Abstract
Transcription in plastids is mediated by a plastid-encoded multimeric (PEP) and a nuclear-encoded single-subunit RNA polymerase (NEP) and a still unknown number of nuclear-encoded factors. By combining gel filtration and affinity chromatography purification steps, we isolated transcriptionally active chromosomes from Arabidopsis thaliana and mustard (Sinapis alba) chloroplasts and identified 35 components by electrospray ionization ion trap tandem mass spectrometry. Eighteen components, called plastid transcriptionally active chromosome proteins (pTACs), have not yet been described. T-DNA insertions in three corresponding genes, ptac2, -6, and -12, are lethal without exogenous carbon sources. Expression patterns of the plastid-encoded genes in the corresponding knockout lines resemble those of Δrpo mutants. For instance, expression of plastid genes with PEP promoters is downregulated, while expression of genes with NEP promoters is either not affected or upregulated in the mutants. All three components might also be involved in posttranscriptional processes, such as RNA processing and/or mRNA stability. Thus, pTAC2, -6, and -12 are clearly involved in plastid gene expression.
INTRODUCTION
Chloroplasts of higher plants possess at least two RNA polymerases with different biochemical properties and phylogenetic origins: a plastid-encoded multimeric RNA polymerase (PEP), which resembles eubacterial RNA polymerases, and a nucleus-encoded phage-type RNA polymerase (NEP) (Hu and Bogorad, 1990; Igloi and Kössel, 1992; Lerbs-Mache, 1993; Pfannschmidt and Link, 1994; Liere and Maliga, 1999). In addition to PEP, the existence of NEP has already been suggested from studies with the parasite Epifagus virginiana, which lacks the plastid rpoBC operon (Morden et al., 1991). Similar results were obtained for the plastid ribosome–deficient barley (Hordeum vulgare) mutant albostrians (Hess et al., 1993) and tobacco (Nicotiana tabacum) rpo deletion mutants (Allison et al., 1996; Hajdukiewicz et al., 1997), both still synthesizing plastid transcripts. NEP genes have been identified in the genome of many plant species (Hedtke et al., 1997; Weihe et al., 1997; Young et al., 1998; Ikeda and Gray, 1999). Arabidopsis thaliana contains three NEPs: RpoTm is located in mitochondria, while RpoTmp and RpoTp are found in plastids (Hedtke et al., 1997, 2000; Liere et al., 2004). RpoTmp is also imported into mitochondria, suggesting a dual function in organellar transcription (Hedtke et al., 2000). NEP genes for plastid proteins might have derived from a gene duplication event of a mitochondrial phage-type RNA polymerase gene (Hedtke et al., 1997).
NEP preferentially transcribes housekeeping genes, while PEP is predominantly involved in the transcription of photosynthesis genes (Allison et al., 1996; Hajdukiewicz et al., 1997). However, in PEP-deficient mutants, spurious transcripts initiated by NEP cover the entire plastome. Besides selective promoter utilization, the transcript pattern of plastids is also determined by transcript stability (Krause et al., 2000; Legen et al., 2002), whereas the RNA stability itself seems to depend on the type of the generating RNA polymerase (Cahoon et al., 2004).
While the subunits αββ′β″ of the core complex of PEP are plastid encoded, the specificity of the holoenzyme (αββ′β″σ) is determined by nuclear-encoded sigma factors (Tiller et al., 1991; Homann and Link, 2003; Suzuki et al., 2004). They are expressed in a light-dependent, developmental, and tissue-specific manner (Allison, 2000; Kasai et al., 2004; Ishizaki et al., 2005). Based on the promoter structures, three classes of plastid genes can be distinguished: class I genes are preferentially transcribed by PEP, class II genes by NEP, and class III genes by both polymerases (Hajdukiewicz et al., 1997; Ishizaki et al., 2005).
Many attempts have been made to identify nuclear-encoded proteins involved in plastid transcription and translation. Subunits of the PEP core are present in two plastid protein preparations, the transcriptionally active chromosome (TAC) and the soluble RNA polymerase (sRNAP). The TAC is membrane attached and consists of several multimeric protein complexes. In in vitro assays, the TAC can transcribe endogenously bound DNA, while the transcriptional activity of sRNAP requires the addition of DNA (Igloi and Kössel, 1992; Krause et al., 2000). TAC fractions have been isolated by gel filtration from different organisms (Briat et al., 1979; Rushlow and Hallick, 1982; Reiss and Link, 1985). The specific activity of the isolated complexes depends on the purification procedure (Suck et al., 1996; Krause et al., 2000). Initially, it was believed that sRNAP transcribes preferentially tRNA genes, while TAC is involved in rRNA transcription (Gruissem et al., 1983; Narita et al., 1985); however, run-on transcription assays uncovered that both highly purified fractions transcribed rRNA, tRNA, and plastid protein-coding genes (Reiss and Link, 1985; Rajashekar et al., 1991; Krupinska and Falk, 1994). Finally, TAC and sRNAP preparations from proplastids, chloroplasts, and etioplasts have different protein compositions, demonstrating their dynamics in response to environmental and developmental changes (Reiss and Link, 1985; Pfannschmidt and Link, 1994; Suck et al., 1996).
Forty to sixty polypeptides appear to be present in the TAC from chloroplasts, along with two core subunits of PEP and ETCHED1 (ET1) with high homology to the nuclear transcription elongation factor TFSII (Suck et al., 1996; Krause and Krupinska, 2000; da Costa e Silva et al., 2004). Together with the findings that the ET1-R mutants exhibit quantitative, but no qualitative, differences in TAC activity, it was suggested that ET1 may be involved in plastid mRNA elongation (da Costa e Silva et al., 2004). Furthermore, analyses of TAC preparations from ribosome-deficient mutants suggest that the fraction also contains a nuclear-encoded RNA polymerase activity (Suck et al., 1996). Analyses of highly purified sRNAP preparations as well as plastid DNA–bound proteins (nucleoids) led to the identification of additional nuclear-encoded components (Murakami et al., 2000; Pfannschmidt et al., 2000; Chi-Ham et al., 2002; Ogrzewalla et al., 2002; Sekine et al., 2002; Jeong et al., 2003; Loschelder et al., 2004; Suzuki et al., 2004). One of the best-characterized components of sRNAP is the transcription kinase cpCK2, which plays a role in the regulation of plastid gene expression via phosphorylation and redox signaling (Baginsky et al., 1997, 1999; Ogrzewalla et al., 2002). The nucleoid-associated protein MFP1 is a substrate of cpCK2, which inhibits the DNA binding activity of MFP1 by phosphorylation (Jeong et al., 2004). In mature chloroplasts, MFP1 is attached to thylakoids; hence, it may possibly anchor the nucleoids to the thylakoid membranes (Jeong et al., 2003). Other well-characterized proteins in nucleoids are a sulfite reductase, which participates in organellar nucleoid organization (Cannon et al., 1999; Chi-Ham et al., 2002; Sekine et al., 2002), and a DNA binding protein CND41 with aspartic protease activity (Murakami et al., 2000), possibly involved in regulation of senescence (Kato et al., 2004).
We isolated highly purified TACs from Arabidopsis and mustard (Sinapis alba) and analyzed their protein composition by electrospray ionization ion trap tandem mass spectrometry. Out of 35 identified components, 18 have not yet been described. In this study, we focused on three TAC components: pTAC2, -6, and -12. Interestingly, pTAC2 and pTAC6 are also present in sRNAP fractions, which have eight additional components besides the four PEP core subunits (Suzuki et al., 2004). While pTAC2 encodes a pentatricopeptide repeat–containing protein, the other two components, pTAC6 and pTAC12, contain no known domain and exhibit no homologies that would allow prediction of their function in plastid gene expression. Analyses of the expression of plastid genes in knockout lines suggest that the three TAC components are required for proper function of the PEP transcription machinery. Based on run on transcription assays, pTAC2 is required for proper transcription of psbA by PEP but not for transcription for atpB and clpP by NEP. All three components might also be involved in posttranscriptional processes, such as RNA processing and/or mRNA stability.
RESULTS
Isolation of TAC from Arabidopsis and Mustard
In order to identify new components involved in plastid gene expression, we purified TACs from Arabidopsis and mustard. Several previously published purification protocols were combined to obtain highly purified TAC preparations (Rushlow and Hallick, 1982; Suck et al., 1996; Krause and Krupinska, 2000). After lyses of chloroplasts obtained by sucrose gradient centrifugation, soluble proteins were applied onto a Sepharose 4B column. Transcriptional active protein fractions were combined, and the TAC in these fractions was concentrated by ultracentrifugation. The precipitated complexes were dissolved and applied to a second column (either Sepharose 2B, Q Sepharose, or Heparin Sepharose CL-6B). Figure 1A demonstrates that the second gel filtration column causes a substantial enrichment of the specific transcriptional activity, similar to results reported previously for other species (Krause and Krupinska, 2000; data shown for Sepharose 2B). After phenol/chloroform extraction and tryptic digestion, these three preparations were directly used for mass spectrometry. Table 1 contains a list of 35 proteins that were identified with cross-correlation factor (xcorr) values >3.5, 2.5, and 1.5 for triple-, double-, and single-charged peptide ions in independent TAC preparations and with at least two independent peptides. Also present in our preparations were light-harvesting chlorophyll a/b binding proteins, D1, D2, and CP47 of photosystem II, α- and β-subunits of the ATP synthase, dihydrolipoamide S-acetyltransferase (At3g25860), and a putative ribulose-bisphosphate carboxylase activase (At1g73110), which were considered as contaminants.
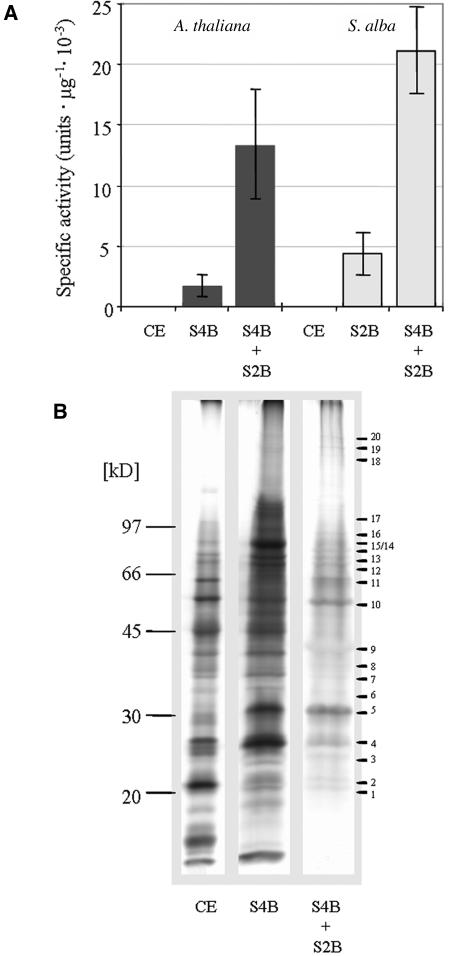
Figure 1.
Purification of the Arabidopsis and Mustard TAC.
(A) The transcriptional activity of the TAC preparations (representative for at least four independent experiments) from Arabidopsis and mustard was measured in the chloroplast lysate, after Sepharose 4B (S4B) and Sepharose 4B plus 2B (S2B) chromatography. The S4B + S2B preparations were used for mass spectrometry. Each point and error bars represent the mean value of three replicate samples and corresponding standard deviation.
(B) SDS-PAGE of TAC preparations from mustard. Lane 1, crude extract of chloroplast (CE, 25 μg); lane 2, after Sepharose 4B (S4B) chromatography; lane 3, after Sepharose 4B and 2B (S4B + S2B) chromatography. Proteins corresponding to equal TAC activity were loaded on lanes 2 and 3. The gel was stained with silver. Mass markers in kilodaltons are at left. Bars at right point to bands that were analyzed by mass spectrometry.
Table 1.
Identified Proteins from Arabidopsis and Mustard in TAC Preparations
| ΔMd
|
xcorre
|
||||||
|---|---|---|---|---|---|---|---|
| NCBI Acc. No./AGI Acc. No.a | Protein | Positionb | Zc | Ara | Sin | Ara | Sin |
| Transcription | |||||||
| gi|7525065 | RNA polymerase α-chain (RpoA) | 40–50 | 2 | 0.7 | 2.60 | ||
| ArthCp055 | 292–310 | 2 | 0.6 | 1.2 | 6.62 | 3.87 | |
| 322–328 | 2 | −0.1 | 2.60 | ||||
| gi|7525025 | RNA polymerase β-chain (RpoB) | 13–31 | 2 | −0.2 | 4.30 | ||
| ArthCp014 | 32–43 | 2 | 0.0 | 3.95 | |||
| 276–290 | 2 | 0.4 | 0.0 | 4.36 | 3.44 | ||
| 322–337 | 2 | 0.1 | 0.5 | 4.98 | 4.08 | ||
| 354–386 | 3 | 0.7 | 3.60 | ||||
| 387–397 | 2 | 0.7 | 3.47 | ||||
| 450–466 | 2 | −0.6 | 4.11 | ||||
| 603–621 | 2 | −0.4 | 3.16 | ||||
| 643–661 | 2 | −0.2 | 3.44 | ||||
| 769–779 | 2 | −0.2 | 3.06 | ||||
| 907–917 | 2 | 0.1 | 3.79 | ||||
| 918–932 | 2 | −1.1 | 4.64 | ||||
| 940–957 | 2 | 0.3 | 0.1 | 3.57 | 3.87 | ||
| 993–1017 | 3 | 0.0 | 0.1 | 4.73 | 4.04 | ||
| 1025–1048 | 2 | 1.4 | 2.72 | ||||
| 1056–1069 | 2 | 0.3 | 0.0 | 5.12 | 4.30 | ||
| gi|7525024 | RNA polymerase β′-chain (RpoC1) | 12–26 | 2 | 0.2 | 3.26 | ||
| ArthCp013 | 173–188 | 2 | 1.0 | 3.34 | |||
| 287–297 | 2 | 0.4 | 3.60 | ||||
| 303–314 | 2 | 0.6 | 3.54 | ||||
| 327–341 | 2 | 0.3 | 2.69 | ||||
| 382–394 | 2 | −0.6 | 3.30 | ||||
| 434–454 | 3 | 0.2 | 5.21 | ||||
| 461–474 | 2 | 0.3 | 0.9 | 3.97 | 3.27 | ||
| 485–510 | 3 | 1.0 | 3.78 | ||||
| 596–605 | 2 | 1.3 | 2.72 | ||||
| gi|7525023 | RNA polymerase β″-chain (RpoC2) | 44–68 | 2 | 0.5 | 0.8 | 3.30 | 3.62 |
| ArthCp012 | 69–84 | 2 | 0.3 | 6.04 | |||
| 241–249 | 2 | −0.8 | 0.1 | 2.98 | 2.57 | ||
| 250–260 | 2 | 0.3 | −1.2 | 3.72 | 2.51 | ||
| 266–276 | 2 | 0.1 | 4.25 | ||||
| 277–288 | 2 | 0.3 | 2.91 | ||||
| 307–338 | 3 | −1.0 | 3.02 | ||||
| 434–454 | 3 | 0.2 | 5.12 | ||||
| 485–510 | 3 | 1.0 | 3.78 | ||||
| 526–535 | 2 | 0.4 | 2.07 | ||||
| 590–608 | 2 | 0.7 | 2.78 | ||||
| 610–620 | 2 | 1.0 | 2.54 | ||||
| 720–730 | 2 | 1.3 | 0.1 | 2.99 | 2.59 | ||
| 779–801 | 2 | 1.1 | 3.12 | ||||
| 810–823 | 2 | −0.6 | 2.96 | ||||
| 824–834 | 2 | 0.7 | −0.3 | 2.98 | 3.98 | ||
| 1152–1162 | 2 | 0.0 | −1.2 | 2.61 | 3.55 | ||
| 1227–1248 | 3 | 0.1 | 5.12 | ||||
| 1309–1325 | 2 | 0.3 | 3.92 | ||||
| 1345–1354 | 2 | 0.3 | 2.92 | ||||
| DNA replication, DNA topology, DNA binding | |||||||
| gi|11994141 | DNA polymerase A, putative (PolA) | 287–302 | 2 | 0.3 | 3.92 | ||
| At3g20540 | 322–337 | 2 | 0.3 | 3.24 | |||
| 342–356 | 2 | 0.6 | 4.12 | ||||
| 377–391 | 2 | 0.5 | 4.11 | ||||
| 448–466 | 2 | −1.1 | 2.72 | ||||
| gi|15228353 | DNA gyrase subunit A, putative (GyrA) | 109–125 | 2 | −0.3 | −1.6 | 2.89 | 2.90 |
| At3g10690 | 194–201 | 1 | 0.7 | 2.19 | |||
| 220–248 | 3 | 0.0 | 4.52 | ||||
| 352–365 | 2 | −0.9 | 3.91 | ||||
| 405–417 | 2 | 0.6 | 3.79 | ||||
| 444–453 | 2 | −0.8 | 2.85 | ||||
| 535–548 | 2 | 0.6 | 0.7 | 4.22 | 3.97 | ||
| 562–571 | 2 | −1.6 | −1.1 | 2.90 | 3.21 | ||
| 675–692 | 2 | −1.7 | 3.91 | ||||
| gi|15228245 | DNA gyrase subunit B, putative (GyrB) | 368–394 | 3 | −0.4 | 3.61 | ||
| At3g10270 | 484–499 | 2 | −1.6 | −0.8 | 5.81 | 4.75 | |
| Translation | |||||||
| gi|15237059 | Elongation factor Tu (EF-Tu) | 127–142 | 2 | 0.5 | 4.22 | ||
| At4g20360 | 205–222 | 2 | 0.5 | 2.97 | |||
| 283–301 | 2 | −0.1 | −0.1 | 4.42 | 3.19 | ||
| 316–326 | 2 | 0.4 | 0.1 | 3.44 | 3.07 | ||
| 342–357 | 2 | −0.2 | 3.53 | ||||
| gi|15232274 | Ribosomal protein L12-1 | 69–82 | 2 | 0.3 | 4.57 | ||
| At3g27830 | 83–91 | 2 | 0.6 | 2.87 | |||
| gi|5881732 | Ribosomal protein S3 | 68–82 | 2 | 0.0 | 4.89 | ||
| ArthCp061 | 92–101 | 2 | 0.5 | 3.61 | |||
| 148–158 | 2 | −0.2 | 4.19 | ||||
| 159–167 | 2 | 0.6 | 3.26 | ||||
| 184–192 | 1 | 0.4 | 1.71 | ||||
| gi|15238369 | Ribosomal protein L29 | 74–86 | 2 | −0.1 | 2.82 | ||
| At5g65220 | 97–103 | 1 | 1.2 | 1.68 | |||
| Detoxification, protein modification | |||||||
| gi|15241373 | Iron superoxide dismutase (FE-SOD1) | 92–113 | 2 | 0.6 | 5.80 | ||
| At5g51100 | 155–165 | 2 | 0.0 | 3.77 | |||
| 168–184 | 2 | 1.1 | 4.03 | ||||
| 188–199 | 2 | −0.8 | 0.2 | 4.28 | 3.67 | ||
| 210–241 | 3 | −0.4 | 0.9 | 4.72 | 4.2 | ||
| 243–252 | 2 | 0.0 | 3.15 | ||||
| 253–262 | 2 | 0.3 | 0.0 | 2.97 | 3.17 | ||
| gi|11358867 | Iron superoxide dismutase (FE-SOD3) | 96–106 | 2 | 0.7 | 3.33 | ||
| At5g23310 | 145–161 | 3 | −1.5 | 3.83 | |||
| 228–245 | 3 | −1.5 | 3.68 | ||||
| gi|15230779 | Thioredoxin, putative | 80–90 | 2 | 0.5 | −1.0 | 3.58 | 3.41 |
| At3g06730 | 132–142 | 2 | 0.3 | 0.4 | 3.14 | 3.40 | |
| 166–183 | 2 | 0.8 | 2.91 | ||||
| 188–199 | 2 | 0.1 | 3.65 | ||||
| Metabolism | |||||||
| gi|15222232 | pfkB-type carbohydrate kinase family protein (PFKB1) | 89–119 | 3 | −1.5 | 4.75 | ||
| At1g69200 | 227–235 | 2 | 0.7 | 2.66 | |||
| 334–342 | 2 | 0.1 | 2.87 | ||||
| 396–412 | 2 | 0.6 | 4.20 | ||||
| 437–454 | 2 | 0.8 | 3.96 | ||||
| 490–504 | 2 | 1.1 | 0.4 | 2.80 | 4.23 | ||
| 505–516 | 2 | 0.6 | −0.6 | 3.22 | 2.52 | ||
| 601–614 | 2 | 0.5 | 3.83 | ||||
| gi|15232415 | pfkB-type carbohydrate kinase family protein (PFKB2) | 146–160 | 3 | −0.5 | 3.05 | ||
| At3g54090 | 223–240 | 2 | −0.8 | 3.46 | |||
| 327–341 | 2 | 0.6 | 4.32 | ||||
| 364–373 | 2 | 0.0 | 3.04 | ||||
| 401–413 | 2 | −0.5 | 3.93 | ||||
| gi|15222509 | Mur ligase family protein | 213–231 | 2 | 0.2 | 3.34 | ||
| At1g63680 | 252–273 | 2 | −1.5 | 2.86 | |||
| 318–338 | 2 | 0.3 | 6.31 | ||||
| 389–409 | 2 | 1.0 | 3.51 | ||||
| 746–767 | 2 | −0.1 | 3.43 | ||||
| DNA/RNA binding, unknown function | |||||||
| gi|15223748 | PTAC1 (DNA binding protein p24-related) | 64–80 | 2 | 0.5 | 3.20 | ||
| At1g14410 | 81–89 | 2 | 0.7 | 2.91 | |||
| 100–112 | 2 | 0.8 | 2.60 | ||||
| 138–156 | 2 | 0.8 | 3.36 | ||||
| 181–199 | 2 | 0.2 | 0.7 | 3.10 | 3.99 | ||
| 200–213 | 2 | 0.6 | 3.64 | ||||
| 214–247 | 3 | 0.1 | 3.73 | ||||
| 248–263 | 2 | 0.0 | −1.4 | 3.56 | 2.68 | ||
| gi|15221411 At1g74850 | PTAC2 (pentatricopeptide repeat-containing protein) | 57–69 | 2 | 0.4 | 3.15 | ||
| 103–113 | 2 | −0.2 | 0.8 | 3.77 | 3.76 | ||
| 192–201 | 2 | 0.3 | −0.5 | 3.37 | 3.18 | ||
| 224–239 | 2 | 0.6 | 4.20 | ||||
| 718–740 | 2 | 0.4 | 2.98 | ||||
| 741–753 | 2 | 0.4 | 3.48 | ||||
| gi|15229259 | PTAC3 (SAP domain-containing protein) | 74–83 | 2 | 0.0 | 3.30 | ||
| At3g04260 | 88–109 | 2 | −0.3 | 4.90 | |||
| 178–189 | 2 | 0.1 | 4.59 | ||||
| 162–171 | 2 | 0.9 | 2.92 | ||||
| 304–319 | 2 | −0.3 | 3.86 | ||||
| 353–376 | 2 | 1.4 | 4.77 | ||||
| 384–395 | 2 | 0.4 | 0.0 | 4.25 | 3.01 | ||
| 414–438 | 2 | 0.6 | 5.10 | ||||
| 459–479 | 2 | 0.8 | 5.30 | ||||
| 496–515 | 2 | 1.1 | 3.55 | ||||
| 547–560 | 2 | 0.6 | 3.43 | ||||
| 618–631 | 2 | 1.0 | 0.7 | 4.27 | 3.82 | ||
| 703–713 | 2 | 1.0 | 4.30 | ||||
| gi|18408237 | PTAC4 (PspA/IM30 family protein) | 84–98 | 2 | 0.7 | 3.10 | ||
| At1g65260 | 99–113 | 2 | −0.4 | 5.19 | |||
| 116–126 | 2 | 0.2 | 3.54 | ||||
| 146–152 | 1 | 0.2 | 1.71 | ||||
| 166–176 | 2 | 0.1 | 3.47 | ||||
| 167–176 | 2 | −0.5 | 3.43 | ||||
| 184–194 | 2 | −0.1 | 3.13 | ||||
| 223–241 | 2 | 0.5 | 5.91 | ||||
| 246–267 | 2 | 1.1 | 5.08 | ||||
| 268–284 | 3 | −0.5 | 4.35 | ||||
| 313–324 | 2 | 0.1 | 4.58 | ||||
| gi|15236331 At4g13670 | PTAC5 (peptidoglycan binding domain-containing protein) | 80–92 | 2 | −0.1 | −0.6 | 4.78 | 4.40 |
| 221–229 | 2 | −0.1 | 2.96 | ||||
| gi|18395008 | PTAC6 (expressed protein) | 83–112 | 2 | 1.1 | 6.16 | ||
| At1g21600 | 113–129 | 2 | −0.3 | 3.21 | |||
| 149–161 | 2 | 1.0 | 3.06 | ||||
| 162–177 | 2 | 1.1 | 2.84 | ||||
| 232–250 | 2 | 0.8 | 5.19 | ||||
| gi|22327025 | PTAC7 (expressed protein) | 64–72 | 2 | 0.6 | 2.73 | ||
| At5g24314 | 80–91 | 2 | −0.8 | 4.50 | |||
| 96–112 | 2 | 0.9 | 2.80 | ||||
| 123–131 | 2 | −0.9 | 2.80 | ||||
| 132–161 | 2 | 0.7 | 4.79 | ||||
| gi|18407178 | PTAC8 (expressed protein) | 64–88 | 2 | 1.0 | 4.80 | ||
| At2g46820 | 127–148 | 2 | −0.2 | 3.96 | |||
| gi|18415421 | PTAC9 (expressed protein) | 121–130 | 2 | 1.5 | 0.0 | 2.63 | 3.59 |
| At4g20010 | 288–299 | 2 | 0.7 | 3.47 | |||
| gi|15228384 | PTAC10 (S1 domain-containing protein) | 41–56 | 2 | 0.7 | −0.4 | 3.36 | 4.10 |
| At3g48500 | 77–93 | 2 | −1.1 | 0.0 | 3.87 | 4.15 | |
| 95–114 | 2 | 0.2 | 4.61 | ||||
| 127–138 | 2 | 0.9 | 3.04 | ||||
| 194–201 | 1 | 0.7 | 2.19 | ||||
| 244–252 | 2 | 0.2 | 2.67 | ||||
| 354–362 | 2 | −0.3 | 2.69 | ||||
| 404–424 | 2 | 0.6 | 5.64 | ||||
| 444–453 | 2 | 0.4 | 3.35 | ||||
| 468–478 | 2 | 0.1 | 3.86 | ||||
| 503–515 | 2 | −0.3 | 4.35 | ||||
| 532–543 | 2 | 0.3 | 0.4 | 3.22 | 3.21 | ||
| 553–571 | 2 | −0.3 | 5.68 | ||||
| gi|15227028 | PTAC11 (DNA binding protein p24-related) | 69–84 | 2 | 0.7 | 3.02 | ||
| At2g02740 | 81–89 | 2 | 0.7 | 2.91 | |||
| 96–103 | 2 | 0.0 | 2.58 | ||||
| 104–116 | 2 | 0.4 | 0.4 | 3.83 | 3.78 | ||
| 120–134 | 2 | 0.1 | −0.6 | 3.73 | 4.33 | ||
| 133–146 | 2 | 0.8 | 3.99 | ||||
| 196–204 | 2 | 0.1 | 0.1 | 3.34 | 3.26 | ||
| 205–218 | 2 | 0.3 | −0.6 | 3.23 | 5.77 | ||
| 219–252 | 3 | 0.6 | 3.76 | ||||
| gi|15226791 | PTAC12 (unknown protein) | 106–121 | 2 | 0.6 | 2.66 | ||
| At2g34640 | 127–142 | 2 | 0.6 | 4.41 | |||
| 190–206 | 2 | 0.1 | 3.03 | ||||
| 225–236 | 2 | 0.4 | 3.28 | ||||
| 250–259 | 2 | 0.4 | 3.28 | ||||
| 269–283 | 2 | −0.1 | 4.55 | ||||
| 280–294 | 2 | 0.3 | 4.79 | ||||
| 304–315 | 2 | −0.4 | 2.70 | ||||
| 329–351 | 3 | 0.2 | 4.88 | ||||
| 340–362 | 3 | 0.2 | 4.65 | ||||
| gi|18398425At3g09210 | PTAC13 (KOW domain-containing transcription factor family protein) | 65–70 | 1 | −0.9 | 1.69 | ||
| 122–138 | 2 | −0.5 | 3.05 | ||||
| 174–185 | 2 | 0.6 | 2.51 | ||||
| 198–209 | 2 | −0.7 | 3.97 | ||||
| 285–298 | 2 | 0.1 | 4.11 | ||||
| gi|42566980 | PTAC14 (SET domain-containing protein) | 118–131 | 2 | 0.8 | 3.29 | ||
| At4g20130 | 224–230 | 1 | 0.6 | 1.70 | |||
| 250–260 | 2 | 0.1 | 2.92 | ||||
| 337–350 | 2 | −0.3 | 3.61 | ||||
| 357–383 | 3 | 0.5 | 5.15 | ||||
| 384–397 | 2 | 0.2 | 4.61 | ||||
| 398–415 | 2 | 1.0 | 2.72 | ||||
| gi|15239573At5g54180 | PTAC15 (mitochondrial transcription termination factor-related) | 222–237 | 2 | −0.2 | 4.15 | ||
| 259–273 | 2 | 1.3 | 3.57 | ||||
| 347–361 | 2 | 0.7 | 3.70 | ||||
| gi|42565672 | PTAC16 (expressed protein) | 120–130 | 2 | 0.2 | 3.10 | ||
| At3g46780 | 149–164 | 2 | 0.3 | 3.13 | |||
| 237–248 | 2 | 0.8 | 2.87 | ||||
| 249–260 | 2 | −1.9 | −0.1 | 4.42 | 2.68 | ||
| gi|15220146At1g80480 | PTAC17 (cobW domain-containing protein) | 89–102 | 2 | 0.4 | 2.66 | ||
| 208–218 | 2 | −1.7 | 3.10 | ||||
| gi|15225202 | PTAC18 (expressed protein) | 75–93 | 2 | 0.7 | −1.1 | 4.00 | 4.64 |
| At2g32180 | 116–130 | 2 | 0.5 | 5.68 | |||
Analysis of tandem MS (MS/MS) spectra is based on the Arabidopsis database (ftp://ftp.arabidopsis.org/home/tair/sequences/blast_datasets/).
National Center for Biotechnology Information/Arabidopsis Genome Initiative accession numbers.
Position of identified peptides within the database protein sequence.
The charge state of the measured ion (z).
The calculated deviation of the experimentally determined mass from the theoretical average mass of the peptide. Ara, Arabidopsis; Sin, S. alba.
xcorr value calculated using the sequest algorithm.
Thirteen proteins identified in this preparation have also been identified in sRNAP preparations (Pfannschmidt et al., 2000; Ogrzewalla et al., 2002; Loschelder et al., 2004; Suzuki et al., 2004). Seventeen of the polypeptides have been described earlier; however, only two of them were identified as components of the TAC. The other 18 proteins reported here have not yet been described. Computer analyses predicted plastid-directing transit sequences for 33 of the identified proteins. No clear information is available for pTAC9 (At4g20010) and pTAC13 (At3g09210).
Seventeen of the 35 polypeptides are involved in replication, transcription, translation, detoxification, protein modifications, or plastid metabolism. For the others, no function has been described. However, the domain structure of eight of them suggests that they might also be involved in replication, transcription, or translation. Besides the four subunits α, β, β′, and β″ of PEP, we identified Fe-dependent superoxide dismutases (SODs) and phosphofructokinases (PFKs). One of the identified SODs (SOD3, At5g22310) and one of the PFK-B-type enzymes (PFKB2, At3g54090) are also present in highly purified sRNAP preparations from mustard and tobacco (Pfannschmidt et al., 2000; Ogrzewalla et al., 2002; Loschelder et al., 2004; Suzuki et al., 2004). Among the known proteins, a putative DNA-polymerase A (At3g20540), two putative subunits of the DNA gyrase (At3g10690 and At3g10270), the elongation factor Tu (At4g20360), three ribosomal proteins (L12-A [At3g27830], S3 [ArthCp061], and L29 [At5g65220]), the UDP-N-acetylmuamoylalanyl-d-glutamate-2,6-diaminopimelate ligase (At1g63680), and a putative thioredoxin (At3g06730) have not yet been described as TAC components. The 18 new components were named plastid TAC proteins 1 to 18 (pTAC1-18). Based on the analyses of their domain structures, some of them contain either DNA or RNA binding domains, domains involved in protein–protein interactions, or epitopes described for other cellular functions (Table 2). However, database searches for four proteins did not reveal known functional domains.
Table 2.
Analysis of pTAC Domains
| AGI Acc. No.a | Protein | cTPb | Domainc |
|---|---|---|---|
| At1g14410 | PTAC1 (DNA binding protein p24-related) | + (47) | ssDNA binding domain |
| At1g74850 | PTAC2 (pentatricopeptide repeat-containing protein) | + (66) | MutSPPRTPR |
| At3g04260 | PTAC3 (SAP domain-containing protein) | + (29) | SAP |
| At1g65260 | PTAC4 (PspA/IM30 family protein) | + (64) | PspA/IM30 |
| At4g13670 | PTAC5 (peptidoglycan binding domain-containing protein) | + (40) | Peptidoglycan binding domainDnaJ |
| At1g21600 | PTAC6 (expressed protein) | + (59) | |
| At5g24314 | PTAC7 (expressed protein) | + (32) | |
| At2g46820 | PTAC8 (expressed protein) | + (45) | |
| At4g20010 | PTAC9 (expressed protein) | − | DUF731 |
| OB fold | |||
| At3g48500 | PTAC10 (S1 domain- containing protein) | + (40) | S1 |
| At2g02740 | PTAC11 (DNA binding protein p24-related) | + (75) | ssDNA binding domain |
| At2g34640 | PTAC12 (unknown protein) | + (16) | |
| At3g09210 | PTAC13 (KOW domain-containing transcription factor family protein) | − | KOW NGN |
| At4g20130 | PTAC14 (SET domain-containing protein) | + (62) | SET |
| At5g54180 | PTAC15 (mitochondrial transcription termination factor-related) | + (64) | mTERF |
| At3g46780 | PTAC16 (expressed protein) | + (19) | Nucleoside-diphosphate-sugar epimerases |
| At1g80480 | PTAC17 (cobW domain-containing protein) | + (68) | cobW |
| At2g32180 | PTAC18 (expressed protein) | + (32) | DUF861 |
Arabidopsis Genome Initiative accession number.
ChloroP prediction of the presence (+) or absence (−) of a transit sequence (cTP). The putative maturation site is given (Emanuelsson et al., 1999).
Protein domains predicted by InterProScan (Apweiler et al., 2000) and BLAST (Altschul et al., 1997).
Twenty-six of the 35 polypeptides were identified in TAC preparations from Arabidopsis and mustard (Tables 1 and 3), four proteins only in Arabidopsis (At3g20540, At5g23310, At2g46820, and At5g54180), and additional five only in mustard preparations (At3g27830, At5g65220, ArthCp061, At1g65260, and At1g80480). We isolated large amounts of TAC from mustard seedlings and solubilized the proteins in Laemmli buffer after sonication. Twenty major bands could be visualized on a silver-stained gel, and they were identified by mass spectrometry (Figure 1B). Seventeen of these bands correspond to proteins also identified in unresolved TAC samples (Table 3). One additional protein not detected by the original mass spectrometry (MS) was found by analysis of the PAGE-derived protein bands (At2g05430). However, the size of protein bands resolved by PAGE does not always correspond to the predicted molecular masses of the polypeptides. This is not surprising considering that complete solubilization of the final TAC in the gel loading buffer could only be achieved by sonication. It appears that the proteins are tightly associated with each other or with nucleic acids. Some identified polypeptides contain RNA or DNA binding motifs, such as pentatricopeptide repeat motif (PPR), small MutS-related motif (SMR), SAP (for SAF-A/B, Acinus, and PIAS), S1, oligonucleotide/oligosaccharide binding (OB) fold, KOW (for Kyrpides, Ouzounis, and Woese), NGN, or mitochondrial transcription termination factor motif (mTERF) (Table 2). Therefore, it seems likely that the proteins identified in this study are components involved in plastid transcription/translation.
Table 3.
Identification of Bands from an SDS Polyacrylamide Gel with Mustard TAC Proteins
| Band | MM (kD) | Protein | Positiona | Zb | ΔMc | xcorrd | AGI Acc. No.e |
|---|---|---|---|---|---|---|---|
| 1 | 19–21 | Thioredoxin, putative | 79–90 | 2 | −0.2 | 4.35 | At3g06730 |
| 80–90 | 2 | −0.5 | 4.04 | ||||
| 132–142 | 2 | −0.3 | 3.83 | ||||
| 148–161 | 2 | −0.1 | 4.16 | ||||
| Ribosomal protein L29p | 97–103 | 1 | 1.1 | 1.68 | At5g65220 | ||
| 2 | 22–23 | Hypothetical protein | 140–156 | 2 | −1.2 | 2.50 | At2g05430 |
| 3 | 24–25 | Ribosomal protein S3 | 92–101 | 2 | −0.3 | 3.19 | ArthCp061 |
| 184–192 | 1 | 0.4 | 1.71 | ||||
| 4 | 26–28 | ? | |||||
| 5 | 30–32 | Hypothetical protein | 140–156 | 2 | −0.4 | 2.51 | At2g05430 |
| 6 | 33–35 | PTAC4 (PspA/IM30 family protein) | 146–152 | 2 | 0.2 | 1.54 | At1g65260 |
| 223–241 | 2 | −0.4 | 3.03 | ||||
| 246–267 | 2 | 1.4 | 2.70 | ||||
| 268–284 | 2 | 0.9 | 3.14 | ||||
| PTAC6 (expressed protein) | 272–279 | 2 | −1.1 | 2.59 | At1g21600 | ||
| 7 | 36–38 | ? | |||||
| 8 | 39–40 | PTAC1 (DNA binding protein p24-related) | 81–89 | 2 | −0.7 | 2.55 | At1g14410 |
| 181–199 | 2 | 1.2 | 3.78 | ||||
| 248–263 | 2 | −1.5 | 2.93 | ||||
| PTAC11 (DNA binding protein p24-related) | 196–204 | 2 | −0.4 | 2.82 | At2g02740 | ||
| 9 | 42–44 | PTAC14 (SET) | 90–96 | 1 | 0.8 | 1.58 | At4g20130 |
| PTAC11 (DNA binding protein p24-related) | 85–93 | 2 | −0.7 | 2.55 | At2g02740 | ||
| 96–103 | 1 | 0.4 | 1.53 | ||||
| 104–116 | 2 | 1.2 | 2.56 | ||||
| 120–134 | 2 | −1.1 | 4.15 | ||||
| 196–204 | 2 | −0.4 | 2.63 | ||||
| 253–259 | 1 | 0.1 | 1.91 | ||||
| 260–268 | 2 | 0.7 | 3.11 | ||||
| 10 | 54–55 | Iron superoxide dismutase (FE-SOD1) | 253–262 | 2 | −0.4 | 2.92 | At5g51100 |
| pfkB-type carbohydrate kinase family protein (PFKB1) | 578–597 | 2 | −0.2 | 2.70 | At1g69200 | ||
| PTAC12 (unknown protein) | 127–142 | 2 | 0.1 | 2.53 | At2g34640 | ||
| 249–259 | 3 | −0.7 | 3.51 | ||||
| 11 | 60–62 | RNA polymerase β″-chain (RpoC2) | 1083–1091 | 2 | −0.7 | 3.22 | ArthCp012 |
| PTAC9 (expressed protein) | 121–130 | 2 | −0.1 | 3.20 | At4g20010 | ||
| 288–299 | 2 | −0.1 | 3.12 | ||||
| 12 | 70–72 | PTAC6 (expressed protein) | 149–161 | 2 | −0.5 | 3.02 | At1g21600 |
| 189–208 | 2 | −0.2 | 3.36 | ||||
| 209–213 | 1 | 0.0 | 1.62 | ||||
| 273–279 | 1 | 0.3 | 1.54 | ||||
| 303–307 | 1 | 0.6 | 1.55 | ||||
| pfkB-type carbohydrate kinase family protein (PFKB2) | 146–160 | 2 | −0.6 | 3.20 | At3g54090 | ||
| 165–170 | 1 | 0.2 | 1.57 | ||||
| 13 | 78–80 | RNA polymerase β′-chain (RpoC1) | 327–341 | 2 | −0.7 | 3.28 | ArthCp013 |
| 461–474 | 2 | −1.4 | 4.02 | ||||
| 595–605 | 2 | −1.0 | 3.01 | ||||
| PTAC6 (expressed protein) | 149–161 | 2 | −0.7 | 2.69 | At1g21600 | ||
| Hypothetical protein | 140–145 | 2 | 0.0 | 2.60 | At2g05430 | ||
| PTAC4 (PspA/IM30 family protein) | 99–113 | 2 | −0.2 | 2.59 | At1g65260 | ||
| 246–267 | 2 | 0.4 | 4.76 | ||||
| 268–284 | 2 | 1.4 | 3.62 | ||||
| Mur ligase family protein | 570–580 | 2 | 0.7 | 2.66 | At1g63680 | ||
| 14 | 82–84 | PTAC10 (S1 domain-containing protein) | 593–606 | 2 | 0.0 | 3.52 | At3g48500 |
| 609–622 | 2 | −1.0 | 2.52 | ||||
| 15 | 85–87 | PTAC5 (peptidoglycan binding domain-containing protein) | 80–92 | 2 | −1.0 | 3.26 | At4g13670 |
| 16 | 88–91 | PTAC5 (peptidoglycan binding domain-containing protein) | 80–92 | 2 | −0.2 | 3.56 | At4g13670 |
| 17 | 95–100 | PTAC11 (DNA binding protein p24-related) | 104–116 | 2 | −0.9 | 3.68 | At2g02740 |
| 120–134 | 2 | −0.1 | 3.48 | ||||
| 196–204 | 2 | −1.3 | 2.55 | ||||
| 18 | 135–145 | PTAC10 (S1 domain-containing protein) | 41–46 | 2 | −0.3 | 3.18 | At3g48500 |
| 72–76 | 1 | 0.1 | 1.53 | ||||
| 95–114 | 2 | 0.7 | 3.47 | ||||
| 172–177 | 1 | 0.2 | 1.61 | ||||
| 383–403 | 2 | 0.2 | 2.84 | ||||
| 484–489 | 1 | 0.2 | 1.62 | ||||
| 19 | 150–160 | PTAC3 (SAP domain-containing protein) | 496–515 | 2 | −0.5 | 3.92 | At3g04260 |
| 703–713 | 2 | −0.3 | 2.85 | ||||
| 618–631 | 2 | 0.2 | 4.22 | ||||
| RNA polymerase β″-chain (RpoC2) | 1083–1091 | 2 | 0.0 | 2.63 | ArthCp012 | ||
| 1144–1151 | 2 | −0.5 | 2.91 | ||||
| 1206–1214 | 2 | 1.1 | 2.53 | ||||
| 20 | 165–175 | RNA polymerase β-chain (RpoB) | 646–977 | 2 | −0.2 | 3.29 | ArthCp014 |
| RNA polymerase β″-chain (RpoC2) | 138–150 | 2 | −0.2 | 3.78 | ArthCp012 | ||
| 266–276 | 2 | −0.2 | 2.53 | ||||
| 511–525 | 2 | −0.1 | 3.09 | ||||
| 1144–1151 | 2 | −0.6 | 2.74 |
Analysis of MS/MS spectra is based on the Arabidopsis database (ftp.arabidopsis.org/home/tair/sequences/blast_datasets/). MM, molecular mass in kilodaltons.
Position of identified peptides within the database protein sequence.
The charge state of the measured ion (z).
The calculated deviation of the experimentally determined mass from the theoretical average mass of the peptide.
xcorr value calculated by using the sequest algorithm.
NCBI/AGI accession numbers.
Identification and Characterization of Knockout Lines with Lesions in TAC Components
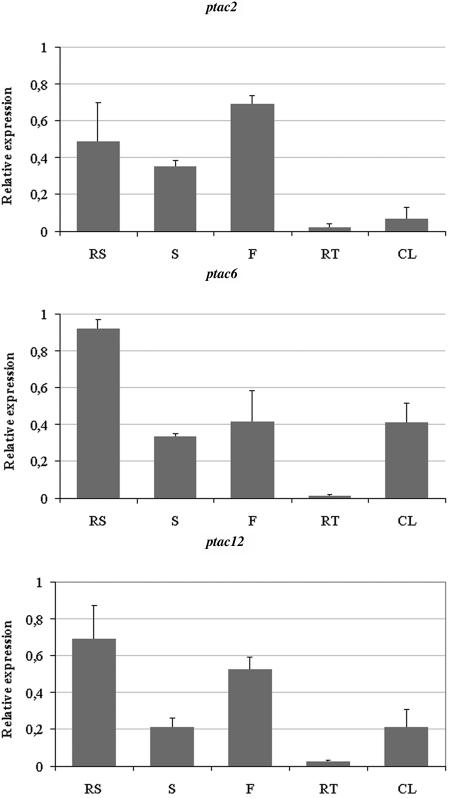
To further substantiate that pTACs are involved in plastid transcription, Arabidopsis knockout lines for pTAC2, -6, and -12 were analyzed. ptac2 (At1g74850) contains two nucleotide binding domains (PPR and SMR) and tetratricopeptide repeat (TPR) domains involved in protein–protein interaction. No obvious protein domains could be detected for ptac6 (At1g21600) and ptac12 (At2g34640). Mutations in homozygote knockout lines for these proteins are lethal when they were grown without exogenous carbon sources. DNA sequence analyses confirmed the positions of the T-DNA insertions provided by the Nottingham Arabidopsis Stock Centre (NASC) [At1g74850, 3rd exon after nucleotide 2571; At1g21600, 1st intron after the nucleotide 518; At2g34640, 8th intron after the nucleotide 2285 downstream of A(+1)TG codon]. RT-PCR with gene-specific primers uncovered that no more transcripts can be detected in ptac2 (Figure 2). The small amounts of transcripts detected in ptac6 and ptac12 mutants might be due to the fact that the insertions are located in introns. However, the phenotype of the mutants clearly demonstrates that the residual transcript levels are too low to allow normal plastid development. A contamination of the RNA with genomic DNA can be excluded because the RT-PCR products do not contain intron sequences (Figure 2). Real-time PCR analyses with RNA from different organs of wild-type seedlings demonstrate that ptac2, -6, and -12 transcripts are present in all tissues and that the mRNA levels are significantly reduced in roots (Figure 3). However, the presence of ptac2, -6, and -12 transcripts in green and nongreen tissue suggests that the gene products are required in chloroplast and other types of plastids. It remains to be determined whether the amounts of these proteins correlate with the number of plastids or nucleoids per cell.
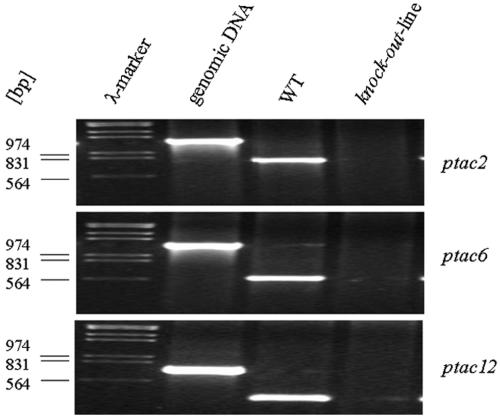
Figure 2.
Mutant Analyses.
The knockout lines ptac2, -6, and -12, which were determined to be homozygotes by PCR, fail to accumulate normal amounts of messages from the inactivated genes. Total RNA was isolated from wild-type and knockout seedlings and used for RT-PCR with gene-specific primers. ptac2 (34 cycles), ptac6 (30 cycles), and ptac12 (45 cycles). DNA: PCR with genomic DNA from wild-type seedlings (23 cycles). Size markers in kilobases are at left.
Figure 3.
Tissue-Specific Expression of ptac2, -6, and -12.
An equal amount of cDNA from the cotyledons (set as 1.0), rosette leaves (RS), stems (S), flowers (F), roots (RT), and cauline leaves (CL) from Arabidopsis was used for real-time PCR with gene-specific primers for the actin gene (Robinson et al., 1999), ptac2, -6, and -12. Fold induction values of genes were calculated with the ΔΔCP equation of Pfaffl (2001) and related to the mRNA level of the genes in cotyledons, which was defined as 1.0. This value is not shown in the graph. The data shown here were obtained from three independent experiments, and the error bars represent standard deviation.
The homozygote seedlings develop white cotyledons, fail to accumulate chlorophyll even under low light intensities, and do not produce primary leaves. On sucrose medium, the mutants reach the rosette stage, but they are much smaller and grow slower than the wild type (Figure 4). While ptac2 develops yellow cotyledons and greenish primary leaves on sucrose medium, the other two mutants stay more yellowish. By contrast, dark-grown ptac plants show the phenotype of the etiolated wild type. A detailed analysis of pigments from low-light-grown ptac plants revealed that chlorophylls and carotenoids accumulate in the mutants, although the overall amounts are quite low. The decrease of total chlorophyll (∼70% in ptac2, ∼99% in ptac6, and ∼95% in ptac12 plants) was accompanied by a noticeable decrease in the chlorophyll a:b ratio in ptac2 and -12 and increase in ptac6 plants (Table 4). These changes might be due to an increase in the photosystem II light-harvesting system and/or a decrease in the photosystem I:II ratio in case of ptac2 and -12 and the opposite for ptac6 plants, respectively. Furthermore, we observed a significant increase in the carotenoid:chlorophyll as well as in the xanthophyll:chlorophyll ratio (Table 4) in mutant seedlings. Interestingly, the antheraxantin level is relatively high and the zeaxanthin level below detectability. Both xanthophyll pigments are not detectable in the wild type, which is known to be the case for plants grown under moderate conditions (Demmig-Adams and Adams, 2002). Fluorescence quenching analysis indicates effects on photochemical efficiency in the mutant lines. As shown in Table 4, the maximum quantum yield (Fv/Fm) and photochemical quenching [(Fm′ − Ft)/Fm′] are significantly lower than those of the wild type. The reduced ratio of variable-to-maximum fluorescence (Fv/Fm) indicates smaller photosystem II antennae and/or fewer functional photosystem II centers (Meurer et al., 1998). In accordance with this observation, ptac2 reveals higher values of nonphotochemical quenching (NPQ; Krause and Weis, 1991). In case of ptac6 and -12, the NPQ values might be caused by the reduced chlorophyll contents.
Figure 4.
Phenotype of ptac2, -6, and -12 Mutant Plants.
Ptac2, -6, and -12 and wild-type seedlings after 12 (A) and 18 (B) d grown on Murashige and Skoog medium under long-day conditions.
Table 4.
Pigment Analysis and Fluorescence Measurements of Wild-Type and Mutant Lines
| Pigment Composition
|
Chlorophyll Fluorescence Measurements
|
|||||
|---|---|---|---|---|---|---|
| Line | Chlorophyll a/b (mol/mol) | Carotenoids/Chlorophyll (mol/mol) | VAZ/Chlorophyll (mmol/mol) | Fv/Fm | (Fm′ − Ft)/Fm′ | NPQ |
| Wild type | 3.2 ± 0.034 | 0.25 ± 0.008 | 40.7 ± 1.4 | 0.854 ± 0.012 | 0.719 ± 0.037 | 0.456 ± 0.030 |
| tac2 | 2.8 ± 0.106 | 0.35 ± 0.012 | 99.1 ± 4.8 | 0.497 ± 0.055 | 0.365 ± 0.039 | 0.890 ± 0.226 |
| tac6 | 4.4 ± 0.286 | 1.48 ± 0.071 | 467.9 ± 18.0 | 0.424 ± 0.089 | 0.371 ± 0.073 | 0.324 ± 0.058 |
| tac12 | 2.8 ± 0.057 | 0.60 ± 0.042 | 216.7 ± 13.5 | 0.546 ± 0.013 | 0.395 ± 0.022 | 0.399 ± 0.044 |
Pigment data are the average of three independent samples. Flourescence quenching analysis is averaged from nine plants. VAZ is the xanthophyll pool size (violaxanthin + antheraxanthin + zeaxanthin).
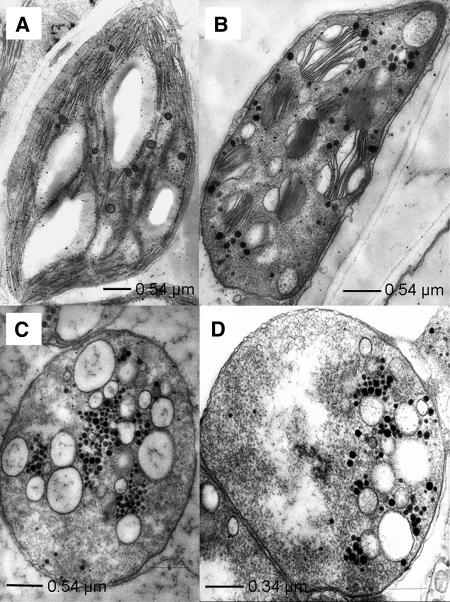
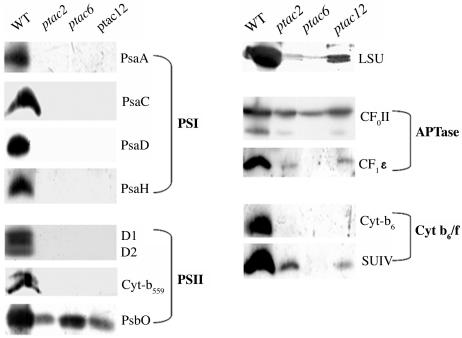
Analysis of the plastid structure in the mutants showed that the organelle development is severely impaired (Figure 5). Compared with the wild type, grana structures in ptac2 plants are unequally expanded. Grana-interlacing stroma thylakoids are completely missing (Figure 5B). Chloroplasts of ptac6 and ptac12 plants do not contain grana thylakoids. They are replaced by oval-shaped vesicles (Figures 5C and 5D). Surprisingly, an accumulation of starch is only observed in the old leaves of ptac6 and ptac12 mutants. Frequently a multiplicity of plastoglobuli is found in close proximity to the above-mentioned vesicles (Figures 5B and 5C). Both plastid- and nuclear-encoded soluble polypeptides and polypeptides associated with the thylakoid membrane are either absent or strongly reduced in the mutants (Figure 6). Since plastid-encoded polypeptides, such as the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase, subunit IV of cytochrome b6/f complex, and CF1ɛ, are still detectable in all three mutants, the mutations do not affect general processes in plastid transcription and/or translation.
Figure 5.
Ultrastructure of Plastids from Wild-Type and Mutant Seedlings.
The wild type (A), ptac2 (B), ptac6 (C), and ptac12 (D).
Figure 6.
Immunoblot Analyses of Plastid Proteins.
Fifteen micrograms of plastid proteins of wild-type and mutant (ptac2, -6, and -12) seedlings were separated on polyacrylamide gels containing SDS. The immunological detection of the proteins occurred with antibodies against the subunits A, C, D, and H of photosystem I (PSI), against D1/2, cytochrome b559 and PsbO of photosystem II (PSII), against CFoII and CF1ɛ of the ATP synthase, against cytochrome b6 and the subunit IV of the cytochrome b6/f complex, and against the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (LSU).
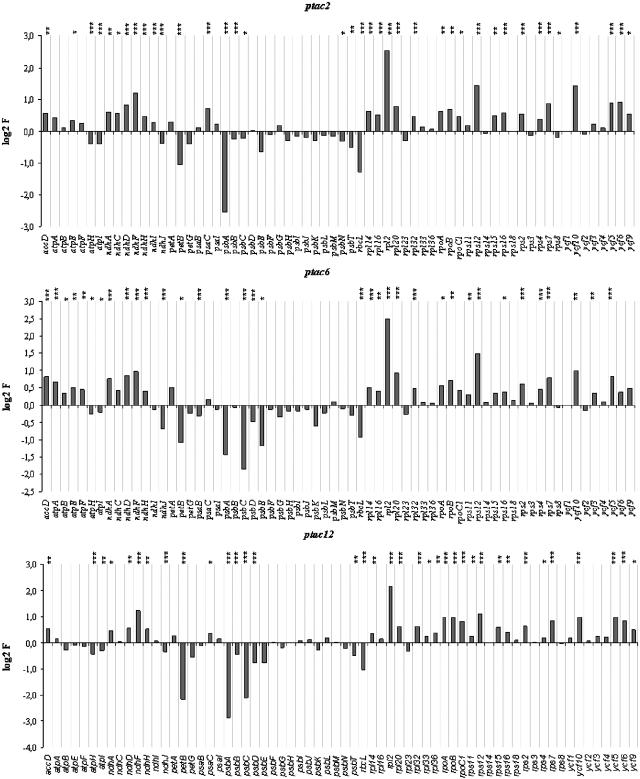
Macroarray analyses demonstrated that all plastid-encoded genes are expressed in the mutants. In most cases, these data confirmed the results obtained by RNA gel blot analyses. Differences in the mRNA results might be caused by radiolabeled antisense RNA, which also hybridized to the probes (Krause et al., 2000; Legen et al., 2002). However, compared with the wild type, we observed remarkable alterations (Figure 7). The transcript levels for components of the two photosystems and for the cytochrome b6/f complex are significantly reduced. For instance, the psbA mRNA level (for the D1 protein) in ptac2 and ptac12 and the psbA, psbC, and psbE mRNA levels (for CP43 and cytochrome b559/2) in ptac6 plants are reduced by 80 and 50%, respectively. In all three mutants, petB (for a subunit of the cytochrom b6/f complex) and rbcL (for the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase) transcripts accumulate to <50%. These mRNAs are probably transcribed from genes with PEP promoters. By contrast, rps, rpl (for ribosomal proteins), ndh (for subunits of the NADH dehydrogenase), atp (for subunits of the ATP synthase), rpo (for subunits of the RNA polymerase), and ycf (for hypothetical chloroplast open reading frames) genes are expressed at higher levels in all three mutants. These messages are transcribed from genes that may contain NEP promoter elements, either alone or in combination with PEP promoter elements. The expression pattern of the plastid genes in the mutants is identical to that described for Δrpo mutants and mutants that do not accumulate PEP. These mutants also contain reduced transcript levels for photosynthesis proteins, while ndh, atp, and rpo genes are expressed at higher levels (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-Maciossek et al., 1999; Krause et al., 2000; Legen et al., 2002). However, for some plastid genes (e.g., for clpP), the expression level in the wild type was not significantly different from the background. Finally, the reduced psbA mRNA level in ptac2 plants is caused by a reduced rate of psbA transcription as shown by run-on transcription assays, while transcription of atpB and clpP appear to be not affected in the mutant (Figure 8). We could not obtain enough chloroplasts from the homozygote mutants to unambiguously confirm this for the other genes and mutants.
Figure 7.
Changes in Transcript Abundance of Plastome-Encoded Genes.
A macroarray representing all protein-coding genes of the plastome was hybridized with probes synthesized from the mutants (ptac2, ptac6, and ptac12) and wild type. The diagram shows the log2 induction factor (log2 IF). IF was calculated from the ratio of the mutant and wild-type signal intensities. Log2 IF is given where 3.32 corresponds to a 10-fold upregulation and −3.32 to a 10-fold downregulation. *** Indicates P < 0.05, ** indicates 0.05 < P < 0.10, and * indicates 0.10 < P < 0.20.
Figure 8.
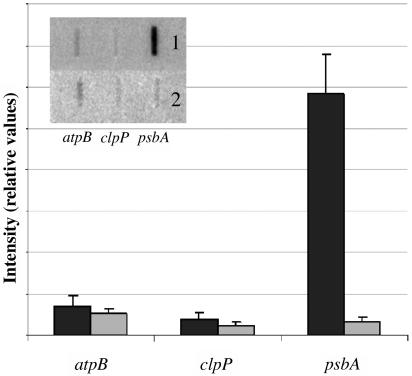
Analyses of Transcription Rates.
For run-on transcription assays of atpB, clpP, and psbA, 2 × 107 chloroplasts were used from wild-type (inset 1) and ptac2 (inset 2) plants. The quantification of signal intensities occurred with a Storm PhosphorImager. The signals were normalized to total signal intensity within mutants and the wild type, respectively. Error bars represent corresponding standard deviation.
To further substantiate the findings derived by macroarray analyses, representative genes were chosen for RNA gel blot analyses (Figure 9). Figure 9A shows genes (psaAB, psaC, psaJ, psbA, rbcL, and rps14) that are downregulated in the mutants, Figures 9B and 9D shows genes (atpB, clpP, and the nuclear-encoded genes psaH, psaE, and psbO) that do not respond significantly to the mutations, and Figure 9C shows genes (accD, atpA, ndhF, ndhB, and ycf3) that are upregulated in the mutants. Genes shown in Figure 9A contain only PEP promoter sequences, and their expression is also downregulated in Δrpo mutants and other mutants impaired in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-Maciossek et al., 1999; Krause et al., 2000; Legen et al., 2002). By contrast, the Figures 9B and 9C show genes that contain NEP promoter elements either alone or in combination with PEP promoter elements. Again, these genes exhibit the same expression behavior in Δrpo mutants and other mutants impaired in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-Maciossek et al., 1999; Krause et al., 2000; Legen et al., 2002). Furthermore, we observed in all mutants defects in mRNA processing, since larger transcripts accumulate for accD, atpB, clpP, ndhB, ndhF, psaAB, rbcL, rps14, and ycf3. This is most obvious for the polycistronic transcript ycf3/psaA/psaB/rps14. Besides mRNAs with 3.2 and 5.2 kb, additional messages with 5.8 and 7.3 kb hybridize to the psaAB probe in the mutants. They represent unprocessed transcripts and include sequences for ycf3, as shown by the hybridization with the ycf3-specific probe (Figure 9C; Summer et al., 2000; Legen et al., 2002). Again, the very same lesions in the processing patterns were observed for Δrpo mutants and other PEP mutants. Taken together, these data suggest that the expression of genes encoding the three novel TAC proteins is required for proper function of PEP transcription machinery. Since messages for all genes studied are present in all three mutants and since at least some of them are also properly processed, the mutations do not completely inhibit crucial steps in plastid transcription or translation. pTAC2 and pTAC6 are also present in sRNAP preparations (Suzuki et al., 2004), suggesting that they are preferentially involved in transcription rather than translation.
Figure 9.
Transcript Accumulation.
RNA gel blot analyses for plastid- and nuclear-encoded genes were performed with RNA from wild-type (lanes 1 to 3) and mutant (lane 4, ptac2; lane 5, ptac6; lane 6, ptac12) seedlings. RNA loaded per lane is as follows: 3.25 μg (lane 1), 7.5 μg (lane 2), and 15 μg RNA (lanes 3 to 6). The gene probes are indicated. The 18S rRNA accumulation is shown in the ethidium bromide–stained gels below each plot to confirm equal loading. Bars mark the position of cotranscripts.
(A) and (C) The transcript levels are reduced (enhanced) in the mutants.
(B) The amount of transcripts is similar in the wild type and mutants.
(D) RNA gel blot analyses of nuclear-encoded genes.
(E) RNA gel blot analyses of 18-d-old dark-grown material.
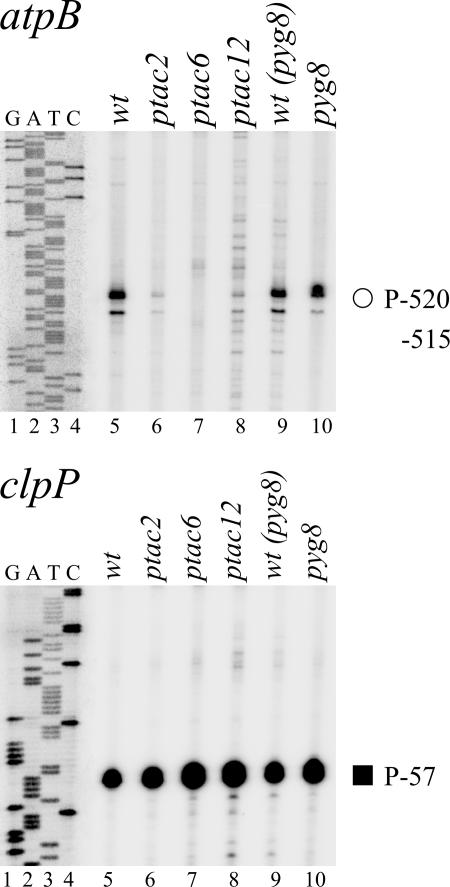
To analyze the effect of the pTAC-2, -6, and -12 mutations on PEP and NEP promoter activity in leaf tissue, we determined transcript levels by primer extension analyses mapping transcript 5′-ends for clpP and atpB, two representative plastid genes with NEP and PEP promoter elements (Figure 10). In these studies, we also included the pale-yellow-green (pyg) mutant 8 (lane 10; J. Stöckel and R. Oelmüller, unpublished data) because it exhibits a very similar phenotype and growth behavior as the ptac mutants. Since signals at the promoter initiation site are dependent on the mRNA level, we included into our investigations only such genes whose mRNA levels are not affected by the mutations. We did not observe any significant difference in the signal intensities at the NEP promoter site P-57 of clpP (panel clpP; Sriraman et al., 1998). This example indicates that the NEP promoter usage is not altered in the ptac and pyg mutants. By contrast, the signal at the P-515/-520 PEP side of atpB is dramatically downregulated in ptac2 and ptac12 mutants (panel atpB, lanes 6 and 8) and below detectability in ptac6 plants (lane 7). Sequence alignments of atpB promoter sequences from different species indicate that the conserved regions represent PEP (-35/-10) elements (data not shown). Since the primer extension signal at this PEP site is nearly equal in the wild type and in the pyg8 mutant (Figure 10, lanes 9 and 10), it appears that the PEP promoter usage is specifically affected in the ptac mutants.
Figure 10.
Primer Extension Analyses of ptac2, ptac6, ptac12, and the Wild Type.
In ptac knockout plants (lanes 6 to 8), transcript accumulation from the PEP atpB-515/520 promoter is negatively affected compared with the wild type (lane 5) but not from the type-II clpP-57 NEP promoter. Primer extension data are shown for the atpB and clpP genes. Mapped PEP (open circle) and NEP type-II (closed square) promoters are identified by their distance between the transcription initiation site and the translation initiation codon in nucleotides (K. Kühn, D. Kaden, and K. Liere, unpublished data; Sriraman et al., 1998). For reference, the same end-labeled primer was used to generate a DNA sequence ladder (lanes 1 to 4). The pyg8 mutant (lane 10; J. Stöckel and R. Oelmüller, unpublished data) and the corresponding wild type [wt(pyg8); lane 9] were used as independent controls for transcriptional activity.
The expression of ptac2, -6, and -12 in organs with different types of plastids (Figure 3) suggests that the gene products are required for general processes of the PEP activity and not restricted to photosynthetically active plastids. To test this further and to exclude the possibility that the phenotypes of the mutants grown in low light (see Methods) are caused by photooxidative damage due to light stress, we repeated the RNA gel blot and primer extension analyses with etiolated material. Segregating populations were germinated in low light for 3 d to identify the homozygote seedlings. They were then transferred to complete darkness for an additional 18 d before harvest of the newly emerged etiolated leaves. RNA gel blot analyses for representative genes of the three classes described above (Figure 9) clearly indicate that the observed differences in the transcript levels in the mutants are not light dependent (Figure 9E). Furthermore, primer extension analysis for atpB also gave comparable results as obtained for seedlings grown in low light (data not shown; Figure 10). Finally, as described for seedlings grown in low light, the atpB transcript levels in etiolated material are similar or even higher in ptac2 and -12 plants (Figure 9E), although the primer extension signals at the PEP sites were downregulated (data not shown). This clearly indicates that ptac2, -6, and -12 function is not related to thylakoid biogenesis, photosynthesis, or photooxidative stress.
DISCUSSION
We hypothesized that nuclear-encoded components required for transcription and translation in plastids might be associated with the TAC. TAC preparations from various organisms were analyzed on SDS gels, and in this way, at least 40 polypeptides associated with TAC have been detected. However, only the α- and β-subunits of the PEP (Suck et al., 1996; Krause and Krupinska, 2000), a homolog of the nuclear transcription elongation factor TFIIS (da Costa e Silva et al., 2004), and few plastid DNA–bound proteins, including PEND (Sato et al., 1993), sulfite reductase (Cannon et al., 1999; Chi-Ham et al., 2002; Sekine et al., 2002), CND41 (Murakami et al., 2000), and MFP1 (Jeong et al., 2003), have been identified until now. We used different gel filtration and affinity chromatography purification steps for the isolation of TACs from mustard and Arabidopsis.
The available sequence information allowed us to identify the Arabidopsis TAC components with high fidelity and to analyze their role in knockout lines. Furthermore, this allowed us to compare the TAC composition of two related organisms. Using the above-mentioned criteria (at least two independent peptides in independent preparations), we identified 35 polypeptides in our TAC preparations, 18 of these components have not yet been described so far (cf. also below). We could demonstrate that three of them influence plastid transcription, RNA accumulation, and processing and therefore are essential for plastid gene expression.
Twenty-five polypeptides could be identified in TAC preparations from Arabidopsis and mustard (Tables 1 and 3). Four additional proteins (At3g20540, At5g23310, At2g46820, and At5g54180) were only detected in Arabidopsis TAC preparations. Only five polypeptides were identified in mustard but not in the Arabidopsis TAC (At3g27830, At5g65220, ArthCp061, At1g65260, and At1g80480). These differences might be caused by the different developmental stages of the plastids, by the different abundance of individual proteins in the two species, or by the fact that the identification of mustard proteins with Arabidopsis databases is not possible in all instances. Based on homology searches, the TAC contains polypeptides involved in replication, transcription, translation, detoxification, protein modification, and plastid metabolism. Eleven identified proteins are DNA or RNA binding proteins or complexes (α-, β-, β′-, and β″-subunits of PEP, a putative DNA polymerase A, two putative subunits of the DNA gyrase, the elongation factor EF-Tu, and three ribosomal proteins, S3, L12-A, and L26). Eight additional proteins contain DNA/RNA binding domains, such as the PPR, SMR, SAP, OB fold, S1, KOW, NGN, mTERF, or single-stranded DNA (ssDNA) binding motifs (Table 2). These motifs are found in proteins involved in chromatin organization, DNA repair, transcription, RNA processing, or translation (Boni et al., 1991; Kyrpides et al., 1996; Fernandez-Silva et al., 1997; Draper and Reynaldo, 1999; Moreira and Philippe, 1999; Aravind and Koonin, 2000; Lurin et al., 2004). Furthermore, pTAC1 and pTAC11 exhibit striking similarities to p24 proteins, which are members of the Whirly proteins of nuclear transcription factors (Desveaux et al., 2002, 2004). All Whirly proteins have target sequences for organelles (Krause et al., 2005); thus, they might be dually targeted to the nucleus and the plastid. The exact functions of the pTAC proteins remain to be determined; however, the striking similarities of the observed phenotypes of the knockout lines for three of these proteins with those of Δrpo mutants and other mutants impaired in PEP function suggests that they are also involved in plastid gene expression. Like Δrpo plants, ptac2, -6, and -12 mutants are defective in plastid gene transcription as well as proper mRNA processing and/or stability, suggesting that they belong to a complex that is involved in different steps of plastid gene expression. This is particularly interesting for pTAC6 and -12, two plastid-localized proteins for which no domains or functions could be predicted. Only the presence of these proteins in our TAC preparations in combination with the observed phenotype of the knockout lines allows us to predict a role of these proteins in plastid gene expression.
In Escherichia coli and bacteriophage λ, different proteins, including subunits of the ribosomes, are components of the transcription and translation machinery, at least during some stages of the cell cycle (Zellars and Squires, 1999). Some RNAP subunits play an important role during transcription initiation, antitermination, elongation, and termination (Squires and Zaporojets, 2000). Moreover, many ribosomal proteins participate in the transcription of their own genes or of rRNA genes (Zengel et al., 1980; Squires and Zaporojets, 2000). Oleskina et al. (1999) also identified ribosomal proteins that are associated with plastid DNA. Studies on phage Q replication demonstrated that S1 and the elongation factors EF-TU and EF-TS are also subunits of the β-RNA replicase (Brown and Gold, 1996). Likewise, EF-Tu, S3, L12, and L29 could have a dual function in plastid gene expression in that they coordinate replication, transcription, and translation. Furthermore, pTAC15 contains a KOW domain, which is characteristic for members of the NusG protein family (Kyrpides et al., 1996). NusG is an essential factor for coupled transcription/translation in E. coli and bacteriophage λ (Zellars and Squires, 1999).
While a putative DNA polymerase (Kimura et al., 2002) and two subunits of the topoisomerase II (Reece and Maxwell, 1991; Cho et al., 2004) are primarily involved in replication and the four plastid-encoded subunits α, β, β′, and β″ of PEP in transcription in the TAC, the exact role of the other components identified in this fraction is less clear. A Fe-SOD has also been detected in the PEP-A complex of a soluble plastid fraction from mustard (Pfannschmidt et al., 2000; Ogrzewalla et al., 2002; Loschelder et al., 2004), together with putative kinases, which we did not find in our preparations. The absence of the cpk2 kinase can be explained by the loose association with the sRNAP (Baginsky et al., 1997). Our data indicate that at least two Fe-SODs are present in our TAC preparations. They might function as protective agents against reactive oxygen species (Pfannschmidt et al., 2000). Hydrogen peroxides, for instance, can stimulate transcription of plastid ndh genes under photooxidative stress (Casano et al., 2001). We also identified two PFKs in the TAC, one of them was also found in sRNAP preparations (Suzuki et al., 2004). PFK might couple the expression of photosynthesis genes to sucrose signals in the plant cell (Oswald et al., 2001). Finally, thioredoxin, which was also identified in our preparations, regulates key enzymes in the plastids via redox signals (Schürmann and Jacquot, 2000). In Rhodobacter sphaeroides, expression of genes from the puf operon is controlled by thioredoxin (Pasternak et al., 1999). Furthermore, redox signals via thioredoxin are able to influence gene expression by affecting gyrase activity (Li et al., 2004). Thus, the association of these components to the TAC might ensure its regulation in response to various signals.
We could not identify the TAC component ET1, which was recently detected immunologically in maize (Zea mays) (da Costa e Silva et al., 2004). However, it cannot be excluded that this protein is present in the TAC fraction. It is conceivable that ET1 was not detected by MS because of simultaneous elution of other abundant peptide ions. Presumably, it is also the case with the nuclear-encoded RNA polymerase RpoTp (RpoT;3; At2g24120), which we could only identify with scores slightly below our standard criteria (xcorr: 2.47 for the double-charged peptide ion 294[KNNAGDSQEELK]305 and 1.59 for the single-charged peptide ion 149[IERKDIDK]156). In future experiments, multdimensional chromatography (e.g., strong cation exchange coupled with reversed-phase liquid chromatography) would probably enable the identification of more peptides by MS, especially low abundant ones. As mentioned above, Table 1 contains only those proteins that were identified by at least two independent peptides in independent preparations. At least 40 other candidate proteins that do not fulfill this criterion were also identified in our preparation, and four of them have been described before as putative candidates of the TAC. This includes PEND (Sato et al., 1993), sulfite reductase (Cannon et al., 1999; Chi-Ham et al., 2002; Sekine et al., 2002), CND41 (Murakami et al., 2000), and MFP1 (Jeong et al., 2003). Some of them are clearly contamination of other plastid multiprotein complexes, such as the small subunit of ribulose bisphosphate carboxylase, PsbS (subunit S of the photosystem II), the α-subunit of carboxyltransferase, and a putative ferritin subunit (At3g11050). Thus, additional work is required to clearly demonstrate which proteins are tightly associated with TAC.
The role of the new components identified in our studies is more enigmatic. Arabidopsis knockout lines for three of these components were analyzed in this study. These components have been chosen because bioinformatic analysis would not allow any prediction of their association with TAC. The knockout lines have severe lesions in plastid transcription and RNA metabolism that lead to almost identical phenotypes reported for Δrpo mutants and mutants that do not accumulate PEP (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-Maciossek et al., 1999; Krause et al., 2000; Legen et al., 2002). While transcripts for all plastid-encoded genes can be detected in the mutants, their expression profiles differ. For some genes, <10% of the wild-type transcript level can be detected. Interestingly, transcription of such genes is only driven from PEP promoters. Expression of genes with NEP promoters is either not altered or upregulated. Similar to the expression profiles in Δrpo mutants and mutants with lesions in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-Maciossek et al., 1999; Krause et al., 2000; Legen et al., 2002), we observe the accumulation of larger amounts of not properly processed transcripts. Primer extension analyses indicate that either the transcriptional activity of the PEP enzyme and/or the mRNA stability of plastid genes are affected in the mutants. Since atpB mRNA levels and transcription rates are not downregulated in the ptac2 mutants (Figures 8 and 9), it is likely that the lower signal at the PEP promoter site of atpB is specific for the PEP activity. We have chosen the atpB gene for this study because the transcript level for this gene is comparable in mutant and wild-type seedlings. A more detailed analysis of NEP and PEP promoter site usages in the ptac mutants requires a better knowledge of the exact initiation sites of plastid genes in Arabidopsis.
Proper expression of plastid genes is essential for the differentiation of proplastids to chloroplasts (Baumgartner et al., 1993; Mache et al., 1997). Hence, defects in plastid gene expression lead to reduced chlorophyll levels and obviously counteract any adverse effects, such as the assembly of thylakoid structures. However, electron micrographs of chloroplasts from wild-type and ptac2, -6, and -12 plants (Figure 5) reveal abnormal membrane structure in mutant plants. This is in accordance with the observation of reduced photochemical efficiency (Table 4) in ptac2, -6, and -12 plants, allowing them to grow only under heterotrophic conditions. The ptac mutants investigated are phenotypically very similar to PEP-deficient tobacco mutants. Both contain undeveloped plastids that have vesiculated internal membranes instead of thylakoids, depending on the stage of the leaf (De Santis-Maciossek et al., 1999). Furthermore, several other mutants affected in plastid gene expression also have severe lesions in chloroplast development (Shirano et al., 2000; Ishizaki et al., 2005).
PTAC2, -6, and -12 exhibit no obvious sequence similarities to other known proteins from prokaryotic organisms, except that pTAC2 contains PPR and TPR motifs, which are characteristic for proteins involved in mRNA processing, stability, and/or translation (Fisk et al., 1999; Boudreau et al., 2000; Yamazaki et al., 2004). Thus, these genes must have newly developed after the establishment of eukaryotism. Several lines of evidence support the idea that these proteins might be involved in plastid transcription/translation. Since ptac2, -6, and ptac12 are expressed in cells with different types of plastids, the proteins should be involved in a more general plastid function. The PEP promoter activity of atpB is specifically downregulated in the mutants, while the NEP promoter in clpP is not (Figure 10). The same alterations in plastid gene expression are observed in etiolated and light-grown seedlings. Thus, neither a block in thylakoid biogenesis or photosynthesis nor photodamage causes the observed phenotype of the organelle and of the plants. The observations that Arabidopsis knockout lines for pTAC2, -6, and -12 exhibit the same phenotype as mutants with lesions in the core subunits of PEP strongly suggests that the three new nuclear-encoded proteins are intrinsic components of the plastid transcription machinery.
METHODS
Plant Material and Growth
Sinapis alba var Albatros and Arabidopsis thaliana (ecotype Columbia) were used for all studies. The T-DNA insertion lines (Salk_075736, Salk_024431, and Salk_025099) were obtained from NASC. Arabidopsis was grown on Murashige and Skoog medium (Murashige and Skoog, 1962) at 21°C with a day/night cycle of 16/8 h (50 to 60 μE m−2 s−1; Osram LW30); if not, other light intensities are mentioned. For propagation, Arabidopsis seedlings were transferred to soil after 18 d, and the seeds were harvested in Aracon tubes (Beta Tech). For analysis of tissue-specific gene expression and biochemical studies, Arabidopsis and mustard plants were grown on soil in a growth chamber (day/night rhythm of 16/8 h; 80 to 100 μE m−2 s−1, 21°C; Osram LW30).
Chlorophyll and Fluorescence Induction Measurements
Chlorophyll and fluorescence measurements were performed with 14-d-old Arabidopsis seedlings (5 to 10 μE m−2 s−1; Osram LW30) grown on Murashige and Skoog medium. For fluorescence measurements, a Fluorcam 700 MF (Photon System Instruments) was used. After dark acclimation (15 min), the fluorescence were measured according to Wagner et al. (2005). The nomenclature of the standard chlorophyll parameters refer to van Kooten and Snel (1990). Chlorophyll and carotenoid pigments were determined by HPLC according to Büch et al. (1994). Samples were ground in liquid nitrogen in Eppendorf tubes and extracted as described by Ensminger et al. (2001).
Isolation of TAC
Chloroplasts were isolated from 2 to 3 kg cotyledons of 5-d-old mustard seedlings or 4-week-old Arabidopsis rosette leaves by sucrose gradient centrifugation (18 gradients at 35 mL; Tiller et al., 1991) at 4°C. After lyses of the chloroplasts in 200 mL buffer A (50 mM Tris-HCl, pH 7.6, 100 mM [NH4]2SO4, 4 mM EDTA, 25% glycerol, 1% Triton X-100, 40 mM 2-mercaptoethanol, and 50 μg/mL phenylmethylsulfonyl fluoride [PMSF]; Rushlow and Hallick, 1982), the insoluble material was removed by centrifugation (20,000g, 4°C, 30 min).
Thirty microliters of the soluble fraction was used for gel filtration on several Sepharose 4B columns (2 cm diameter, 100 cm length, column volume 380 mL) at 4°C with buffer A. Fractions with transcriptional activity were combined and the TAC precipitated by ultracentrifugation (5 h, 200,000g, 4°C). The pellets were resuspended in 15 mL of buffer B (50 mM Tris-HCl, pH 7.6, 100 mM [NH4]2SO4, 4 mM EDTA, 25% glycerol, 40 mM 2-mercaptoethanol, and 50 μg/mL PMSF), the insoluble material removed by centrifugation (10 min, 20,000g, 4°C), and the soluble proteins applied to a Sepharose 2B column (diameter 1.5 cm, length 170 cm, and volume 301 mL). After elution of the transcriptional active fractions with buffer B, the TAC was obtained by ultracentrifugation as described above. Final resuspension occurred in 1 mL of buffer C (50 mM Tris-HCl, pH 7.6, 50 mM [NH4]2SO4, 4 mM EDTA, 10% glycerol, 40 mM 2-mercaptoethanol, and 50 μg/mL PMSF). Alternatively, the eluate of the first column was subjected to either Heparin Sepharose CL-6B or Q Sepharose chromatography. After resuspension of the TAC in 50 mM Tris-HCl, pH 7.6, 50 mM [NH4]2SO4, 5 mM MgCl2; 10% glycerol, 40 mM 2-mercaptoethanol, and 50 μg/mL PMSF, and DNase/RNase treatment at 37°C for 1 h, it was loaded onto a Heparin Sepharose CL-6B column (diameter 1.0 cm, length 10 cm, and volume 7 mL). The proteins were eluted with 50 mM Tris-HCl, pH 7.6, 2 M [NH4]2SO4, 10% glycerol, 4 mM EDTA, 40 mM 2-mercaptoethanol, and 50 μg/mL PMSF, dialyzed and concentrated against the buffer C, and used for MS. For Q Sepharose chromatography (diameter 1.0 cm, length 10 cm, and volume 7 mL), the DNase/RNase treatment was omitted.
Transcription Assay
In vitro transcription was analyzed by measuring [3H]UTP incorporation into RNA under conditions as described by Reiss and Link (1985). One unit was defined as incorporation of 1 fmol [3H] into UTP in 30 min at 30°C (Suck et al., 1996).
Preparation of Samples and MS
After phenol/chloroform extraction of the transcriptionally active fraction, proteins were precipitated with ethanol, dried, and resuspended in 50 μL 50 mM NH4HCO3 and 0.05% β-dodecylmaltoside. After denaturation (95°C for 10 min), proteins were digested with trypsin according to manufacturer's instructions (Promega) at 37°C for 16 h. The peptides were then purified with POROS-R2 (Applied Biosystems) on a C18 Zip-Tip column (Millipore; Stauber et al., 2003). Silver-stained bands from protein gels were analyzed according to Stauber et al. (2003). MS and MS/MS spectra were recorded with a LCQ Deca XP IONTRAP mass spectrometer (Thermo) equipped with a nano-HPLC system (Ultimate; Dionex) according to Stauber et al. (2003). Liquid chromatography was performed with a reverse-phase column (PepMap C18 column; LC-Packings, Dionex). Identification of the proteins occurred with the Finnigan Sequest/Turbo Sequest software (revision 2.0; ThermoQuest) by comparing the recorded masses with those calculated for trypsin digestion products of all Arabidopsis peptides available in the databases (ftp://ftp.arabidopsis.org/sequences/blast_datasets/; The Arabidopsis Information Resource).
Array Hybridization and Quantification
Theme-specific plastome macroarrays were produced by spotting gene-specific PCR products of all 78 plastome-encoded proteins (100 to 500 bp in length, amplified from the 3′ end of the cDNA) in duplicates on Hybond N+ membranes (Amersham Pharmacia Biotech). DIG-labeled cDNA of the wild type and mutants were synthesized from 5 μg of RNA and quantified as described by Kandlbinder et al. (2004). Each cDNA from three independent experiments was hybridized with an array filter of the plastome macroarrays. Array hybridization and data evaluation were performed as described in detail by Kandlbinder et al. (2004). The spot intensities were quantified with AIDA Array Vision software (Raytest) and normalized according to whole intensity of all spots on the array. Data management excluded all spots that gave signal intensities below 120% of the background or whose signals varied >50% in duplicates in at least one experiment. The normalized spot intensities were tested for each treatment against the wild type by the use of Student's t test analysis (P < 0.05, P < 0.1, and P < 0.2). To quantify differential expression of the mutant lines, IF was calculated by the ratio of the average normalized signal intensities of the mutant lines and the wild type.
Real-Time PCR
Real-time quantitative RT-PCR was performed using an iCycler iQ real-time PCR detection system and iCycler software version 2.2 (Bio-Rad). Total RNA was isolated from three independent replicates of seedlings. For the amplification of PCR products, iQ SYBR Green Supermix (Bio-Rad) was used according to the manufacturer's protocol in a final volume of 25 μL. The iCycler was programmed to 95°C 2 min; 40 × (95°C 30 s, 55°C 40 s, 72°C 45 s), 72°c 10 min followed by a melting curve program (55 to 95°C in increasing steps of 0.5°C). All reactions were performed in triplicate. The mRNA levels for each cDNA probe were normalized with respect to the actin mRNA level. Fold induction values of target genes were calculated with the ΔΔCP equation of Pfaffl (2001) and related to the mRNA level of target genes in cotyledons, which was defined as 1.0.
Primer Extension Analysis
Seedlings were either kept in low light (5 to 10 μE m−2 s−1; Osram LW30) or 3-d-old low-light grown seedlings were transferred to darkness for an additional 18 d. Primer extension reactions were performed with 10 μg of total leaf RNA according to standard protocols (Sambrook et al., 1989). Briefly, primer PE3AthatpB (5′-CGGTTATGCGTCCCATTTATTCATC-3′; position 54,537) and PE104AthclpP (5′-GGTACTTTTGGAACGCCAATAGGC-3′; position 71,857) were end-labeled with [γ-32P]-ATP and T4 polynucleotide kinase. Primer extensions were performed using Superscript III MMLV reverse transcriptase (Gibco BRL) at 55°C and the resulting products analyzed on 5% sequencing gels. DNA sequences were generated by sequencing plasmid pMS2 and pMS3 using the same primer. Plasmid pMS2 harbors the Arabidopsis rbcL-atpB intergenic region (position 54,079 to 55,090), and pMS3 contains the Arabidopsis clpP upstream region (position 71,774 to 72,897; M. Swiatecka and K. Liere, unpublished data).
Run-On Transcription
Run-on assays were performed with 2 × 107 chloroplasts isolated from 12 g of leaves of 24-d-old plants in 100 μL of 50 mM Hepes-KOH, pH 8.0, 33 mM KCl, 10 mM MgCl2, 125 μM ATP, 125 μM CTP, 125 μM GTP, 10 μM UTP, 50 μCi α-32P-UTP (110 TBq/mmol), and 55 μg heparin (Mullet and Klein, 1987). Incorporation of α-32P-UTP into RNA was determined according to Rushlow and Hallick (1982). Four picomoles plasmid DNA, denatured in 0.5 M NaOH, was immobilized on nylon membranes and hybridized to the radiolabeled transcripts. Analyses occurred with the Storm PhosphorImager (Molecular Dynamics).
Miscellaneous
Methods not specified here were performed as described by Sambrook et al. (1989). The following primers were used for the amplification of genomic or plastid DNA fragments from wild-type or knockout lines (SALK) or for cDNA fragments: left border of inserted T-DNA in the SALK lines, 5′-TGGTTCACGTAGTGGGCCATCG-3′; Salk_075736-f, 5′-TGTTTATGGGAAGACGGAGAG-3′; Salk_075736-r, 5′-GGTCGCATTATGTGCCAGC-3′; Salk_024431-f, 5′-ATGGCGTCTTCCGCCGCTTCTC-3′; Salk_024431-r, 5′-GATGTGCAATGAGTGACTATGGATG-3′; Salk_025099-f, 5′-GCGGTTATGGATTTCAGAGGACC-3′; Salk_025099-r, 5′-GAAGAGGAGACTGATCCTTAA-3′; ptac2-3f, 5′-AAGCGTGGACTTTTCCCTGAG-3′; At1g74850-r, 5′-TTAAGCTGTGCTCCCTGCTAGTTCTT-3′; ptac6-2f, 5′-ACCGGAACAGAAGGACAAACG-3′; At1g21600-r, 5′-TTCAGTGTCTCAAATTGGTTCTAG-3′; ptac12-2f, 5′-CAAAAGAGAAAACCGAGCAGC-3′; At2g34640-r, 5′-TTAAGGATCAGTCTCCTCTTC-3′; actin-f, 5′-GGTAACATTGTGCTCAGTGGTGG-3′; actin-r, 5′-CTCGGCCTTGGAGATCCACATC-3′; accD-f, 5′-TGGATAGTTTTGCTCCTGGTGA-3′; accD-r, 5′-TCACGAATCTTTCCGCTTTCA-3′; clpP-f, 5′-CCTGGAGAAGGAGATACATCTTGG-3′; clpP-r, 5′-CTTGGGCTTCTGTTGCTGAC-3′; atpA-f, 5′-TGGTAACCATTAGAGCCGACG-3′; atpA-r, 5′-CGACGATTGGTAAGGCAGTCA-3′; atpB-f, 5′-GACGTATCGCCCAAATCATTG-3′; atpB-r, 5′-CGCTGGATAGATACCTTTGGCA-3′; ndhB-f, 5′-CTTCAGCTTCAGCCACTCGAA-3′; ndhB-r, 5′-CAATCGCAATAATCGGGTTCA-3′; ndhF-f, 5′-TCCTTTTTCCGACAGCAACAA-3′; ndhF-r, 5′-CTCCATGGCATCAGGTAACCA-3′; rbcL-f, 5′-GTATGGACGTCCCCTATTAGGATG-3′; rbcL-r, 5′-GGTAGTGAGACCCAATCTTGAGTG-3′; rps14-f, 5′-TCATTTGATTCGTCGATCCTCA-3′; rps14-r, 5′-AACCATTTCCCGAAGGATGTG-3′; ycf3-f, 5′-ATGTCGGCTCAATCTGAAGGA-3′; ycf3-r; 5′-CGGTAATGACAGATCACAGCCA-3′. Insertions were checked by amplifying DNA fragments with gene-specific and T-DNA–specific primers. After cloning into the pGem-T vector, the insertions were sequenced. For RNA gel blot analyses and macroarray analyses, total RNA from 18-d-old wild-type or mutant plants or the tissue indicated in the figure was isolated with Trizol reagent (Gibco BRL). Etiolated material (see above) was used for the results shown in Figure 9E. For RNA gel blot analyses, RNA was isolated according to Sambrook et al. (1989), and 15 μg was loaded per lane. RT-PCR was performed with RevertAid H Minus M-MuLV reverse transcriptase according to the manufacturer's instruction (MBI-Fermentas).
For the immunological detection of proteins on membranes, plant material was homogenized with liquid nitrogen and dissolved in a fivefold excess of homogenization buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 2 mM EGTA, and 10 mM DTT). After separation of soluble and membrane-bound proteins by centrifugation (10 min, 17,000g), soluble proteins were precipitated with trichloroacetic acid and resuspended in gel loading buffer (100 mM NaCO3, 50 mM DTT, and 10% saccharose). The pellet containing membrane proteins was resuspended in gel loading buffer. The primary antibodies used for protein gel blot analyses have been described (Stöckel and Oelmüller, 2004). All procedures of electron microscopy were also described (Kusnetsov et al., 1994).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Table 2.
Acknowledgments
Research was supported by Friedrich-Schiller-University. We thank W. Fischer (General Botany, Jena, Germany) for the electron micrographs and T. Pfannschmidt for critically reading the manuscript. Knockout lines were obtained from NASC.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ralf Oelmüller (b7oera@uni-jena.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036392.
References
- Allison, L.A. (2000). The role of sigma factors in plastid transcription. Biochimie 82 537–548. [DOI] [PubMed] [Google Scholar]
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., et al. (2001). The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (2000). SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25 112–114. [DOI] [PubMed] [Google Scholar]
- Baginsky, S., Tiller, K., and Link, G. (1997). Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol. Biol. 34 181–189. [DOI] [PubMed] [Google Scholar]
- Baginsky, S., Tiller, K., Pfannschmidt, T., and Link, G. (1999). PTK, the chloroplast RNA polymerase-associated protein kinase from mustard Sinapis alba L.), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 39 1013–1023. [DOI] [PubMed] [Google Scholar]
- Baumgartner, B.J., Rapp, J.C., and Mullet, J.E. (1993). Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol. 101 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni, I.V., Isaeva, D.M., Musychenko, M.L., and Tzareva, N.V. (1991). Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.D., Ossenbuhl, F., and Rochaix, J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.F., Laulhere, J.P., and Mache, R. (1979). Transcription activity of a DNA-protein complex isolated from spinach plastids. Eur. J. Biochem. 98 285–292. [DOI] [PubMed] [Google Scholar]
- Brown, D., and Gold, L. (1996). RNA replication by Q beta replicase: A working model. Proc. Natl. Acad. Sci. USA 93 11558–11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büch, K., Stransky, H., Bigus, H.J., and Hager, A. (1994). Enhancement by artificial electron acceptors of thylakoid lumen acidification and zeaxanthin formation. J. Plant Physiol. 144 641–648. [Google Scholar]
- Cahoon, A.B., Harris, F.M., and Stern, D.B. (2004). Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 5 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, G.C., Ward, L.N., Case, C.I., and Heinhorst, S. (1999). The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol. Biol. 39 835–845. [DOI] [PubMed] [Google Scholar]
- Casano, L.M., Martin, M., and Sabater, B. (2001). Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 125 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi-Ham, C.L., Keaton, M.A., Cannon, G.C., and Heinhorst, S. (2002). The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol. Biol. 49 621–631. [DOI] [PubMed] [Google Scholar]
- Cho, H.S., Lee, S.S., Kim, K.D., Hwang, I., Lim, J.S., Park, Y.I., and Pai, H.S. (2004). DNA gyrase is involved in chloroplast nucleoid partitioning. Plant Cell 16 2665–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa e Silva, O., Lorbiecke, R., Garg, P., Müller, L., Wassmann, M., Lauert, P., Scanlon, M., Hsia, A.P., Schnable, P.S., Krupinska, K., and Wienand, U. (2004). The Etched1 gene of Zea mays (L.) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. Plant J. 38 923–939. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B., and Adams, W.W. (2002). Antioxidants in photosynthesis and human nutrition. Science 298 2149–2153. [DOI] [PubMed] [Google Scholar]
- De Santis-Maciossek, G., Kofer, W., Bock, A., Schoch, S., Maier, R.M., Wanner, G., Rudiger, W., Koop, H.U., and Herrmann, R.G. (1999). Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: Molecular biology, biochemistry and ultrastructure. Plant J. 18 477–489. [DOI] [PubMed] [Google Scholar]
- Desveaux, D., Allard, J., Brisson, N., and Sygusch, J. (2002). Crystallization and preliminary X-ray crystallographic analysis of p24, a component of the potato nuclear factor PBF-2. Acta Crystallogr. D Biol. Crystallogr. 58 296–298. [DOI] [PubMed] [Google Scholar]
- Desveaux, D., Subramaniam, R., Despres, C., Mess, J.N., Levesque, C., Fobert, P.R., Dangl, J.L., and Brisson, N. (2004). A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 6 229–240. [DOI] [PubMed] [Google Scholar]
- Draper, D.E., and Reynaldo, L.P. (1999). RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 27 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ensminger, I., Xyländer, M., Hagen, C., and Braune, W. (2001). Strategies providing success in a variable habitat: III. Dynamic control of photosynthesis in Cladophora glomerata. Plant Cell Environ. 24 769–779. [Google Scholar]
- Fernandez-Silva, P., Martinez-Azorin, F., Micol, V., and Attardi, G. (1997). The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J. 16 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem, W., Narita, J.O., Greenberg, B.M., Prescott, D.M., and Hallick, R.B. (1983). Selective in vitro transcription of chloroplast genes. J. Cell. Biochem. 22 31–46. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T.J., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277 809–811. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, W.R., Prombona, A., Fieder, B., Subramanian, A.R., and Börner, T. (1993). Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann, A., and Link, G. (2003). DNA-binding and transcription characteristics of three cloned sigma factors from mustard (Sinapis alba L.) suggest overlapping and distinct roles in plastid gene expression. Eur. J. Biochem. 270 1288–1300. [DOI] [PubMed] [Google Scholar]
- Hu, J., and Bogorad, L. (1990). Maize chloroplast RNA polymerase: The 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc. Natl. Acad. Sci. USA 87 1531–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi, G.L., and Kössel, H. (1992). The transcriptional apparatus of chloroplast. CRC Crit. Rev. Plant Sci. 10 525–558. [Google Scholar]
- Ikeda, T.M., and Gray, M.W. (1999). Identification and characterization of T7/T3 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol. Biol. 40 567–578. [DOI] [PubMed] [Google Scholar]
- Ishizaki, Y., Tsunoyama, Y., Hatano, K., Ando, K., Kato, K., Shinmyo, A., Kobori, M., Takeba, G., Nakahira, Y., and Shiina, T. (2005). A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42 133–144. [DOI] [PubMed] [Google Scholar]
- Jeong, S.Y., Peffer, N., and Meier, I. (2004). Phosphorylation by protein kinase CKII modulates the DNA-binding activity of a chloroplast nucleoid-associated protein. Planta 219 298–302. [DOI] [PubMed] [Google Scholar]
- Jeong, S.Y., Rose, A., and Meier, I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31 5175–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandlbinder, A., Finkemeier, I., Wormuth, D., Hanitzsch, M., and Dietz, K.J. (2004). The antioxidant status of photosynthesizing leaves under nutrient deficiency: Redox regulation, gene expression and antioxidant activity in Arabidopsis thaliana. Physiol. Plant 120 63–73. [DOI] [PubMed] [Google Scholar]
- Kasai, K., Kawagishi-Kobayashi, M., Teraishi, M., Ito, Y., Ochi, K., Wakasa, K., and Tozawa, Y. (2004). Differential expression of three plastidial sigma factors, OsSIG1, OsSIG2A, and OsSIG2B, during leaf development in rice. Biosci. Biotechnol. Biochem. 68 973–977. [DOI] [PubMed] [Google Scholar]
- Kato, Y., Murakami, S., Yamamoto, Y., Chatani, H., Kondo, Y., Nakano, T., Yokota, A., and Sato, F. (2004). The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 220 97–104. [DOI] [PubMed] [Google Scholar]
- Kimura, S., et al. (2002). A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 30 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 313–349. [Google Scholar]
- Krause, K., Kilbienski, I., Mulisch, M., Rodiger, A., Schafer, A., and Krupinska, K. (2005). DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 579 3707–3712. [DOI] [PubMed] [Google Scholar]
- Krause, K., and Krupinska, K. (2000). Molecular and functional properties of highly purified transcriptionally active chromosomes from spinach chloroplasts. Physiol. Plant 109 188–195. [Google Scholar]
- Krause, K., Maier, R.M., Kofer, W., Krupinska, K., and Herrmann, R.G. (2000). Disruption of plastid-encoded RNA polymerase genes in tobacco: Expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol. Gen. Genet. 263 1022–1030. [DOI] [PubMed] [Google Scholar]
- Krupinska, K., and Falk, J. (1994). Changes in RNA polymerase activity during development and senescence of barley chloroplasts. Comparative analysis of transcripts synthesized either in run-on assays or by transcriptionally active chromosomes (TAC). J. Plant Physiol. 143 298–305. [Google Scholar]
- Kusnetsov, V., Oelmüller, R., Sarwat, M.I., Porfirova, S.A., Cherepneva, G.N., Herrmann, R.G., and Kualeva, O.N. (1994). Cytokinins, abscisic acid and light affect accumulation of chloroplast proteins in Lupinus luteus cotyledons without notable effect on steady-state mRNA levels. Specific protein response to light/phytohormone interaction. Planta 194 318–327. [Google Scholar]
- Kyrpides, N.C., Woese, C.R., and Ouzounis, C.A. (1996). KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21 425–436. [DOI] [PubMed] [Google Scholar]
- Legen, J., Kemp, S., Krause, K., Profanter, B., Herrmann, R.G., and Maier, R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31 171–188. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache, S. (1993). The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: Single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc. Natl. Acad. Sci. USA 90 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K., Pasternak, C., Hartig, E., Haberzettl, K., Maxwell, A., and Klug, G. (2004). Thioredoxin can influence gene expression by affecting gyrase activity. Nucleic Acids Res. 32 4563–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere, K., Kaden, D., Maliga, P., and Börner, T. (2004). Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Res. 32 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere, K., and Maliga, P. (1999). In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J. 18 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschelder, H., Homann, A., Ogrzewalla, K., and Link, G. (2004). Proteomics-based sequence analysis of plant gene expression–The chloroplast transcription apparatus. Phytochemistry 65 1785–1793. [DOI] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mache, R., Zhou, D.X., Lerbs-Mache, S., Harrak, H., Villain, P., and Gauvin, S. (1997). Nuclear control of early plastid differentiation. Plant Physiol. Biochem. 35 199–203. [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden, C.W., Wolfe, K.H., dePamphilis, C.W., and Palmer, J.D. (1991). Plastid translation and transcription genes in a non-photosynthetic plant: Intact, missing and pseudo genes. EMBO J. 10 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, D., and Philippe, H. (1999). Smr: A bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem. Sci. 24 298–300. [DOI] [PubMed] [Google Scholar]
- Mullet, J.E., and Klein, R.R. (1987). Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 6 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, S., Kondo, Y., Nakano, T., and Sato, F. (2000). Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett. 468 15–18. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15 473–497. [Google Scholar]
- Narita, J.O., Rushlow, K.E., and Hallick, R.B. (1985). Characterization of a Euglena gracilis chloroplast RNA polymerase specific for ribosomal RNA genes. J. Biol. Chem. 260 11194–11199. [PubMed] [Google Scholar]
- Ogrzewalla, K., Piotrowski, M., Reinbothe, S., and Link, G. (2002). The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur. J. Biochem. 269 3329–3337. [PubMed] [Google Scholar]
- Oleskina, Yu.P., Yurina, N.P., Odintsova, T.I., Egorov, T.A., Otto, A., Wittmann-Liebold, B., and Odintsova, M.S. (1999). Nucleoid proteins of pea chloroplasts: Detection of a protein homologous to ribosomal protein. Biochem. Mol. Biol. Int. 47 757–763. [DOI] [PubMed] [Google Scholar]
- Oswald, O., Martin, T., Dominy, P.J., and Graham, I.A. (2001). Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak, C., Haberzettl, K., and Klug, G. (1999). Thioredoxin is involved in oxygen-regulated formation of the photosynthetic apparatus of Rhodobacter sphaeroides. J. Bacteriol. 181 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., and Link, G. (1994). Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol. Biol. 25 69–81. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Ogrzewalla, K., Baginsky, S., Sickmann, A., Meyer, H.E., and Link, G. (2000). The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur. J. Biochem. 267 253–261. [DOI] [PubMed] [Google Scholar]
- Rajashekar, V.K., Sun, E., Meeker, R., Wu, B.W., and Tewari, K.K. (1991). Highly purified pea chloroplast RNA polymerase transcribes both rRNA and mRNA genes. Eur. J. Biochem. 195 215–228. [DOI] [PubMed] [Google Scholar]
- Reece, R.J., and Maxwell, A. (1991). DNA gyrase: Structure and function. Crit. Rev. Biochem. Mol. Biol. 26 335–375. [DOI] [PubMed] [Google Scholar]
- Reiss, T., and Link, G. (1985). Characterization of transcriptionally active DNA-protein complexes from chloroplasts and etioplasts of mustard (Sinapis alba L.). Eur. J. Biochem. 148 207–212. [DOI] [PubMed] [Google Scholar]
- Robinson, N.J., Procter, C.M., Connolly, E.L., and Guerinot, M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697. [DOI] [PubMed] [Google Scholar]
- Rushlow, K.E., and Hallick, R.B. (1982). The isolation and purification of a transcriptionally active chromosome from chloroplast of Euglena gracilis. In Methods in Chloroplast Molecular Biology, M. Edelman, R.B. Hallick, and N.-H. Chua, eds (Amsterdam: Elsevier), pp. 543–550.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sato, N., Albrieux, C., Joyard, J., Douce, R., and Kuroiwa, T. (1993). Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J. 12 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann, P., and Jacquot, J.P. (2000). Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 371–400. [DOI] [PubMed] [Google Scholar]
- Sekine, K., Hase, T., and Sato, N. (2002). Reversible DNA compaction by sulfite reductase regulates transcriptional activity of chloroplast nucleoids. J. Biol. Chem. 277 24399–24404. [DOI] [PubMed] [Google Scholar]
- Shirano, Y., et al. (2000). Chloroplast development in Arabidopsis thaliana requires the nuclear-encoded transcription factor sigma B. FEBS Lett. 485 178–182. [DOI] [PubMed] [Google Scholar]
- Silhavy, D., and Maliga, P. (1998). Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 33 340–344. [DOI] [PubMed] [Google Scholar]
- Squires, C.L., and Zaporojets, D. (2000). Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54 775–798. [DOI] [PubMed] [Google Scholar]
- Sriraman, P., Silhavy, D., and Maliga, P. (1998). The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res. 26 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber, E.J., Fink, A., Markert, C., Kruse, O., Johanningmeier, U., and Hippler, M. (2003). Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 5 978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel, J., and Oelmüller, R. (2004). A novel protein for photosystem I biogenesis. J. Biol. Chem. 12 10243–10251. [DOI] [PubMed] [Google Scholar]
- Suck, R., Zeltz, P., Falk, J., Acker, A., Kossel, H., and Krupinska, K. (1996). Transcriptionally active chromosomes (TACs) of barley chloroplasts contain the alpha-subunit of plastome-encoded RNA polymerase. Curr. Genet. 30 515–521. [DOI] [PubMed] [Google Scholar]
- Summer, H., Pfannschmidt, T., and Link, G. (2000). Transcripts and sequence elements suggest differential promoter usage within the ycf3-psaAB gene cluster on mustard (Sinapis alba L.) chloroplast DNA. Curr. Genet. 37 45–52. [DOI] [PubMed] [Google Scholar]
- Suzuki, J.Y., Jimmy Ytterberg, A., Beardslee, T.A., Allison, L.A., Wijk, K.J., and Maliga, P. (2004). Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 40 164–172. [DOI] [PubMed] [Google Scholar]
- Tiller, K., Eisermann, A., and Link, G. (1991). The chloroplast transcription apparatus from mustard (Sinapis alba L.). Evidence for three different transcription factors which resemble bacterial sigma factors. Eur. J. Biochem. 198 93–99. [DOI] [PubMed] [Google Scholar]
- van Kooten, O., and Snel, J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25 147–150. [DOI] [PubMed] [Google Scholar]
- Wagner, R., Fey, V., Borgstädt, R., Kruse, O., and Pfannschidt, T. (2005). Screening for Arabidopsis thaliana mutants deficient in acclimatory long-term to varying light qualities using chlorophyll fluorescence imaging. Photosynthesis: Fundamental aspects to global perspectives. In Proceedings of the 13th International Congress of Photosynthesis, A. van der Est and D. Bruce, eds (Montreal, Canada: Allen Press), pp. 693–696.
- Weihe, A., Hedtke, B., and Börner, T. (1997). Cloning and characterization of a cDNA encoding a bacteriophage-type RNA polymerase from the higher plant Chenopodium album. Nucleic Acids Res. 25 2319–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Tasaka, M., and Shikanai, T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38 152–163. [DOI] [PubMed] [Google Scholar]
- Young, D.A., Allen, R.L., Harvey, A.J., and Lonsdale, D.M. (1998). Characterization of a gene encoding a single-subunit bacteriophage-type RNA polymerase from maize which is alternatively spliced. Mol. Gen. Genet. 260 30–37. [DOI] [PubMed] [Google Scholar]
- Zellars, M., and Squires, C.L. (1999). Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32 1296–1304. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M., Mueckl, D., and Lindahl, L. (1980). Protein L4 of the E. coli ribosome regulates an eleven gene r-protein operon. Cell 21 523–535. [DOI] [PubMed] [Google Scholar]