOI] [PubMed] [Google Scholar]
- Bundock, P., and Hooykaas, P. (2002). Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12 1138–1144. [DOI] [PubMed] [Google Scholar]
- Carles, C.C., and Fletcher, J.C. (2003). Shoot apical meristem maintenance: The art of a dynamic balance. Trends Plant Sci. 8 394–401. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Henderson, I.R., and Jacobsen, S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A., and Rine, J. (1997). Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics 147 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P. (2003). Plant meristems: A merry-go-round of signals. Curr. Biol. 13 R368–R374. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., and Matzke, M.A. (2003). The small RNA world. J. Cell Sci. 116 4689–4693. [DOI] [PubMed] [Google Scholar]
- Fox, C.A., Ehrenhofer-Murray, A.E., Loo, S., and Rine, J. (1997). The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276 1547–1551. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., and de Jong, J.H. (2002). Chromatin dynamics in plants. Curr. Opin. Plant Biol. 5 560–567. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 525–535. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., Strezoska, Z., and Waggener, J.M. (2002). Initiation of DNA replication in multicellular eukaryotes. J. Struct. Biol. 140 17–30. [DOI] [PubMed] [Google Scholar]
- Gong, Z., Morales-Ruiz, T., Ariza, R.R., Roldan-Arjona, T., David, L., and Zhu, J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111 803–814. [DOI] [PubMed] [Google Scholar]
- Guyomarc'h, S., Vernoux, T., Traas, J., Zhou, D.X., and Delarue, M. (2004). MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J. Exp. Bot. 55 673–684. [DOI] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10 30–35. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Jensen, M.B., Dalmay, T., and Baulcombe, D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308 118–120. [DOI] [PubMed] [Google Scholar]
- Huang, Y. (2002). Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W., Fanti, L., Pak, D.T., Botchan, M.R., Pimpinelli, S., and Kellum, R. (1998). Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: Their phosphorylation levels and associations with origin recognition complex proteins. J. Cell Biol. 142 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, T., Kimura, S., Furukawa, T., Hatanaka, M., Hashimoto, J., and Sakaguchi, K. (2001). Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: Molecular cloning, characterization, and cellular & tissue distribution. Gene 272 335–343. [DOI] [PubMed] [Google Scholar]
- Ishibashi, T., Koga, A., Yamamoto, T., Uchiyama, Y., Mori, Y., Hashimoto, J., Kimura, S., and Sakaguchi, K. (2005). Two types of replication protein A in seed plants. FEBS J. 272 3270–3281. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Sakai, H., Finnegan, E.J., Cao, X., and Meyerowitz, E.M. (2000). Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr. Biol. 10 179–186. [DOI] [PubMed] [Google Scholar]
- Jacqmard, A., Gadisseur, I., and Bernier, G. (2003). Cell division and morphological changes in the shoot apex of Arabidopsis thaliana during floral transition. Ann. Bot. (Lond.) 91 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22 94–97. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Huettel, B., Mette, M.F., Aufsatz, W., Jaligot, E., Daxinger, L., Kreil, D.P., Matzke, M., and Matzke, A.J. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37 761–765. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Mette, M.F., Kreil, D.P., Aufsatz, W., Matzke, M., and Matzke, A.J. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14 801–805. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131–142. [DOI] [PubMed] [Google Scholar]
- Kotake, T., Takada, S., Nakahigashi, K., Ohto, M., and Goto, K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44 555–564. [DOI] [PubMed] [Google Scholar]
- Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410 116–120. [DOI] [PubMed] [Google Scholar]
- Larsson, A.S., Landberg, K., and Meeks-Wagner, D.R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., et al. (2004). Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23 4286–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20 251–257. [DOI] [PubMed] [Google Scholar]
- Marwedel, T., Ishibashi, T., Lorbiecke, R., Jacob, S., Sakaguchi, K., and Sauter, M. (2003). Plant-specific regulation of replication protein A2 (OsRPA2) from rice during the cell cycle and in response to ultraviolet light exposure. Planta 217 457–465. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., and Bender, J. (2004). RNA-directed DNA methylation. J. Cell Sci. 117 4881–4888. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Mourrain, P., Beclin, C., and Vaucheret, H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10 1591–1594. [DOI] [PubMed] [Google Scholar]
- Nakajima, K., and Benfey, P.N. (2002). Signaling in and out: Control of cell division and differentiation in the shoot and root. Plant Cell 14 (suppl.), S265–S276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Allshire, R.C., Klar, A.J., and Grewal, S.I. (2001). A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 20 2857–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Klar, A.J., and Grewal, S.I. (2000). A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101 307–317. [DOI] [PubMed] [Google Scholar]
- Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., Hause, G., Aurich, A.C., Dorn, R., Jenuwein, T., and Reuter, G. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., Allis, C.D., and Grewal, S.I. (2001). Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293 1150–1155. [DOI] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Pak, D.T., Pflumm, M., Chesnokov, I., Huang, D.W., Kellum, R., Marr, J., Romanowski, P., and Botchan, M.R. (1997). Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91 311–323. [DOI] [PubMed] [Google Scholar]
- Pineiro, M., Gomez-Mena, C., Schaffer, R., Martinez-Zapater, J.M., and Coupland, G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst, A.V., Fagard, M., Proux, F., Mourrain, P., Boutet, S., Earley, K., Lawrence, R.J., Pikaard, C.S., Murfett, J., Furner, I., Vaucheret, H., and Scheid, O.M. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J., and Elgin, S.C. (2002). Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 108 489–500. [DOI] [PubMed] [Google Scholar]
- Rocha, P.S., Sheikh, M., Melchiorre, R., Fagard, M., Boutet, S., Loach, R., Moffatt, B., Wagner, C., Vaucheret, H., and Furner, I. (2005). The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 17 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale, C., Neecke, H., Longhese, M.P., Lucchini, G., and Plevani, P. (1995). Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J. Mol. Biol. 254 595–607. [DOI] [PubMed] [Google Scholar]
- Scheid, O.M., Probst, A.V., Afsar, K., and Paszkowski, J. (2002). Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 13659–13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, N.U., Stronghill, P.E., Dengler, R.E., Hasenkampf, C.A., and Riggs, C.D. (2003). Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130 3283–3295. [DOI] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Scheid, O.M., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Inagaki, S., Nakajima, S., Akashi, T., Ohto, M.A., Kobayashi, M., Seki, M., Shinozaki, K., Kato, T., Tabata, S., Nakamura, K., and Morikami, A. (2004). A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 38 673–684. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Scheid, O.M., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M., Habu, Y., and Paszkowski, J. (2002). Depletion of MOM1 in non-dividing cells of Arabidopsis plants releases transcriptional gene silencing. EMBO Rep. 3 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M., and Paszkowski, J. (2004). DNA and histone methylation in plants. Trends Genet. 20 244–251. [DOI] [PubMed] [Google Scholar]
- Tompa, R., McCallum, C.M., Delrow, J., Henikoff, J.G., van Steensel, B., and Henikoff, S. (2002). Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12 65–68. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wold, M.S. (1997). Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66 61–92. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., Johansen, L.K., Xie, Z., Carrington, J.C., and Jacobsen, S.E. (2004). Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14 1214–1220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.