Abstract
We report the identification and characterization of a low tocopherol Arabidopsis thaliana mutant, vitamin E pathway gene5-1 (vte5-1), with seed tocopherol levels reduced to 20% of the wild type. Map-based identification of the responsible mutation identified a G→A transition, resulting in the introduction of a stop codon in At5g04490, a previously unannotated gene, which we named VTE5. Complementation of the mutation with the wild-type transgene largely restored the wild-type tocopherol phenotype. A knockout mutation of the Synechocystis sp PCC 6803 VTE5 homolog slr1652 reduced Synechocystis tocopherol levels by 50% or more. Bioinformatic analysis of VTE5 and slr1652 indicated modest similarity to dolichol kinase. Analysis of extracts from Arabidopsis and Synechocystis mutants revealed increased accumulation of free phytol. Heterologous expression of these genes in Escherichia coli supplemented with free phytol and in vitro assays of recombinant protein produced phytylmonophosphate, suggesting that VTE5 and slr1652 encode phytol kinases. The phenotype of the vte5-1 mutant is consistent with the hypothesis that chlorophyll degradation-derived phytol serves as an important intermediate in seed tocopherol synthesis and forces reevaluation of the role of geranylgeranyl diphosphate reductase in tocopherol biosynthesis.
INTRODUCTION
Tocopherols represent a group of important nutrients for animals and humans and are lipophilic antioxidants synthesized by some photosynthetic bacteria and all green plants (for reviews, see Grusak and DellaPenna, 1999; Sattler et al., 2004b). The biopotency of tocopherols for animals and humans is expressed as vitamin E activity (IUPAC-IUP Joint Commission on Biochemical Nomenclature, 1982; Cunniff, 1990). It is believed that the beneficial effects of tocopherols are derived from their radical scavenging activity in lipophilic environments, resulting in the stabilization of polyunsaturated fatty acids in membrane lipids and oils. Recent studies using Arabidopsis thaliana mutants have provided experimental evidence supporting the protective role of tocopherols against oxidative stress (Kanwischer et al., 2005) as well as beneficial effects on seed longevity and germination as a result of lipid stabilization by tocopherols in plants (Sattler et al., 2004a).
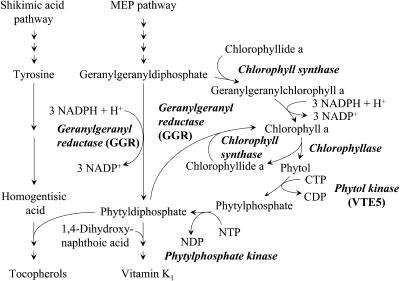
While most biochemical reactions leading to tocopherol biosynthesis were identified more than two decades ago (Soll and Schultz, 1980; Soll et al., 1980, 1983, 1985), genes encoding enzymes for tocopherol biosynthesis were identified relatively recently (Shintani and DellaPenna, 1998; Collakova and DellaPenna, 2001; Porfirova et al., 2002; Savidge et al., 2002; Cheng et al., 2003; Rohmer, 2003; Van Eenennaam et al., 2003). Homogentisic acid and phytyldiphosphate (PDP) serve as precursors for tocopherol biosynthesis (Figure 1). In plants, homogentisic acid is formed from Tyr via Tyr transaminase and p-hydroxyphenylpyruvate dioxygenase (Norris et al., 1998). PDP for tocopherol biosynthesis is thought to originate from direct reduction of geranylgeranyldiphosphate (GGDP) by geranylgeranyl diphosphate reductase (GGR) (Keller et al., 1998).
Figure 1.
Schematic Drawing of the Tocopherol Biosynthetic Pathway.
The metabolic fate of GGDP is shown in detail. Enzyme designations are shown in italicized bold text. GGDP is proposed either to be directly reduced by GGR to PDP or to serve as substrate for chlorophyll synthase to form geranylgeranyl chlorophyll a. GGR reduces geranylgeranyl chlorophyll a to form chlorophyll a. Hydrolysis of chlorophyll by the action of chlorophyllase releases phytol, which can be reassimilated through two subsequent phosphorylation reactions with phytol kinase (VTE5) catalyzing a CTP-dependent phosphorylation of free phytol. Another enzyme, phytylphosphate kinase, is proposed for the second phosphorylation reaction to obtain PDP. PDP can enter the tocopherol biosynthetic pathway, be utilized for chlorophyll biosynthesis, or enter vitamin K (phylloquinone) biosynthesis. MEP, methylerythritol phosphate pathway.
Phytol, which constitutes approximately one-third of the mass of chlorophyll, is generally considered to be the most abundant acyclic isoprenoid compound in the biosphere (Volkman and Maxwell, 1986). Serving as the hydrophobic membrane-anchor of chlorophyll in the photosynthetic membrane, it plays essential roles in the assembly, structure, and function of both plant and microbial photosynthetic reaction centers (Ben-Shem et al., 2003). Phytol is also a constituent of both tocopherols, which collectively comprise vitamin E (Grusak and DellaPenna, 1999; Bramley et al., 2000) and vitamin K (phylloquinone; Shibata et al., 2004) in plants and cyanobacteria.
In photosynthetic organisms, free phytol is generated during chlorophyll catabolism by chlorophyllase (Figure 1). While the subsequent fate of the chlorophyll breakdown product chlorophyllide a and its derived pigments has been well characterized, little is known about the metabolic fate of phytol (for general reviews on chlorophyll catabolism, see Hörtensteiner, 1999; Matile et al., 1999; Kräutler, 2002). Phytol derived from chlorophyll breakdown is reesterified to acetic acid or other fatty acids (Peisker et al., 1989; Patterson et al., 1993), while a portion is also irreversibly destroyed by photodegradation (Rontani et al., 1996). Recycling of catabolized phytol into intermediates of central metabolism has not yet been demonstrated in plants. It has been suggested that chlorophyll-derived phytol may be a precursor for the biosynthesis of tocopherols, based on the temporal patterns of chlorophyll degradation and leaf and seed tocopherol accumulation during leaf senescence (Rise et al., 1989) and seed maturation (Goffman et al., 1999). However, this possibility was suggested as being unlikely by Hörtensteiner (1999) since it would require the activation of phytol by phosphorylation, and no such enzymatic activity had been demonstrated.
An Arabidopsis mutant screen for plants with altered seed total tocopherol levels or composition (Van Eenennaam et al., 2003) identified the mutant isolate low total tocopherol 1 (lt1), which accumulates ∼20% of the wild-type seed tocopherol levels. In this report, we describe the identification of the gene responsible for this trait, Arabidopsis gene locus At5g04490 (http://arabidopsis.org/servlets/TairObject?id=136046&type=locus). Data are presented strongly supporting the hypothesis that this gene encodes a chloroplast-targeted membrane-bound enzyme with phytol kinase activity. Homologs of this gene are found in other plants as well as in cyanobacteria, and we demonstrate that slr1652, the Synechocystis sp PCC 6803 homolog, is essential for normal tocopherol accumulation in vivo and that At5g04490 and slr1652 proteins have phytol kinase activity. On the basis of these results, we rename the Arabidopsis gene vitamin E pathway gene5 (VTE5). These findings identify an enzymatic pathway for the activation and reutilization of phytol from chlorophyll degradation in plant metabolism, including tocopherol biosynthesis.
RESULTS
Isolation and Identification of an Arabidopsis Low Tocopherol Mutant
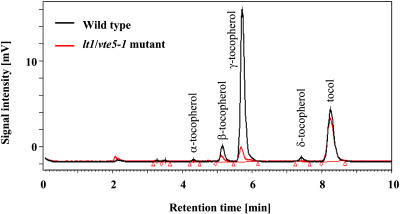
A screen for mutants with altered seed tocopherols (Van Eenennaam et al., 2003) revealed four classes of mutants: those with altered composition (high α- or high δ-tocopherol) and those with increased or decreased total tocopherol. We focused on low tocopherol (lt) lines, reasoning that recessive mutations leading to this phenotype would reveal genes that play a role in regulating flux through the pathway, potentially including novel biosynthetic enzymes and pathways. One such mutant, lt1, was chosen for further analysis because it showed an 80% reduction in total seed tocopherols, as shown in Figure 2. There was a 65% reduction in leaf tocopherol in this mutant: 6.0 ± 0.3 ppm (n = 55) for the mutant versus 17.4 ± 0.8 ppm (n = 9) for the wild type, expressed on a fresh weight basis (±se of the mean). These data demonstrate that the phenotype was not restricted to the seed.
Figure 2.
HPLC Analysis of Tocopherol Content in Seed Extracts from Wild-Type Arabidopsis and the Arabidopsis lt1 Mutant.
Wild-type trace is shown in black and lt1 in red. Tocol was used as an internal standard.
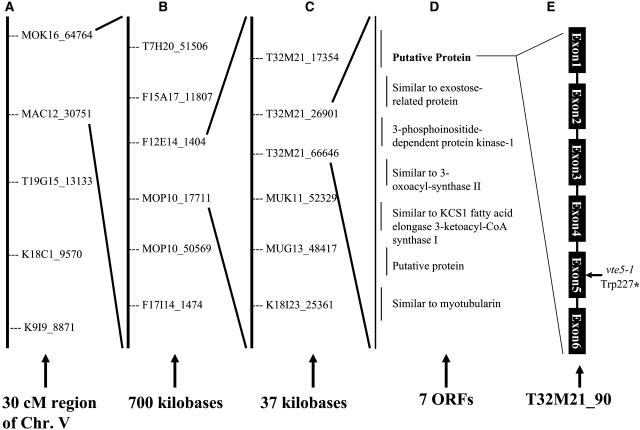
As shown in Figure 3, genetic mapping narrowed this mutation to a 37-kb region of BAC T32M21 on chromosome 5, unlinked to previously published VTE loci (Sattler et al., 2004b). DNA sequence analysis revealed a single mutation compared with wild-type Columbia-0 (Col-0) in the previously unannotated gene T32M21_90 (Arabidopsis gene locus At5g04490). This G→A transition mutation at base pair 681 relative to the ATG for this protein is predicted to convert a Trp residue (TGG) to an opal stop codon (TGA) and would cause production of a 24.4-kD truncated protein in place of the predicted 33.1-kD full-length protein. Based upon the genetic results and data described below, we propose the allele designation vte5-1.
Figure 3.
Map-Based Identification of the vte5-1 Mutation.
(A) Low-resolution mapping. Four hundred F3 plants were genotyped to show linkage to the genetic markers MOK16_64764 and MAC12_30751 on chromosome V. cM, centimorgan.
(B) Narrowing the interval between two core markers. Additional markers were assayed to identify a 700-kb region containing the mutation.
(C) High-resolution mapping. An additional 2000 F3 plants were scored using single nucleotide polymorphism markers within the 700-kb region. Markers T32M21_26901 and T32M21_66646 were found to be the closest flanking markers based on the available recombinants, narrowing the interval to 37 kb.
(D) The entire 37-kb region containing seven open reading frames (ORFs) was sequenced from the lt1 mutant.
(E) A single G→A point mutation was detected in exon 5 of T32M21_90. It is predicted to convert a Trp codon to an opal nonsense codon.
Transgenic Expression of VTE5 in Arabidopsis Wild Type and vte5-1 Mutant
To test whether the mutation in At5g04490 caused the low total tocopherol phenotype observed in the mutant lt1, both mutant and parental Col-0 wild type were transformed with pMON69914, harboring wild-type VTE5 flanked by the napin promoter and the napin 3′-untranslated region on its 5′- and 3′-ends, respectively (Norris et al., 2004), or with an empty vector control. Seed tocopherol levels in vte5-1 transformed with the seed-specific VTE5 expression construct increased more than threefold compared with mutant seed tocopherol levels and reached nearly wild-type seed tocopherol levels in R2 seed populations (Table 1); the small difference from the wild type is likely to be caused by segregation of homozygous vte5-1 mutant progeny without the complementing transgene and by variation of VTE5 expression levels among different events due to the tDNA integration loci. These results confirm that the vte5-1 mutation caused the low tocopherol phenotype in lt1. Transformation of wild-type Arabidopsis with a binary vector harboring the seed-specific T32M21_90 expression cassette (pMON69914) resulted in slightly increased seed tocopherol levels in the population of R2 seed (Table 1); however, the level of tocopherol increase was not significant at P-values below 0.05. In R3 seed, only one out of five events (S10154N) chosen from this population had a small (6%) but significant increase in seed tocopherol levels, at the 0.05 level (see Supplemental Table 1 online).
Table 1.
Tocopherol Content of Transgenic and Mutant Arabidopsis Seed
| Line | Generation | Event No. | nb | Mean Tocopherol Levelsa (ng/mg seed) | Significancea | ||
|---|---|---|---|---|---|---|---|
| vte5-1 mutant parent | Mean of population | 17 | 100 ± 2.7 | C | |||
| Wild type + vector control | R2 | Mean of all events | 4 | 458 ± 14.3 | A | ||
| Wild type + pMON69914 | R2 | Mean of all events | 28 | 476 ± 5.4 | A | ||
| vte5-1 mutant + pMON69914 | R2 | Mean of all events | 20 | 365 ± 6.4 | B | ||
| vte5-1 mutant + vector control | R2 | Mean of all events | 4 | 87 ± 14.3 | C | ||
Means, standard error, and significance were calculated using Tukey-Kramer's honestly significant difference (HSD) test. Means followed by the same letters are not significantly different from each other (α = 0.05).
Number of individual plants (for vte5-1 mutants) or events (for transgenic lines) analyzed.
Generation of a Synechocystis Mutant
To test the general involvement of VTE5 orthologs in tocopherol biosynthesis, we evaluated the phenotype of a Synechocystis sp PCC 6803 slr1652 deletion-insertion mutant. The slr1652 gene is the closest Synechocystis homolog of VTE5. This mutant was generated through homologous recombination as described in Methods. Cells were grown to OD730 2.0 to 2.5 and tested for total tocopherols. The wild-type culture and two representative mutant cultures (Δslr1652-1 and Δslr1652-2) revealed average tocopherol contents of 80.5 ± 0.5, 42.0 ± 0.9, and 21.0 ± 0.1 ng per OD unit (OD730) for two independent measurements, respectively. This represents a tocopherol reduction of 50 to 75% in the mutants.
Bioinformatic Analysis
Bioinformatic analysis of the deduced VTE5 amino acid sequence was undertaken to seek hypotheses regarding possible roles for this gene in tocopherol metabolism and revealed homology to the PFAM domain of cytidylyltransferases (CTP_transf_1, PFAM accession number PF01148). The members of this family are integral membrane proteins and include the archetype phosphatidate cytidylyltransferase (EC 2.7.7.41) as well as SEC59 from yeast, which is a dolichol kinase (EC 2.7.1.108). A likely membrane localization of the putative VTE5 protein product was reinforced when the deduced amino acid sequence was analyzed using the TMHMM algorithm (Krogh et al., 2001), which predicted six transmembrane helices corresponding to amino acids 29 to 51, 66 to 88, 105 to 122, 127 to 149, 168 to 187, and 226 to 248.
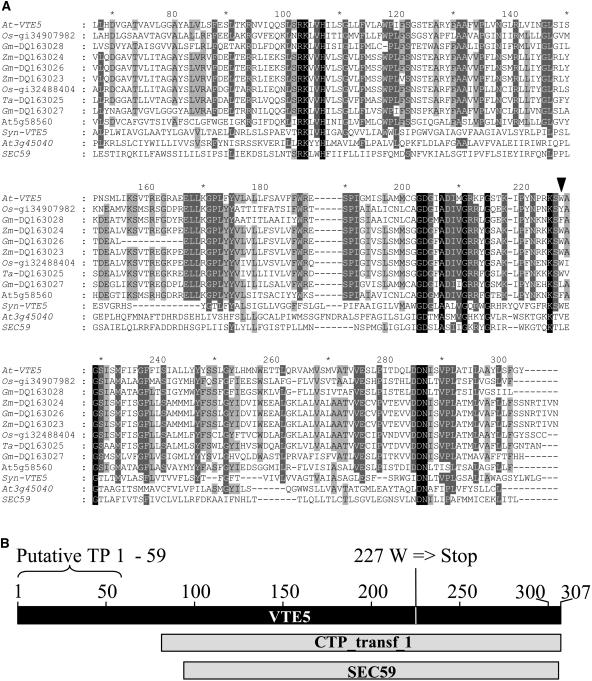
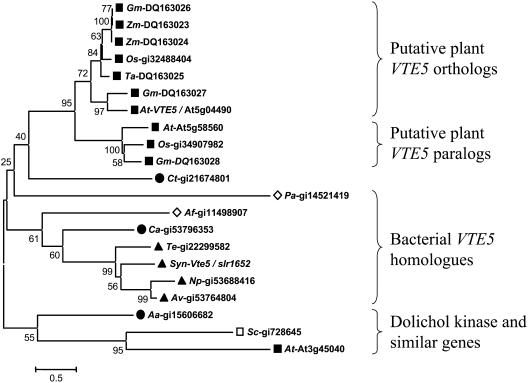
A BLAST search of VTE5 against the Arabidopsis genome identified a putative paralog (gi number 51970322, ATG ID At5g58560). The deduced amino acid sequences of these two genes were 38.7% identical (using GAP algorithm from the GCG Wisconsin Package; http://www.accelrys.com/products/gcg_wisconsin_package/), as shown in Figure 4A. At5g58560 was also found to contain the PFAM domain of integral membrane protein cytidylyltransferases (CTP_transf_1) and also includes six TMHMM-predicted transmembrane helices. It is interesting to note that the non-sense protein fragment predicted to be synthesized in vte5-1 would delete approximately one-third of the conserved C-terminal sequence, including the last transmembrane domain of Arabidopsis VTE5 (Figure 4B). A second BLAST search comparing VTE5 with the GenBank nonredundant amino acid database and Monsanto proprietary databases identified homologous sequences in archaea, eubacteria, cyanobacteria, and plants. A multiple sequence alignment comparing putative plant and Synechocystis orthologs to each other and Arabidopsis and yeast dolichol kinases is shown in Figure 4A. Sequence analysis of the deduced amino acid sequences of plant-derived VTE5 orthologs using ChloroP (Emanuelsson et al., 1999) predicted chloroplast targeting for all plant genes. Phylogenetic analysis of these genes identifies two distinct clades in plants, each represented by VTE5 or At5g58560, its related Arabidopsis gene. A neighbor-joining tree constructed using representative sequences is shown in Figure 5.
Figure 4.
Multiple Alignments of VTE5 Orthologs and Dolichol Kinases.
(A) Multiple alignments were visualized and edited using GeneDoc (Nicholas et al., 1997). The residues are shaded using conserved residue shading mode set to level 4 using default settings. Similar amino acid residues conserved in all columns have dark shading. Amino acids with 100, 80, and 60% conservation are shaded as white letters on black background, white letters on dark-gray background, and black letters on light-gray background, respectively. N-terminal nonconserved amino acids encoding putative chloroplast target sequences were deleted from the alignment. Numbers above the sequence refer to amino acid positions within the Arabidopsis VTE5 sequence. Arrowhead indicates the location of vte5-1 mutation, which is predicted to cause truncation of the remaining amino acid residues. Reference numbers are as follows: At3g45040, putative Arabidopsis dolichol kinase; Arabidopsis VTE5, At5g04490; numbers starting with the prefix DQ represent accession numbers; SEC59, S. cerevisiae dolichol kinase, accession number gi|728645; Synechocystis vte5, nucleotides 3,452,461 to 3453162, accession number gi|47118304. Aa, Aquifex aeolicus VF5; Af, Archaeoglobus fulgidus; At, Arabidopsis thaliana; Av, Anabaena variabilis; Ca, Chloroflexus aurantiacus; Ct, Chlorobium tepidum; Gm, Glycine max; Np, Nostoc punctiforme; Os, Oryza sativa; Pa, Pyrococcus abyssi; Sc, Saccharomyces cerevisiae; Syn, Synechocystis sp PCC 6803; Te, Thermosynechococcus elongatus; TP, chloroplast target peptide; Zm, Zea mays.
(B) Shematic drawing of Arabidopsis VTE5 aligned to the conserved PFAM domain of cytidylyltransferases (CTP_transf_1) and dolichol kinase (SEC59).
Figure 5.
Neighbor-Joining Tree of VTE5 Orthologs.
The tree was constructed from representative VTE5 orthologous sequences using a p-distance amino acid distance model and pairwise deletion. For details, see Methods. The alignment used to generate this tree is provided in Supplemental Figure 1 online. Closed squares represent plant sequences, the open square indicates yeast sequence, closed triangles represent cyanobacterial sequences, closed circles show eubacterial sequences, and open diamonds indicate archaeal sequences. The relationship of yeast (Sc) and Arabidopsis (At) dolichol kinases to the VTE5 family is also shown. The numbers on the left side indicate percent bootstrap values. The branch lengths calculated based on 5% amino acid substitutions per site are shown by the bar labeled 0.5. Reference numbers are as follows: numbers following the species abbreviations represent accession numbers; Synechocystis vte5, nucleotides 3,452,461 to 3453162, accession number gi|47118304. Aa, Aquifex aeolicus VF5; Af, Archaeoglobus fulgidus; At, Arabidopsis thaliana; Av, Anabaena variabilis; Ca, Chloroflexus aurantiacus; Ct, Chlorobium tepidum; Gm, Glycine max; Np, Nostoc punctiforme; Os, Oryza sativa; Pa, Pyrococcus abyssi; Sc, Saccharomyces cerevisiae; Syn, Synechocystis sp PCC 6803; Te, Thermosynechococcus elongatus; Zm, Zea mays.
Accumulation of Tocopherol Pathway Intermediates and Chlorophyll
Dolichol is a long-chain isoprenoid molecule consisting of nine or more isoprenoid units and thus is similar in structure to phytol, a putative precursor for tocopherol biosynthesis (Collakova and DellaPenna, 2003). Based on the similarity of VTE5 to known dolichol kinases, we developed a working hypothesis that VTE5 encodes a phytol kinase. If PDP is synthesized from phytol, a phytol kinase loss-of-function mutant would be expected to accumulate free phytol. Consistent with our hypothesis, Arabidopsis seed from the vte5-1 mutant contained threefold more free phytol than wild-type seed, as shown in Table 2. In Synechocystis, this effect was much stronger: while free phytol was below the limit of detection (<1 ng/OD730 unit) in wild-type cells, Δslr1652-1 and -2 mutant cells contained >200 ng free phytol/OD730 unit (Table 2), representing a >200-fold increase.
Table 2.
Free Phytol Levels in Mature Arabidopsis Seed and Synechocystis Cells
| Phytol Content [ng/mg] |
||
|---|---|---|
| Source | na | Meanb |
| Arabidopsis Col-0 | 2 | 101.7 ± 6.5 |
| Arabidopsis vte5-1 | 2 | 297.4 ± 24.2 |
| Phytol Content (ng/OD unit) | ||
| Synechocystis sp PCC 6803 | 2 | ndc |
| Synechocystis sp PCC 6803 Δslr1652-1 | 2 | 229.5 ± 4.0 |
| Synechocystis sp PCC 6803 Δslr1652-2 | 2 | 252.5 ± 12.5 |
Number of individual samples originating from separately grown cultures analyzed.
Means and standard error were calculated using Tukey-Kramer's HSD test.
Below the limit of detection (<1 ng/OD730 unit in bacterial cell samples).
The accumulation of free phytol in Synechocystis cultures and Arabidopsis seed gave rise to the question of how chlorophyll levels might be affected. To elucidate this question, total leaf chlorophyll levels in nine wild-type plants and 55 vte5-1 mutants were analyzed. The average chlorophyll levels were 1.25 ± 0.05 and 1.17 ± 0.02 μg/mg fresh leaf mass for the Arabidopsis wild type and the vte5-1 mutant, respectively. Although the chlorophyll content of the mutants was on average lower than in wild-type plants, the difference was not significant within the 95% confidence interval (α = 0.05).
Heterologous Expression of Arabidopsis VTE5 and Synechocystis Vte5 Proteins in Escherichia coli
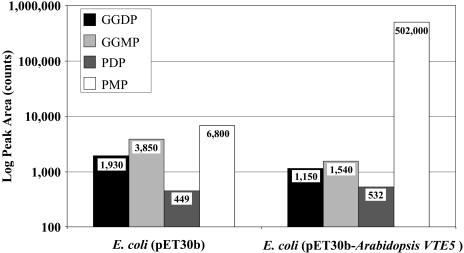
Additional confirmation that Arabidopsis VTE5 encodes a protein with phytol kinase activity was obtained by heterologous expression of the protein in Escherichia coli in the presence of exogenously added phytol. Extracts from treated cells were subjected to liquid chromatography–mass spectrometry (LC-MS) analysis for the presence of phytolmonophosphate (PMP), PDP, geranylgeranylmonophosphate (GGMP), and GGDP (see Methods for details regarding these assays). As shown in Figure 6, E. coli cells transformed with the plasmid expressing Arabdiopsis VTE5 (pET30b-VTE5) contained >70-fold more PMP than the vector control (note the log scale on the ordinate of Figure 6). Supplementation of the incubation medium with 0.2% toluene further increased PMP accumulation in the VTE5-expressing strain by >10%, presumably due to improved phytol uptake (data not shown). PDP, GGMP, or GGDP levels detected in these E. coli extracts were each <1% of PMP levels and did not increase as a result of VTE5 expression.
Figure 6.
Contents of Isoprenoid Monophosphates and Diphosphates in Extracts of E. coli Expressing Arabidopsis VTE5 and E. coli Harboring the Empty Plasmid Control.
Isoprenoid phosphates were measured on lyophilized E. coli cells obtained from 50 mL of overnight-induced LB cultures that had been harvested and resuspended in 5 mL of fresh LB medium containing 5 mM phytol, followed by 3 h incubation at 22°C and 220 rpm shaking.
Further confirmation that VTE5 encodes a phytol kinase was obtained by enzyme assay of washed E. coli membrane fractions from strains expressing Synechocystis Vte5 or Arabidopsis VTE5. As shown in Table 3, both enzymes exhibited highest activity using CTP as cosubstrate (Table 3). ATP and GTP did not support the phytol kinase reaction.
Table 3.
Phytol Kinase Activity in Membrane Fractions from Transformed E. coli Expressing Arabidopsis VTE5 or Synechocystis vte5
| Enzyme Activity (nmol·mg−1·min−1) |
|||||
|---|---|---|---|---|---|
| Cosubstrate |
|||||
| Vector Designation | 0.5 mM ATP | 0.5 mM GTP | 0.5 mM CTP | 0.5 mM UTP | 0.15 mM NTPs |
| pET30b Arabidopsis VTE5 | nda | nd | 36.7 ± 2.5 | 21.1 ± 4.2 | 33.1 ± 0.6 |
| pET30b Synechocystis vte5 | nd | nd | 16.9 ± 1.6 | 12.5 ± 1.2 | 12.8 ± 0.2 |
| pET30b | nd | nd | nd | nd | nd |
Average enzyme activities are based on four independent measurements.
nd, below the limit of detection (<32 pmol·mg−1·min−1).
DISCUSSION
We describe the identification of VTE5, a gene encoding a novel enzyme required for seed tocopherol biosynthesis using Arabidopsis mutant screening and map-based gene identification. The starting Arabidopsis mutant has reduced seed and leaf tocopherol, and a knockout mutation of the Synechocystis homolog of Arabidopsis VTE5 also led to reduced tocopherol accumulation in Synechocystis, reinforcing the general relevance of these genes for tocopherol biosynthesis.
We reasoned that the Arabidopsis VTE5 and Synechocystis vte5 genes might encode phytol kinases based on homology of the deduced amino acid sequences to dolichol kinases. Two lines of in vivo evidence are consistent with this hypothesis. First, gas chromatography–mass spectrometry analysis of the Arabidopsis and Synechocystis mutants demonstrated accumulation of free phytol. Second, expression of Arabidopsis VTE5 protein in E. coli in the presence of free phytol led to the formation of PMP. These data were confirmed in enzyme assays using washed E. coli membranes from strains expressing Arabidopsis VTE5 or Synechocystis Vte5 proteins: both enzymes produced PMP in the presence of CTP or UTP, consistent with observations of a preference for CTP by dolichol kinase and farnesyl kinase (Heller et al., 1992; Thai et al., 1999). While published data indicate that farnesyl kinase also utilized UTP and GTP as cosubstrates (Thai et al., 1999), phytol kinase only utilized UTP as an alternative cosubstrate. Finally, both dolichol kinase and farnesyl kinase appear to produce the respective isoprenoid monophosphates (Heller et al., 1992; Thai et al., 1999). Taken together, the mutant phenotypes, in vitro activities, and analogy to related enzymes strongly suggest that Arabidopsis VTE5 and Synechocystis vte5 encode phytol kinases.
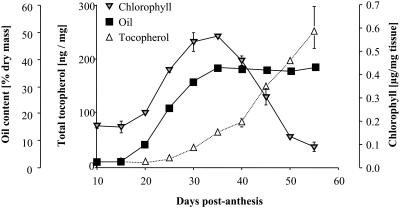
While the presence of an isoprenoid kinase for phytol activation was previously discussed (Soll et al., 1980; Hörtensteiner, 1999), it appears to be generally accepted that direct reduction of GGDP to PDP by GGR is the main pathway for the formation of the tocopherol precursor PDP (Soll et al., 1983; Keller et al., 1998; Grusak and DellaPenna, 1999; Bramley et al., 2000; Collakova and DellaPenna, 2003; Hofius and Sonnewald, 2003; Rippert et al., 2004). By contrast, our study strongly suggests that the majority of tocopherol that accumulates in Arabidopsis seed and leaf and in Synechocystis cells is synthesized via a phytol intermediate and that PMP is converted to PDP by a second, as yet unidentified, kinase. Recycling of phytyl chains during chlorophyll degradation is a logical source of substrate for phytol kinase as shown in Figure 1, and there are several lines of evidence in support of this hypothesis. For example, Rise et al. (1989) showed that accumulation of α-tocopherol was negatively correlated with chlorophyll content in several plant species during natural and artificial leaf senescence. We (Figure 7) and others (Goffman et al., 1999) have also observed an inverse correlation between seed chlorophyll and tocopherol levels during canola seed development, indicating that this phenomenon is not restricted to leaf. Analysis of ATGENEXPRESS 22K Affymetrix ATH1 transcript profiling data (http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm) using GENEVESTIGATOR software (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch/) revealed that VTE5 mRNA expression is relatively high in 6-week-old senescent Arabidopsis leaves compared with slightly younger vegetative plants (ATGENEXPRESS experiment 25). The gene is also highly expressed early in seed development (silique stages 3 to 5; ATGENEXPRESS experiments 76 to 78). Taken together, the combination of Arabidopsis VTE5 and Synechocystis vte5 mutant phenotypes, the results from expression of VTE5 proteins in E. coli, the plausible central role for phytol kinase in chlorophyll phytyl tail recycling, and correlations between conditions that enhance chlorophyll breakdown and increases in tocopherol levels and VTE5 transcript strongly suggest that phytol kinase plays an important role in tocopherol biosynthesis and phytol recycling. This role is also consistent with the observation of nearly unchanged chlorophyll levels in the vte5-1 mutant compared with the wild type, assuming chlorophyll formation from GGDP is not limiting and GGDP is the primary source for chlorophyll biosynthesis via geranylgeranyl chlorophyll as suggested previously (Rüdiger et al., 1980).
Figure 7.
Profile of Tocopherol, Chlorophyll, and Oil Accumulation in Developing Brassica napus Seed.
Seeds were harvested every 5 d from 10 to 55 d after pollination and were assayed for tocopherol (open triangles) and chlorophyll (shaded triangles) content on a fresh weight basis. Oil content (squares) measured by percentage of fatty acid methyl ester was measured on a dry weight basis. Each data point represents the average of two independent measurements, and error bars represent standard deviation.
The relative importance of synthesis of PDP from GGDP versus from free phytol remains an open question for several reasons. First, although the vte5-1 allele is predicted to encode a truncated peptide missing 9.7 out of 33.1 kD at the conserved C terminus, including phylogenetically conserved regions, it is unknown whether the nonsense fragment has residual enzymatic activity. A second complication is due to the possibility that other isoprenoid kinases, such as the paralog At5g58560, might partially substitute for VTE5 activity in the mutant. Substitution by farnesyl kinase is unlikely, as this enzyme was localized in the microsomal fraction (Thai et al., 1999), whereas VTE5 appears to be targeted to the plastids. Unfortunately, published data for tobacco (Nicotiana tabacum) antisense GGR lines (Tanaka et al., 1999) and a Synechocystis GGR mutant (Shpilyov et al., 2005) with reduced tocopherols do not provide clear evidence in favor of either model. This is because of the possible dual role for GGR in PDP formation via a gerenylgeranyl chlorophyll intermediate from chlorophyll recycling and in conversion of GGDP to PDP (Figure 1).
While PDP is commonly thought to be an intermediate in the biosynthesis of phytyl chain–containing molecules, such as tocopherols and phylloquinone, and potentially in chlorophyll synthesis and recycling, less attention has been paid to a role for PMP. Our data suggest that PMP is indeed an intermediate in tocopherol biosynthesis and that Arabidopsis and Synechocystis presumably contain an activity that converts PMP to PDP. An alternative hypothesis is that the Arabidopsis VTE5 and Synechocystis Vte5 proteins normally convert phytol directly to PDP, and the PMP detected in our experiments is an assay artifact. Indeed, LC-MS analysis of E. coli extracts revealed the unexpected presence of substantial amounts of GGMP in addition to GGDP (Figure 6), raising the possibility of an interfering isoprenoid diphosphate phosphatase in the E. coli extracts.
Three lines of evidence argue against the phosphatase hypothesis. First, incubation of exogenous radiolabeled GGDP with the washed E. coli membrane fractions used for the phytol kinase assay did not result in the detection of GGMP (data not shown), arguing against the presence of a contaminating isoprenoid diphosphate phosphatase activity in the VTE5 enzyme assay. Second, the isoprenoid kinases characterized to date, SEC59 and farnesyl kinase, produce isoprenoid monophosphates, as we observed for VTE5 (Heller et al., 1992; Thai et al., 1999). Third, E. coli (pET30b) produced low levels of PDP and PMP when incubated with free phytol, indicating that E. coli harbors an enzyme or enzymes that can produce these intermediates at low levels. While the PMP level in E. coli expressing Arabidopsis VTE5 protein increased by >70-fold when incubated with phytol, PDP levels were equivalent to the empty vector control. If PDP was the primary reaction product of Arabidopsis VTE5 and was subsequently broken down to PMP, the PDP:PMP ratio might be expected to be equivalent to the empty vector control. The results shown in Figure 6 indicate that this is not likely to be the case; the substantial PMP increase observed in Arabidopsis VTE5 expressing E. coli in the presence of free phytol was not accompanied by a comparable change in PDP. Taken together, these arguments are consistent with the hypothesis that the Arabidopsis VTE5 and slr1652 genes encode proteins that catalyze the conversion of phytol to PMP.
If PMP is a bona fide intermediate in tocopherol biosynthesis, both Arabidopsis and Synechocystis must contain PMP kinase activity. Indeed, an isoprenoid kinase that converts GGMP to GGDP has been previously reported in etioplast membranes (Rüdiger et al., 1980). The high degree of structural similarity between GGMP and PMP might allow for both of these substrates to be phosphorylated by this enzyme. One possibility is that the Arabidopsis VTE5 paralog At5g58560 might encode such an enzyme, though we found no clear homolog to slr1652 in the Synechocystis sp PCC 6803 genome. In fact there is precedence in the tocopherol biosynthetic pathway for related enzyme activities being encoded by genes with limited amino acid similarity in cyanobacteria and Arabidopsis: the 2-methyl-6-phytylbenzoquinol methyltransferases of Anabena and Synechocystis share limited sequence similarity to Arabidopsis VTE3 (Cheng et al., 2003; Van Eenennaam et al., 2003).
More distant VTE5 homologs are also identifiable in archaea as well as in fungi and animals, where all of these genes, including VTE5 and At5g58560, have been classified as members of COG0170 in the Clusters of Orthologous Genes classification system (Tatusov et al., 2001). COG0170 has been designated as dolichol kinase, based on alignment with the parent archetype, the SEC59 dolichol kinase of Saccharomyces cerevisiae (Heller et al., 1992). Based on the data presented in this article, we propose COG0170 to be renamed with the more inclusive name of “polyprenol kinase,” with members functioning as dolichol kinases, phytol kinases, and polyprenol kinases with as yet unidentified substrate specificities.
The results described above demonstrate an important role for phytol as a substrate in tocopherol biosynthesis. The most significant immediate impact of this study is in focusing attention on the importance of chlorophyll breakdown in producing tocopherol phytyl tails. However, our data leave open significant questions. First, given that Arabidopsis GGR enzyme is proposed to use both GGDP and geranylgeranyl chlorophyll a as substrate based upon assays using E. coli lysates (Keller et al., 1998), it is important to determine the substrate specificities using purified enzyme and to use genetic approaches to assess the relative in vivo contributions of GGDP and phytol in seed tocopherol accumulation. Although our studies focused on the impact of phytol kinase on seed tocopherol accumulation, this enzyme is likely to play important roles in photosynthetic tissues in tocopherol biosynthesis, recycling of chlorophyll, and synthesis of phylloquinone. Finally, it will be important to identify the kinase that is responsible for conversion of PMP to PDP. These studies reinforce the continued value of forward genetic screens in detailed characterization of biosynthetic pathways and the enzymes that regulate flux and accumulation of small molecules.
METHODS
Mutant Isolation and Gene Identification
Tocopherol mutants were screened as described by Van Eenennaam et al. (2003), and individual plant lines with decreased total tocopherol levels were selected as putative lt mutants. To both confirm the mutant phenotype and attempt to isolate true breeding lines, tocopherol levels were analyzed in M4 families.
Genetic Mapping and Gene Identification
A homozygous mutant line (Col-0 background) with an 80% reduction in tocopherol levels, referred to as lt1 was selected for map-based cloning. Genetic mapping and gene identification were done according to Jander et al. (2002). DNA extraction, genotyping, and sequencing of the 37-kb region containing vte5-1 were performed according to Van Eenennaam et al. (2003).
Bacterial Strains and Growth Conditions
Synechocystis sp PCC 6803 (ATCC 27184) was obtained from the American Type Culture Collection. Synechocystis cultures were grown as described (Savidge et al., 2002), and mutants were grown in medium supplemented with 5 μg/mL kanamycin.
Generation of Synechocystis sp PCC 6803 Mutant Strains
A BLAST search of Cyanobase (http://www.kazusa.or.jp/cyanobase/) using the deduced amino acid sequence of T32M21_90 against the Synechocystis sp PCC 6803 genomic sequence (Kaneko et al., 1996) identified slr1652, which we designated Synechocystis vte5 as the Synechocystis ortholog of Arabidopsis thaliana VTE5. To generate Synechocystis sp PCC 6803 vte5 deletion-insertion mutants (Synechocystis sp PCC 6803 Δslr1652), two separate ∼0.45-kb fragments corresponding to sequences surrounding the 5′- and 3′-regions of the Synechocystis vte5 coding sequence were amplified from Synechocystis genomic DNA by PCR using primer pairs 1652-e-1-f (5′-CCGAGCGGCCGCATTATCCCAAGATCACTGG-3′) plus 1652-i-2-r (5′-GCCGAGGATCCACCAAGCAATCAGCACC-3′), and 1652-i-3-f (5′-GCCGAGGATCCTGGTGGGACAAAGGTG-3′) plus 1652-e-4-r (5′-GCCGAGCTCGAGCCCAATTCCGGGTATTG-3′), respectively. The PCR products were cloned into pBluescript KSII+ (Stratagene) as NotI-BamHI and BamHI-XhoI fragments, resulting in the creation of a 236-bp deletion from Synechocystis vte5 starting 186 bp downstream of the ATG codon. Subsequently, the 1.25-kb kanamycin resistance cassette from pKISS (Amersham Pharmacia Biotech) was excised as an Ecl136II fragment and cloned into the T4-DNA polymerase–blunted BamHI site of the Synechocystis vte5 deletion plasmid. The resulting plasmid was designated pMON78621. It contained the kanamycin resistance gene in the same orientation as the internally truncated Synechocystis vte5 gene.
Knockout mutants were obtained as described by Williams (1988). Two kanamycin-resistant transconjugants (Synechocystis sp PCC 6803 Δslr1652-1 and Δslr1652-2) were selected for further experimentation. For confirmation that the deletion allele had cleanly replaced the wild-type Synechocystis vte5 gene in both transconjugants, genomic DNA from those mutants was subjected to PCR with the primer pair 1652-e-1-f and 1652-e-4-r.
Bioinformatic Analysis
Sequence homology searches were performed using the GenBank nonredundant database (www.ncbi.nlm.nih.gov). Assignment of T32M21_90 (Arabidopsis VTE5) and slr1658 (Synechocystis vte5) to existing protein families was performed using the PFAM database (http://pfam.wustl.edu). Multiple alignments of representative Arabidopsis VTE5 homolog sequences and dolichol kinase sequence from yeast and Arabidopsis was performed using ClustalX (Thompson et al., 1997) and visualized and edited using Genedoc (Nicholas et al., 1997). Sequences from the N-terminal region of yeast dolichol kinase and Arabidopsis not conserved in Arabidopsis VTE5 were deleted from multiple alignments. In addition, the first 10 to 70 N-terminal amino acids presumed to function as cellular localization signals based on ChloroP analysis and not well conserved among Arabidopsis VTE5 homologs were deleted from the multiple alignments. Phylogenetic analysis was performed using MEGA2 phylogenetic analysis software (Kumar et al., 2001). A neighbor-joining tree was constructed using a p-distance amino acid distance model and pairwise deletion. Bootstrap values calculated from 500 replications are shown. ChloroP 1.1 was used for predicting chloroplast target peptides (Emanuelsson et al., 1999). Transmembrane helices were predicted using TMHHM 2.0 software (Krogh et al., 2001).
Expression of Arabidopsis VTE5 and Synechocystis Vte5 Proteins in E. coli
The full-length coding sequence of Synechocystis vte5 (nucleotides 3,452,461 to 3,453,162, accession number gi|47118304) and the ChloroP predicted mature Arabidopsis VTE5 protein coding sequence (5′-ATGTCAGCAGTTGCGACGAATTC-3′ to 5′-ATTTAAGTTTCGGATATTAG-3′) were amplified by PCR and subcloned in the expression plasmid pET30b (Novagen), and the sequences were confirmed by DNA sequence analysis, yielding plasmids pET30b Synechocystis vte5 (pMON78631) and pET30b Arabidopsis VTE5 (pMON69926), respectively. These plasmids were transformed into Escherichia coli Tuner (DE3) cells (Novagen) and transformants selected on Luria-Bertani (LB) medium supplemented with 25 μg/mL kanamycin. For induction of gene expression, transformants selected from single colonies were grown in liquid LB supplemented with 25 μg/mL kanamycin to ∼0.8 A600, and 0.5 mM isopropylthio-β-galactoside (Gold Biotechnologies) was added, followed by 18 h of incubation at 30°C and 220 rpm shaking.
For in vivo phytol feeding experiments, 50 mL of induced cells were harvested by 10 min centrifugation at 7000g and 30°C. The cell pellet was resuspended in 5 mL fresh LB-kanamycin medium (22°C) containing 5 mM phytol (Sigma-Aldrich), with or without 0.2% toluene to facilitate phytol uptake, and the culture was transferred to 50-mL Erlenmeyer flasks. The cultures were shaken at 220 rpm at 30°C for 3 h. At the end of the incubation period, cells were harvested by 10-min centrifugation at 7000g at 4°C and washed once with ice-cold LB medium supplemented with 10 mM phosphatase inhibitor sodium fluoride. The washed cells were subsequently frozen at −80°C and lyophilized to dryness. Dried cell powders were used for extraction and analysis of phytol phosphates.
Isolation of E. coli Membrane Fractions
Purified membrane fractions from induced E. coli cells were isolated as described by Moshiri et al. (1991), with the following modifications: cell pellets from 150 mL induced E. coli Tuner (DE3) cultures were washed once with 20 mM Tris-HCl and 150 mM NaCl, pH 7.4, and resuspended in 6 mL of breaking buffer (20 mM Tris-HCl, containing 150 mM NaCl, 2 mM MgCl2, and 200 μM phenylmethane sulfonyl fluoride, pH 7.4) supplemented with 50 units of Benzonase protease inhibitor (Novagen). After ultracenrifugation, the membrane pellet was washed once with the same buffer lacking Benzonase. Washed membranes were resuspended in 3 mL of 20 mM Tris-HCl, pH 7.4, containing 10 mM MgCl2, 5 mM mercaptoethanol, and 200 μM phenylmethane sulfonyl fluoride and stored in aliquots at −80°C until use.
Tocopherol Analysis
Tocopherol analysis of Arabidopsis seed and Synechocystis cultures was performed as described by Savidge et al. (2002) and Van Eenennaam et al. (2003), respectively. For leaf tocopherol analysis, whole plants were collected and immediately frozen on dry ice. The leaf material was ground in medium Delrin tubes using a precooled three-quarter-inch steel ball (cooled in liquid nitrogen) by vigorously shaking for 60 s at 1000 strokes per minute at an amplitude of 3.81 cm, using a machine built in house. Approximately 25 to 40 mg of frozen ground leaf material was weighed into 1.4-mL Screen Mate tubes and proceeded as described (Savidge et al., 2002). Statistical data analysis was performed using JMP statistical discovery software (SAS Institute; www.jmp.com).
Determination of Free Phytol
Phytol levels in liquid extracts of various biological materials, such as plants, was performed by use of gas chromatography (GC) using an Agilent 6890 (Agilent Technologies) coupled with a Pegasus III (LECO) time-of-flight mass spectrometer. The quantification was accomplished using an external standard curve constructed using a unique mass that represents phytol and using the retention time of the phytol eluting from the gas chromatograph. The identity of the phytol was further confirmed by comparing the mass spectra at this retention time with the reference standard. The detection limit of the instrument was 0.05 ng × μL−1. The method detection limit depends on the extract solution and the amount of noise created by the extract sample itself. Thus, method detection limits were determined on a sample-by-sample basis. The analysis time was <4 min per sample.
The GC column was a DB5. The dimensions of the column were 10 m in length, with an internal diameter of 180 μm, and a film thickness of 0.18. The carrier gas was helium and flows through the column at a rate of 1.5 mL/min. A constant flow was maintained throughout the programmed temperature ramp of the chromatograph. The initial temperature of the column was 130°C with no hold time. The temperature was increased at a rate of 30°C per minute until the column reached 270°C and was held at 270°C for 2 min. One microliter of extract was injected into the injection port of the gas chromatograph. The mode of injection was splitless. The temperature of the injector was 250°C.
The outlet of the GC column was placed through a heated transfer line (250°C) into the time-of-flight mass spectrometer. The instrument was operated in electron impact mode with ionization energy of 70 eV. The source was operated at a temperature of 200°C, and the detector voltage was 1600 V. The mass-to-charge (m/z) range was set to be between 45 and 305.
The standard curve was constructed using commercial standards (Sigma-Aldrich). The standard curve was an external curve and ranged in concentration from 7 ng × μL−1 to 0.14 ng × μL−1. The r2 value for the curve was 0.9993. The m/z ratio used for quantification was 71. The retention time was 178 s for the cis-isomer and 183 s for the trans-isomer.
Extraction of GGDP/PDP for LC-MS Analysis
Isoprenoid phosphates were extracted by adding 1.5 mL of 80:20 (v:v) methanol:ethyl ether to each sample vial containing 12 to 15 mg of lyophilized E. coli material. A spatula was used to break up the cell pellet to aid dissolution. The samples were treated for 10 min at speed 7 on a plate vortexer (VX-2500 Multi-Tube Vortexer; VWR). Samples were then centrifuged for 4 min in an Eppendorf tabletop centrifuge 5804 R at 2500g, and the supernatants were removed for analysis.
Determination of GGDP, PDP, and Their Monophosphates Using LC-MS
The LC-MS system consisted of an HP1100 series HPLC (Agilent Technologies) connected to an API QSTAR Pulsar-i (Applied Biosystems) mass spectrometer using their TurboIonspray (i.e., electrospray) interface. The system was operated using the Analyst QS software (Applied Biosystems). The output of the LC column was connected to the TurboIonspray source utilizing a split of ∼1:5 (MS:waste) at a HPLC flow rate of 1 mL/min. The mass spectrometer was operated in negative ion TOF MS mode, scanning from 360 to 460 atomic mass units to detect the various masses for GGDP, PDP, GGMP, and PMP. A Zorbax Eclipse XDB C8 Rapid Resolution column (4.6 × 50 mm, 3.5 μM; Agilent Technologies) was used to separate the analytes using gradient conditions. The HPLC mobile phase components used were (1) water, (2) acetonitrile, (3) 0.5% formic acid, and (4) 2% ammonium hydroxide. The gradient conditions are detailed in Supplemental Table 2 online. The complex gradient employed the use of both acid and base modifiers to achieve good analyte retention in HPLC as well as maintain ionization efficiency for MS. An injection volume of 40 μL was used for all standards and samples. The total run time was 7 min.
The relative quantitation was based on the peak areas derived from extracted ion chromatograms centered around the appropriate quasimolecular ion for each analyte: GGDP = 449.05 ± 0.05; GGMP = 369.10 ± 0.05; PDP = 455.10 ± 0.05; and PMP = 375.15 ± 0.05. The approximate retention times of the analytes were as follows: GGDP, 2.6 min; GGMP, 2.8 min; PDP, 2.8 min; and PMP, 2.9 min. Although GGMP and PDP coeluted from the HPLC, they could be distinguished by mass.
GGDP standard was purchased from Sigma-Aldrich, and PDP was synthesized in house according to Joo et al. (1973). Based on LC-MS analysis, both commercial GGDP and in-house-synthesized PDP contained small amounts of the respective monophosphates. Representative chromatograms for GGDP, GGMP, PDP, and PMP standards and for extracts from E. coli (pET30b Arabidopsis VTE5) that had been incubated with free phytol for 3 h are shown in Supplemental Figure 2 online. Supplemental Figure 3 online represents an LC/MS/MS chromatogram of such an E. coli extract (A) and two representative product ion spectra observed during the MS/MS (product ion mode) scans, set to fragment the parent ions of 369.1 m/z (B) and 375.1 m/z (C) to verify the presence of GGMP and PMP, respectively.
Phytol Kinase Enzyme Assay
Membrane protein was quantified with Bradford reagent (Bio-Rad) using BSA as standard. The phytol kinase assays were performed according to a modified procedure from Thai et al. (1999). The enzymatic reaction mixture contained 25 mM Tris-HCl, pH.7.4, 0.1 to 0.5 mM CTP, 5 mM MgCl2, 0.05% CHAPS, 10 mM sodium orthovanadate, 0.5 μCi [3H]-phytol (20 Ci/mmol; Moravek Biochemicals), 5 μM nonradiolabeled phytol (Sigma-Aldrich), and 100 to 250 μg membrane protein in a total volume of 100 μL. Where indicated, CTP was replaced with ATP, GTP, UTP, or an NTP mixture. The reaction was incubated for 25 min at 37°C and terminated by the addition of 1.0 mL of chloroform:methanol (2:1, v/v). The samples were centrifuged for 5 min at 3000g, and the chloroform and methanol/aqueous fractions were taken to dryness under N2 gas. The enzyme assay products were resolved in 200 μL methanol and analyzed on an HP1100 series HPLC system consisting of an HP G1329A auto sampler, an HP G1311A quaternary pump, an HP G1315A diode array detector, an HP G1321A fluorescence detector (Agilent Technologies), and a Packard Radiomatic 500TR flow scintillation analyzer (Hewlett-Packard). Phytol kinase reaction products were separated by reverse-phase HPLC using a Vydac model 201TP54 C18 HPLC column (4.6 × 250 mm, 5 μm) coupled with an All-tech C18 guard column (P.J. Cobert Associates) and solvent system A (25 mM NaHCO3 in water) and B (100% acetonitrile). The solvent gradient raised buffer B from 30 to 100% in the time period 0 to 20 min, held buffer B constant at 100% from 20 to 39 min, and then reduced buffer B from 100 to 30% from 39 to 40 min. Compounds of interest were detected by a diode array detector set at 210, 254, and 365 nm and by the flow scintillation analyzer set at 156 keV of upper level discriminator to detect 3H compounds. The sample injection volume was 20 μL, and the flow rate was set at 1.0 mL/min. The retention times of phytyl diphosphate, phytyl monophosphate, and phytol are 10.2, 11.5, and 26.4 min, respectively, under these HPLC conditions.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers provided in Figure 5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Tocopherol Content of Transgenic Arabidopsis Seed from the R3 Generation.
Supplemental Table 2. LC-MS Gradient for the Separation of Isoprenoidphosphates.
Supplemental Figure 1. Multiple Alignments of All VTE5 Homologs and Dolichol Kinases Used to Generate the Tree Shown in Figure 5.
Supplemental Figure 2. Total and Extracted Ion Chromatograms from LC-MS Analysis of Standard Mix and E. coli Cell Extract.
Supplemental Figure 3. LC/MS/MS Verification of Isoprenoidmonophosphate Formation in E. coli.
Supplementary Material
Acknowledgments
We thank Rod Croteau, Dean DellaPenna, and Yossi Hirschberg for comments on the manuscript. We wish to thank our colleagues in biochemistry, plant transformation, and the plant growth facilities for their expert assistance and support. We also wish to thank our colleagues at Renessen for their support and advice.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Henry E. Valentin (henry.e.valentin@monsanto.com).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037077.
References
- Ben-Shem, A., Frolow, F., and Nelson, N. (2003). Crystal structure of plant photosytem I. Nature 426 630–635. [DOI] [PubMed] [Google Scholar]
- Bramley, P.M., Elmadfa, I., Kafatos, A., Kelly, F.J., Manios, Y., Roxborough, H.E., Schuch, W., Sheehy, P.J.A., and Wagner, K.-H. (2000). Vitamin E. J. Sci. Food Agric. 80 913–938. [Google Scholar]
- Cheng, Z., Sattler, S., Maeda, H., Sakuragi, Y., Bryant, D.A., and DellaPenna, D. (2003). Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15 2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova, E., and DellaPenna, D. (2001). Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127 1113–1124. [PMC free article] [PubMed] [Google Scholar]
- Collakova, E., and DellaPenna, D. (2003). The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol. 133 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunniff, P.A., ed (1990). Nomenclature rules for vitamin E (972.31). In Official Methods of Analysis, 15th ed. (Arlington, VA: AOSC International), pp. 1070–1071.
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman, F.D., Velasco, L., and Becker, H.C. (1999). Tocopherols accumulation in developing seeds and pods of rapeseed (Brassica napus). Lipid-Fett 101 400–403. [Google Scholar]
- Grusak, M.A., and DellaPenna, D. (1999). Improving the nutrient composition of plants to enhance human nutrition and health. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 133–161. [DOI] [PubMed] [Google Scholar]
- Heller, L., Orlean, P., and Adair, W.L., Jr. (1992). Saccharomyces cerevisiae sec59 cells are deficient in dolichol kinase activity. Proc. Natl. Acad. Sci. USA 89 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D., and Sonnewald, U. (2003). Vitamin E biosynthesis: Biochemistry meets cell biology. Trends Plant Sci. 1 6–8. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner, S. (1999). Chlorophyll breakdown in higher plants and algae. Cell. Mol. Life Sci. 56 330–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUPAC-IUP Joint Commission on Biochemical Nomenclature (1982). Nomenclature of tocopherols and related compounds. Recommendation 1981. Eur. J. Biochem. 123 473–475. [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, C.N., Park, C.E., Kramer, J.K.G., and Kates, M. (1973). Synthesis and acid hydrolysis of monophosphate and pyrophosphate esters of phytanol and phytol. Can. J. Biochem. 51 1527–1536. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3 109–136. [DOI] [PubMed] [Google Scholar]
- Kanwischer, M., Porfirova, S., Bergmüller, E., and Dörmann, P. (2005). Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol. 137 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, P., Bouvier, F., D'Harlingue, A., and Camara, B. (1998). Metabolic compartmentation of plastid prenyllipid biosynthesis. Evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251 413–417. [DOI] [PubMed] [Google Scholar]
- Kräutler, B. (2002). Unravelling chlorophyll catabolism in higher plants. Biochem. Soc. Trans. 30 625–630. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17 1244–1245. [DOI] [PubMed] [Google Scholar]
- Matile, P., Hörtensteiner, S., and Thomas, H. (1999). Chlorophyll degradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 67–95. [DOI] [PubMed] [Google Scholar]
- Moshiri, F., Chawla, A., and Maier, R.J. (1991). Cloning, characterization, and expression in Escherichia coli of the genes encoding the cytochrome d oxidase complex from Azotobacter vinelandii. J. Bacteriol. 173 6230–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K.B., Nicholas, H.B., Jr., and Deerfield II, D.W. (1997). GeneDoc: Analysis and visualization of genetic variation. EMBnet.news 4 1–4. [Google Scholar]
- Norris, S.R., Lincoln, K., Abad, M.S., Eilers, R., Hartsuyker, K., Kindle, K., Hirshberg, J., Karunanandaa, B., Moshiri, F., Stein, J.C., Valentin, H.E., and Venkatesh, T.V. (2004). Plant genes for sequence homologs of phytol kinase of tocopherol biosynthesis and their use in engineering plant tocopherol profiles and drought resistance. International patent application WO 2004013312 A2.
- Norris, S.R., Shen, X., and DellaPenna, D. (1998). Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 117 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.W., Hugly, S., and Harrison, D. (1993). Sterols and phytyl esters of Arabidopsis thaliana under normal and chilling temperatures. Phytochemistry 33 1381–1383. [Google Scholar]
- Peisker, C., Düggelin, T., Rentsch, D., and Matile, P. (1989). Phytol and the breakdown of chlorophyll in senescent leaves. J. Plant Physiol. 135 428–432. [Google Scholar]
- Porfirova, S., Bergmüller, E., Tropf, S., Lemke, R., and Dörmann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99 12495–12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert, P., Scimemi, C., Dubald, M., and Matringe, M. (2004). Engineering plant shikimate pathway for production of tocotrienol and improving herbicide resistance. Plant Physiol. 134 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rise, M., Cojocaru, M., Gottlieb, H.E., and Goldschmidt, E.E. (1989). Accumulation of α-tocopherol in senescing organs as related to chlorophyll degradation. Plant Physiol. 89 1028–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, M. (2003). Mevalonate-independent methylerythritol phosphate pathway for isoprenoid biosynthesis. Elucidation and distribution. Pure Appl. Chem. 75 375–388. [Google Scholar]
- Rontani, J.-F., Cuny, P., and Grossi, V. (1996). Photodegradation of chlorophyll phytol chain in senescent leaves of higher plants. Phytochemistry 42 347–351. [Google Scholar]
- Rüdiger, W., Benz, J., and Guthoff, C. (1980). Detection and partial characterization of activity of chlorophyll synthetase in etioplast membranes. Eur. J. Biochem. 109 193–200. [DOI] [PubMed] [Google Scholar]
- Sattler, S.E., Cheng, Z., and DellaPenna, D. (2004. b). From Arabidopsis to agriculture: Engineering improved vitamin E content in soybean. Trends Plant Sci. 9 365–367. [DOI] [PubMed] [Google Scholar]
- Sattler, S.E., Gilliland, L.U., Magallanes-Lundback, M., Pollard, M., and DellaPenna, D. (2004. a). Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge, B., Weiss, J.D., Wong, Y.-H.H., Lassner, M.W., Mitsky, T.A., Shewmaker, C.K., Post-Beittenmiller, D., and Valentin, H.E. (2002). Isolation and characterization of tocopherol phytyltransferase genes from Synechocystis PCC 6803 and Arabidopsis. Plant Physiol. 129 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, M., Tsuyama, M., Takami, T., Shimizu, H., and Kobayashi, Y. (2004). Accumulation of menaquinones with incompletely reduced side chains and loss of α-tocopherol in rice mutants with alternations in the chlorophyll moiety. J. Exp. Bot. 55 1989–1996. [DOI] [PubMed] [Google Scholar]
- Shintani, D., and DellaPenna, D. (1998). Elevating the vitamin E content of plants through metabolic engineering. Science 282 2098–2100. [DOI] [PubMed] [Google Scholar]
- Shpilyov, A.V., Zinchenko, V.V., Shestakov, S.V., Grimm, B., and Lokstein, H. (2005). Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1706 195–203. [DOI] [PubMed] [Google Scholar]
- Soll, J., Kemmerling, M., and Schultz, G. (1980). Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch. Biochem. Biophys. 204 544–550. [DOI] [PubMed] [Google Scholar]
- Soll, J., and Schultz, G. (1980). 2-Methyl-6-phytylquinol and 2,3-dimethyl-5-phytylquinol as precursors of tocopherol synthesis in spinach chloroplasts. Phytochemistry 19 215–218. [Google Scholar]
- Soll, J., Schultz, G., Joyard, J., Douce, R., and Block, M.A. (1985). Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch. Biochem. Biophys. 238 290–299. [DOI] [PubMed] [Google Scholar]
- Soll, J., Schultz, G., Rüdiger, W., and Benz, J. (1983). Hydrogenation of geranylgeraniol. Plant Physiol. 71 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, R., Oster, U., Kruse, E., Rüdiger, W., and Grimm, B. (1999). Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. (2001). The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 29 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai, L., Rush, J.S., Maul, J.E., Devarenne, T., Rodgers, D.L., Chappell, J., and Waechter, C.J. (1999). Farnesol is utilized for isoprenoid biosynthesis in plant cells via farnesyl pyrophosphate formed by successive monophosphorylation reactions. Proc. Natl. Acad. Sci. USA 96 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eenennaam, A.L., et al. (2003). Engineering vitamin E content: From Arabidopsis mutant to soy oil. Plant Cell 15 3007–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman, J.K., and Maxwell, J.R. (1986). Acyclic isoprenoids as biological markers. In Biological Markers in the Sedimentary Record, R.B. Johns, ed (Amsterdam, The Netherlands: Elsevier), pp. 1–46.
- Williams, J.G.K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. In Methods in Enzymology, A. Glazer, ed (New York: Academic Press), 766–778.
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.