Abstract
Plant resistance to disease is controlled by the combination of defense response pathways that are activated depending on the nature of the pathogen. We identified the Arabidopsis thaliana BOTRYTIS-INDUCED KINASE1 (BIK1) gene that is transcriptionally regulated by Botrytis cinerea infection. Inactivation of BIK1 causes severe susceptibility to necrotrophic fungal pathogens but enhances resistance to a virulent strain of the bacterial pathogen Pseudomonas syringae pv tomato. The response to an avirulent bacterial strain is unchanged, limiting the role of BIK1 to basal defense rather than race-specific resistance. The jasmonate- and ethylene-regulated defense response, generally associated with resistance to necrotrophic fungi, is attenuated in the bik1 mutant based on the expression of the plant defensin PDF1.2 gene. bik1 mutants show altered root growth, producing more and longer root hairs, demonstrating that BIK1 is also required for normal plant growth and development. Whereas the pathogen responses of bik1 are mostly dependent on salicylic acid (SA) levels, the nondefense responses are independent of SA. BIK1 is membrane-localized, suggesting possible involvement in early stages of the recognition or transduction of pathogen response. Our data suggest that BIK1 modulates the signaling of cellular factors required for defense responses to pathogen infection and normal root hair growth, linking defense response regulation with that of growth and development.
INTRODUCTION
Plant resistance to potential pathogens involves an elaborate system of signal perception and transduction to activate cellular defense. At least some host responses appear to be mediated by receptor molecules either associated with or embedded in the plant cell membrane, with the receptor domains on either the inner or outer surface of the membrane. These receptors may interact with various pathogen factors. These could include so-called pathogen-associated molecular patterns that are invariant between and are shared by many related pathogens (e.g., flg22, a conserved peptide domain in a bacterial flagella protein) and race-specific elicitors that are widespread among biotrophic plant pathogens (Nurnberger et al., 2004). After recognition, various host molecules, including protein kinases and transcription factors, relay the signal to activate downstream defense responses, including the synthesis of antimicrobial compounds. The speed and intensity of the activation of such responses are crucial for plant resistance to microbial infection. The signal recognition and early signal relay mechanisms appear to be distinct depending on the nature of the pathogen, whereas downstream effector molecules show a large degree of overlap between different pathogens.
Plant resistance to pathogens is controlled by the combination of pathogen defense response pathways that are triggered depending on the nature of the pathogen and its modes of pathogenesis. Gene-for-gene resistance is based on the direct or indirect recognition of a race-specific pathogen elicitor molecule by the cognate plant resistance gene (Flor, 1971). These interactions frequently culminate in the activation of the hypersensitive response, which correlates with resistance to biotrophic fungal, nematode, viral, and bacterial pathogens. Basal defense pathways, which are activated by pathogen-associated molecular patterns (also called general elicitors), generally result in resistance without the hypersensitive response and appear to be effective against a broad range of pathogens (Asai et al., 2002; Nurnberger et al., 2004). General induced and systemic defense responses, such as systemic acquired resistance (SAR), jasmonate/ethylene-mediated resistance, and induced systemic resistance, have also been described (Nimchuk et al., 2003; Glazebrook, 2005). SAR is a form of resistance induced systemically in the plant after a localized pathogen challenge that provides protection against secondary infection by a broad spectrum of primarily biotrophic pathogens (Cao et al., 1997; Reuber et al., 1998; Dewdney et al., 2000). SAR can be activated by chemical agents or pathogens that cause cell death and correlates with the expression of a subset of pathogenesis-related proteins, some with antimicrobial activity. In dicotyledonous plants, salicylic acid (SA) is necessary and sufficient for SAR induction (Vernooij et al., 1994).
Necrotrophic pathogens are a destructive group of plant pathogens that have pathogenesis strategies distinct from biotrophic pathogens. Whereas necrotrophic pathogens induce cell death in their hosts by secreting toxic substances into host tissue before and during colonization, biotrophic pathogens require living cells to complete their life cycle. The necrotroph Botrytis cinerea produces cell wall–degrading enzymes (Prins et al., 2000), toxic levels of reactive oxygen intermediates (Edlich et al., 1989; Deighton et al., 1999; Muckenschnabel et al., 2002), and toxins (Tiedemann, 1997; Colmenares et al., 2002) that result in the death and maceration of tissue, leading to plant decay. Evidence to date strongly suggests a limited role for SAR and gene-for-gene (race-specific) resistance against necrotrophic pathogens in Arabidopsis thaliana. Mutants that are impaired in signaling required for the activation of SAR or disrupted in the biosynthesis of SA retain wild-type levels of resistance to Botrytis (Ferrari et al., 2003). Chemical or biological activation of SAR failed to contain Botrytis growth in Arabidopsis (Govrin and Levine, 2002). In addition, neither a race-specific fungal elicitor nor an R gene required for gene-for-gene resistance has been identified in any Botrytis–plant interactions. It also appears that resistance to Botrytis requires the concerted action of many genes, as opposed to the more usual single gene–mediated recognition-dependent resistance to biotrophic pathogens.
Induced general responses regulated by the plant hormones ethylene (ET) and jasmonate (JA) are required for defense responses to necrotrophic pathogens in some plant species (Thomma et al., 1999; Diaz et al., 2002; Ferrari et al., 2003). Arabidopsis and tomato (Lycopersicon esculentum) mutants altered in ET and JA signaling or biosynthesis show increased susceptibility to Botrytis and other necrotrophic pathogens (Thomma et al., 1999; Diaz et al., 2002). The expression of the plant defensin gene PDF1.2 is activated in response to Botrytis and Alternaria brassicicola in an ET- and JA-dependent manner and correlates with resistance (Penninckx et al., 1998). However, susceptible responses to Botrytis and A. brassicicola have also been observed during normal activation of PDF1.2, suggesting that PDF1.2 may not be sufficient to provide full plant protection (Ferrari et al., 2003; Mengiste et al., 2003). Here, we describe the identification and characterization of the BOTRYTIS-INDUCED KINASE1 (BIK1) gene of Arabidopsis as a critical component of the Arabidopsis pathogen response pathway required for full resistance to at least two necrotrophic pathogens, Botrytis and A. brassicicola. Although BIK1 is required for resistance to necrotrophic pathogens, our data show that it is a negative regulator of basal defense responses to virulent strains of the bacterial pathogen Pseudomonas syringae pv tomato (Pst) but has no role in race-specific and nonhost resistance.
RESULTS
Identification of BIK1 and Characterization of Its Role in Disease Resistance
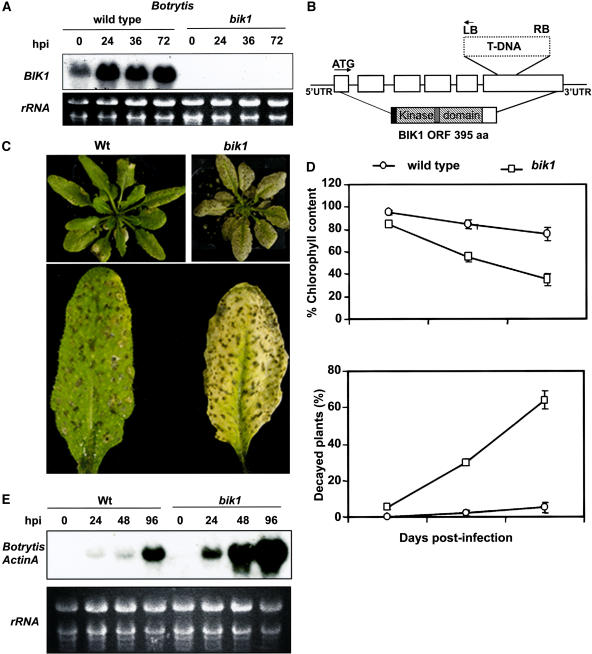
To identify genes that contribute to Botrytis resistance, we compared transcript profiles of Botrytis-infected and healthy Arabidopsis leaves using an Arabidopsis microarray. We selected genes encoding potential components of signal transduction pathways that were significantly induced during Botrytis infection and focused on a putative Ser/Thr kinase (At2g39660) that we named BIK1. In the microarray experiments, the transcript of the BIK1 gene increased an average of 6.5-fold in Botrytis-inoculated leaves compared with mock-inoculated controls (data not shown). This induction of BIK1 expression after Botrytis inoculation was confirmed using RNA gel blots (Figure 1A). Healthy wild-type plants express low basal levels of BIK1 RNA, but the transcript accumulates to high levels within 24 h of inoculation with Botrytis (Figure 1A).
Figure 1.
Botrytis-Induced Expression of BIK1, Structure of the bik1 Mutant Allele, and Responses to Botrytis.
(A) RNA blot showing the induction of the BIK1 gene in wild-type plants in response to Botrytis and the lack of the BIK1 transcript in the bik1 mutant. hpi, hours after inoculation.
(B) Genomic organization of the bik1 mutant allele and the cDNA encoded by the BIK1 open reading frame (ORF). The BIK1 open reading frame is shown with the N-terminal myristoylation motif (black), the protein kinase domain (striped), and the Ser/Thr kinase active site signature (gray). aa, amino acids; LB, left border; RB, right border; UTR, untranslated region.
(C) Wild-type (left) and bik1 (right) plants 5 d after inoculation with Botrytis. Four-week-old soil-grown plants were inoculated by spraying a fungal spore suspension (2 × 105 spores/mL).
(D) Susceptibility of bik1 plants as measured by the loss of chlorophyll and plant decay. Top, chlorophyll content in inoculated plants is expressed as a percentage of the chlorophyll content of the corresponding mock-inoculated plants. Chlorophyll was extracted from entire rosette leaves of six mock-inoculated and six Botrytis-inoculated plants for each genotype. The data are means ± se from two independent experiments. Bottom, percentage of decayed wild-type and mutant plants caused by Botrytis infection as a function of time after inoculation. Plants were scored as dead when their centers were completely rotten. Data represent means ± se of three different experiments performed with 36 plants per genotype.
(E) Accumulation of the BOTRYTIS ACTIN A mRNA in inoculated plants. In (A) and (E), 20 μg of total RNA was loaded per lane.
To determine the function of BIK1 in defense and nondefense response pathways, we searched for a loss-of-function mutant. A mutant allele of the BIK1 gene containing a T-DNA insertion in the BIK1 coding region was identified from the SALK T-DNA insertion collection (Alonso et al., 2003). The T-DNA inserted into the last exon of the BIK1 gene and resulted in the loss of detectable BIK1 transcript, indicating that bik1 is a loss-of-function mutant (Figures 1A and 1B). To determine the role of BIK1 in resistance to Botrytis, homozygous bik1 plants were inoculated by spraying a suspension containing 2 × 105 Botrytis spores per milliliter, as described previously (Mengiste et al., 2003; Veronese et al., 2004). In wild-type (ecotype Columbia) plants, Botrytis infection caused restricted disease symptoms, with no significant damage to the plant (Figure 1C). Necrotic spots at the sites of primary infection were clearly visible, but the necrosis remained restricted and did not spread. On lower leaves, this infection occasionally led to the collapse of an entire leaf, although the infection rarely became systemic. In bik1 plants, disease symptoms increased rapidly, showing chlorosis, necrosis, and complete plant decay compared with wild-type plants (Figures 1C and 1D). At 8 d after infection, Botrytis infection of wild-type plants caused a loss of chlorophyll ranging from 20 to 30% compared with the corresponding mock-inoculated control plants (Figure 1D, top). At the corresponding time point, bik1 plants had lost 60 to 70% of their chlorophyll. In wild-type plants, Botrytis infection rarely moved to tissues that were not present at the time of infection, and <5% of the plants were killed. In the mutant, infection became systemic and resulted in the complete decay of 60 to 70% of the inoculated plants over a period of 8 to 12 d after infection, suggesting a severely impaired resistance mechanism (Figure 1D, bottom). These data suggest a clear role for BIK1 in resistance to Botrytis infection.
The growth of the pathogen in inoculated plants was measured to determine whether the enhanced tissue death displayed by the bik1 mutant was attributable to increased pathogen growth or to increased symptom development in response to normal pathogen growth. As a measure of fungal growth in planta, blots of RNA extracted from inoculated plants were hybridized with a probe made from the constitutively expressed BOTRYTIS ACTIN A gene as described (Benito et al., 1998). Fungal RNA accumulated in both wild-type and mutant plants as infection advanced and correlated well with the disease symptoms, including loss of chlorophyll (Figures 1C and 1E). In the bik1 mutant, however, Botrytis RNA accumulated to significantly higher levels at 2 to 4 d after infection. At later stages, Botrytis proliferated further, with the fungal hyphae becoming clearly visible on macerated leaf surfaces of bik1, whereas hyphae were not visible on the surfaces of wild-type leaves. Similar results were obtained by staining infected leaves with trypan blue to visualize the growth of the fungus in planta (data not shown). These data indicate that bik1 is unable to restrict fungal growth, and in this respect it is phenotypically similar to the Botrytis-susceptible1 (bos1) mutant (Mengiste et al., 2003).
The loss of BIK1 function resulted in enhanced susceptibility to a second necrotrophic pathogen, A. brassicicola, the causal agent of early blight in cruciferous plants. As with Botrytis, the bik1 mutation resulted in increased disease symptoms and pathogen growth in plants inoculated with A. brassicicola (Figures 2A and 2B). In wild-type plants, disease symptoms were limited to a lesion at the point of inoculation. In bik1, infection resulted in slightly larger lesions surrounded by extensive chlorosis. In addition, the fungus proliferated significantly more in mutant plants, with the production of spores on inoculated bik1 leaves approximately fivefold greater than on wild-type leaves (Figure 2B). The loss of both Botrytis and A. brassicicola resistance in bik1 plants suggests a broad role for BIK1 in resistance to microbial necrotrophy.
Figure 2.
bik1 Plants Show Increased Susceptibility to the Necrotrophic Pathogen A. brassicicola.
(A) Leaves of drop-inoculated wild-type (top row) and bik1 (bottom row) plants at 6 d after infection.
(B) Mean number of spores in A. brassicicola–inoculated plants at 6 d after infection. Data represent means ± se from three independent experiments. Each experiment contained the average spore counts from 20 inoculated leaves per genotype. Col-0, ecotype Columbia (wild type).
Plants were challenged with strains of the bacterial pathogen Pst to determine whether the resistance function of BIK1 extends to bacterial pathogens. Interestingly, bik1 plants showed increased resistance to the normally virulent strain PstDC3000. No visible disease symptoms developed on inoculated bik1 leaves, and bacterial growth was reduced significantly compared with that in wild-type leaves (Figure 3A). Resistance to an avirulent bacterial strain, PstDC3000(avrRpm1), was unchanged relative to that in wild-type plants, suggesting a role for BIK1 in basal defense response rather than in R gene–mediated resistance (Figure 3B). The bik1 plants were not affected in their resistance to the nonhost pathogen Pseudomonas syringae pv phaseolicola (data not shown). These results suggest that BIK1 acts as a negative regulator of basal resistance to virulent pathogens but has no role in resistance to the nonhost and avirulent pathogens tested.
Figure 3.
Responses of bik1 to Pst Inoculation.
Representative leaves showing disease symptoms (left panels) and bacterial growth (right panels) after infiltration with PstDC3000 (A) or PstDC3000 (avrRpm1) (B). Bacterial growth (colony forming units [CFU]/cm2 leaf area) at 0, 2, and 4 d after infection is plotted for wild-type and bik1 plants. Plants were infiltrated with a bacterial suspension (OD600 = 0.001). Photographs of diseased leaves are from 3 d after infection. Data represent average values ± se from three experiments.
The BIK1 cDNA Rescues the Disease Responses of bik1 to Wild-Type Levels
The observations that BIK1 expression is upregulated during pathogen infection and that bik1 plants exhibit altered responses to Botrytis, A. brassicicola, and Pst infection support the hypothesis that BIK1 functions in a pathogen response pathway. To confirm that the observed phenotypes conferred by bik1 are specifically attributable to the mutation in BIK1, we performed complementation experiments. Expression of the BIK1 cDNA under the control of the cauliflower mosaic virus 35S (CaMV35S) promoter in bik1 resulted in transgenic plants with wild-type resistance levels to Botrytis, A. brassicicola, and Pst infection (Figures 4A and 4B). These data show that the mutation in BIK1 is the sole cause of the bik1 disease responses and confirm the identity of the BIK1 gene.
Figure 4.
The BIK1 cDNA Rescues the bik1 Disease Phenotypes.
The BIK1 cDNA expressed in bik1 plants under the control of the CaMV35S promoter rescued susceptibility to Botrytis and A. brassicicola (A) and resistance to PstDC3000 (B) to wild-type levels. The bottom panel in (B) shows bacterial growth. All assays were repeated three times on five independent lines. Data points for bacterial growth in (B) represent averages ± se from three experiments. Photographs were taken at 5 d after inoculation with Botrytis or A. brassicicola and at 3 d after inoculation with PstDC3000. bik1+BIK1, bik1 plants expressing the wild-type BIK1 cDNA under the control of the CaMV35S promoter.
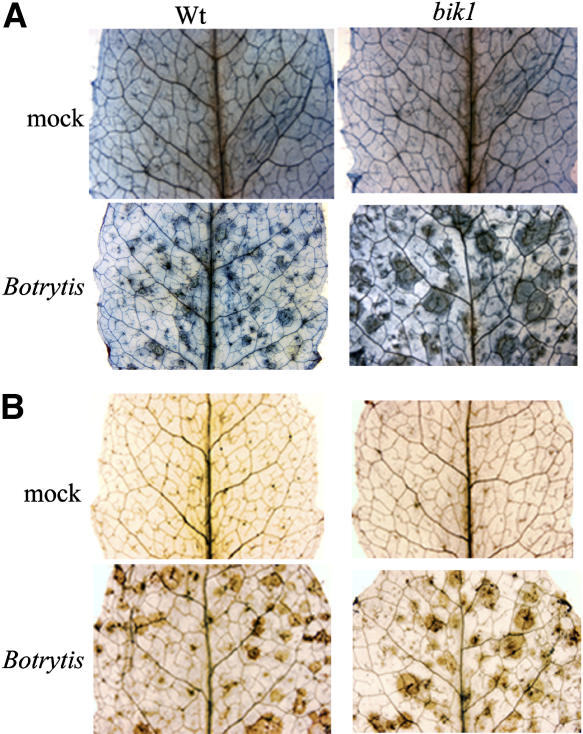
bik1 Is Not a Lesion-Mimic Mutant
The presence of dead cells and necrotic tissues is thought to promote Botrytis infection by providing an initial saprophytic growth base from which the fungus further invades healthy tissue. Dead cells could provide nutrients for the growth of Botrytis, which is unable to obtain nutrients from living cells. However, no cell death lesions were observed before infection in bik1 plants (Figure 5A), suggesting that the susceptibility of bik1 to Botrytis is not attributable to the presence of spontaneous cell death lesions before inoculation. After Botrytis infection, patches of dead cells were visible in both wild-type and mutant plants. This is consistent with previously described Arabidopsis mutants that have increased Botrytis susceptibility in the absence of a lesion-mimic phenotype (Thomma et al., 1999; Mengiste et al., 2003; Veronese et al., 2004).
Figure 5.
Cell Death and Production of H2O2 in Botrytis-Inoculated Plants.
(A) Trypan blue staining of leaves from mock-inoculated (top row) or Botrytis-inoculated (bottom row) plants at 3 d after infection showing cell death triggered by pathogen infection.
(B) Production of H2O2 in mock-inoculated (top row) or Botrytis-inoculated (bottom row) plants at 2 d after infection. Leaves were stained with DAB as described in Methods. The brown precipitate shows DAB polymerization at the site of H2O2 production.
In both (A) and (B), representative leaves detached from inoculated plants are shown. The experiments were repeated three times with similar results.
Accumulation and Sensitivity to Reactive Oxygen Intermediates in bik1
Reactive oxygen intermediates (ROIs) have been implicated in both susceptible and resistance responses, depending on the nature of the pathogen (Levine et al., 1994; Lamb and Dixon, 1997; Govrin and Levine, 2000). ROIs are required for cell death and resistance to biotrophic pathogens (Torres et al., 2002). In the case of necrotrophic pathogens, cell death exacerbates susceptibility (Govrin and Levine, 2000). We determined the levels of H2O2 by 3,3′-diaminobenzidine (DAB) staining (Thordal-Christensen et al., 1997) 2 d after inoculation with Botrytis. Inoculated plants generated brown precipitate indicative of H2O2 production that was absent in noninoculated plants (Figure 5B). There was a small but consistent increase in the intensity and size of the stained area in infected bik1 plants. This may come from the increased Botrytis growth in the mutant, resulting in increased H2O2 levels.
The previously described bos1 mutant showed increased susceptibility to Botrytis and sensitivity to oxidative stress (Mengiste et al., 2003; Veronese et al., 2004). In the case of bik1, sensitivity to oxidative stress generated by paraquat (methyl viologen) was comparable to that in wild-type plants, suggesting that susceptibility to Botrytis is not linked to altered plant sensitivity to oxidative stress (data not shown).
The bik1 Mutation Affects SA Accumulation but Not Camalexin
To determine whether the altered disease responses of bik1 are associated with altered levels of SA, we determined free SA levels in Botrytis-inoculated and noninoculated plants (see Methods). In noninoculated bik1 plants, basal SA levels were more than twofold greater than those in the corresponding wild-type plants. In both cases, SA levels increased after Botrytis infection, with the level of SA in the mutant being higher than that in the corresponding wild-type plants at 2 d after infection (Figure 6A). These data implicate BIK1 as a negative regulator of SA accumulation and hence also of the downstream responses to SA. To test whether BIK1 also regulates phytoalexin accumulation, we determined the levels of the phytoalexin camalexin after Botrytis infection. Despite the high degree of susceptibility to Botrytis, bik1 mutant plants accumulated normal levels of camalexin during Botrytis infection (data not shown), indicating that BIK1 is not involved in regulating camalexin levels during Botrytis infection.
Figure 6.
bik1 Plants Show Increased SA Accumulation and Altered Defense Gene Expression.
(A) SA levels before and after Botrytis infection in wild-type and bik1 plants. FW, fresh weight.
(B) Expression of the PDF1.2 gene in response to Botrytis, paraquat, MeJA, and ACC.
(C) Expression of PR-1 in response to Botrytis infection and SA treatment.
SA levels shown in (A) represent means ± se from three independent experiments. In (B) and (C), the experiments were repeated three times with similar results. Each lane contained 10 μg of total RNA.
The bik1 Mutation Affects the Expression of Defense-Related Genes
To relate the disease susceptibility of bik1 to defects in defense response pathways, we studied the expression of molecular markers of JA/ET- and SAR-mediated disease response pathways. The expression of the plant defensin gene PDF1.2 correlates with the activation of the ET/JA defense response pathway and is implicated in resistance to necrotrophic pathogens (Penninckx et al., 1996, 1998). The bik1 mutation significantly reduced the expression of PDF1.2 in response to Botrytis infection, paraquat, and methyl jasmonate (MeJA) relative to wild-type plants (Figure 6B). This finding implicates BIK1 as a positive regulator of a defense response pathway that controls PDF1.2 expression. The transcript of the SAR marker gene PR-1 increased through the first 48 h after Botrytis inoculation in wild-type plants. In bik1, this expression was significantly higher at 24 and 48 h after inoculation than in wild-type plants (Figure 6C). This may mean that bik1 plants are more sensitized and respond faster in terms of PR-1 expression, although the increased PR-1 levels may be an indirect consequence of the increased rate of fungal growth in bik1 plants. By contrast, the SA-induced expression of PR-1 was comparable in wild-type and bik1 plants, suggesting that BIK1 does not mediate SA signaling (Figure 6C). Limited PR-1 expression under uninduced conditions was observed when >10 μg of RNA was loaded on an RNA gel (data not shown). In summary, the induction of some defense responses and the inhibition of others in bik1 suggest that BIK1 is a regulatory factor that modulates defense responses.
BIK1 Encodes a Ser/Thr Protein Kinase
The BIK1 genomic region is composed of six exons and five introns (Figure 1B). The cDNA sequence of BIK1 contains an open reading frame encoding a protein of 395 amino acid residues with an estimated molecular mass of 44.09 kD and a pI of 9.36 (see Supplemental Figure 1 online). The BIK1 open reading frame contains a consensus plant N-myristoylation motif, Met-Gly-XXX-Ser/Thr (Arg, where X is any amino acid [MGXXXS/T(R)] (Thompson and Okuyama, 2000), suggesting membrane localization. Protein N-myristoylation refers to the covalent attachment of myristic acid by an amide bond to the N-terminal Gly residue of a nascent polypeptide. Myristoylation is involved in directing and anchoring proteins to membranes. Membrane-anchored proteins can have various functions, including cellular regulation, signal transduction, and translocation (Podell and Gribskov, 2004). The central part of the BIK1 protein (residues 67 to 355) contains a kinase catalytic domain and harbors all 11 conserved subdomains of protein kinases (Hanks and Quinn, 1991; Hardie, 1999). Two consensus sequences, the Lys (K) residue in the DIKASN motif and the G-T/S-XX-Y/F-X-APE motif, identify BIK1 as a potential Ser/Thr kinase rather than a Tyr kinase (Stone and Walker, 1995). The protein kinase active site signature (YRDIKASNILL) and the conserved amino acid triplets D219FG (subdomain VII) and APE248 (subdomain VIII) that define the borders of the activation domain segment of BIK1 are consistent with other protein kinases (Johnson et al., 1996) (see Supplemental Figure 1 online). Activation of protein kinases occurs by regulatory phosphorylation in this region (Morgan and De Bondt, 1994). There are six putative phosphorylatable residues within the activation segment of the BIK1 kinase (residues 219 to 248): Ser (S), Thr (T), or Tyr (Y).
Sequence comparisons and database searches revealed that BIK1 shares sequence similarity with a large number of putative receptor-like cytoplasmic kinases in subfamily VII of the receptor-like cytoplasmic kinase family of proteins (RLCK VII). These proteins have a common monophyletic origin with receptor-like kinases but have no apparent transmembrane domain (Shiu and Bleecker, 2001a, 2001b). The function of a large number of these protein kinases is unknown, including 46 members of the Arabidopsis RLCK VII subfamily. BIK1 is most similar to two putative proteins (At3g55450 and At5g02290) with 77 and 63% identity, respectively, over a stretch of >233 amino acids covering the kinase domain. In addition, two closely related proteins with Ser/Thr kinase activities, APK1a and APK1b (Hirayama and Oka, 1992), are similar to BIK1. The M locus protein kinase (MLPK) involved in Brassica self-incompatibility is closely related to BIK1, and APK1a and APK1b are the Arabidopsis homologs of MLPK (Murase et al., 2004). The Arabidopsis disease resistance protein kinase PBS1 (Swiderski and Innes, 2001) is 51% identical to BIK1 in the kinase domain. These sequence similarities are further confirmed by the phylogenetic analysis of the relationship between BIK1 and other related plant protein kinases (see Supplemental Figures 2 and 3 online). BIK1 is closely related to Arabidopsis kinases, including APK proteins (Hirayama and Oka, 1992), Brassica rapa MLPK, and rice (Oryza sativa; XP_4943889 and NP_910058) and Solanum demissum (AAT40481) putative kinases (see Supplemental Figures 2 and 3 online). A comprehensive classification of Arabidopsis protein kinases, including BIK1 and related subfamilies, is available at http://plantsp.genomics.purdue.edu/plantsp/family/class.html.
BIK1 Is Induced by Paraquat but Not by SA, JA, or the ET Precursor 1-Aminocyclopropane-1-Carboxylate
BIK1 is expressed at low but detectable levels in leaf tissues. Expression of the gene was not induced in response to the plant defense–mediating hormones SA, JA, or 1-aminocyclopropane-1-carboxylate (ACC), the natural precursor of ET (Figure 7A). However, paraquat (methyl viologen), an herbicide that generates ROIs, strongly induced BIK1 gene expression. In addition, inoculation with both virulent and avirulent strains of P. syringae induced BIK1 transcript accumulation (Figure 7B). The ein2, coi1, nahG, and pad2 genotypes of Arabidopsis are impaired in ET, JA signaling, SA, and camalexin accumulation, respectively, and show altered defense responses to Botrytis and/or other pathogens (Thomma et al., 1999). The BIK1 transcript was induced at levels comparable to wild-type levels in these genotypes, suggesting that BIK1 may be upstream of these genes or may function independently (see Supplemental Figure 4 online).
Figure 7.
BIK1 Is a Functional Protein Kinase That Localizes to the Plasma Membrane.
(A) Accumulation of the BIK1 transcript in response to Botrytis, SA, MeJA, and ACC.
(B) Accumulation of the BIK1 transcript in response to P. syringae and paraquat.
(C) Subcellular localization of the BIK1-GFP fusion protein in onion epidermal cells. Control GFP only (top) and BIK1-GFP (bottom) constructs were transiently expressed after particle bombardment. The micrographs were taken 24 h after bombardment of the cells. The experiment was repeated three times with similar results.
(D) GST-BIK1 phosphorylates the mylein basic protein (MBP) in vitro. Left, Coomassie blue staining; right, autoradiogram of the gel shown at left.
(E) Expression of selected transcription factor and MAPK genes in response to Botrytis in wild-type and bik1 plants. The RNA gel blots were repeated in three independent infection assays with similar results. Each lane contained 20 μg of total RNA.
The sensitivity of root and/or hypocotyl elongation of bik1 to the plant hormones JA, ACC, and auxins (2,4-D and indole-3-acetic acid [IAA]) was comparable to that of wild-type plants, suggesting that the phenotype conferred by bik1 is not attributable to altered responses to any of these phytohormones (data not shown).
BIK1 Is a Functional Protein Kinase That Is Localized to the Plasma Membrane
To better understand the mechanisms of BIK1 function, we determined the subcellular localization of the BIK1 protein. Full-length BIK1 was translationally fused with green fluorescent protein (GFP), and the chimeric protein was expressed under the control of the CaMV35S promoter. Because N-myristoylated proteins are often translocated to the membrane, we expected the BIK1 protein to be localized to the membrane. Onion (Allium cepa) epidermal cells transiently expressing the BIK1-GFP fusion showed the GFP signal predominantly at the plasma membrane, whereas cells expressing GFP alone (control) exhibited signal around the nucleus and in the cytosol (Figure 7C). To confirm that BIK1 is a functional protein kinase, we performed a phosphorylation assay using BIK1 produced in Escherichia coli as a glutathione S-transferase (GST) fusion protein. BIK1 exhibited autophosphorylation activity and also phosphorylated myelin basic protein as an artificial substrate (Figure 7D).
The structure of the BIK1 kinase suggests possible involvement in the early steps of recognition or signal transduction after contact with the pathogen. We hypothesized that BIK1 may function as an upstream component of the plant mitogen-activated protein kinase (MAPK) pathway. This hypothesis is consistent with the activation of the Arabidopsis MAPK pathway by bacteria, fungi, and ROIs, suggesting that signaling events initiated by diverse pathogens and external signals converge into a conserved MAPK cascade (Nühse et al., 2000; Asai et al., 2002). In addition, MPK4 and MPK6 were implicated as negative regulators of defense responses based on mutant phenotypes (Petersen et al., 2000; Menke et al., 2004). Recently, the OXIDATIVE SIGNAL-INDUCIBLE1 (OXI1) Ser/Thr protein kinase was described to function in the MAPK pathway upstream of MPK3 and MPK6 (Rentel et al., 2004). Loss of OXI1 confers opposite disease resistance and root hair growth phenotypes (see below) to those of BIK1. To investigate the involvement of BIK1 in the early steps of pathogen recognition upstream of the plant MAPK pathways, we tested for MPK3, MPK6, MPK4, and OXI1 kinase activities using immunocomplex kinase assays. Although Botrytis infection induced the kinase activities of MPK3 and MPK6, no consistent differences between bik1 and wild-type plants were observed (see Supplemental Figure 5 online). The bik1 seedlings grown in tissue culture did not have significant constitutive activities of MPK3, MPK6, MPK4, and OXI1 relative to wild-type plants (data not shown). In addition, no direct physical interaction was observed between BIK1 and these MAPKs or OXI1 proteins in yeast two-hybrid assays (data not shown). There was also no difference detected in transcript amounts of MPK4 and MPK6 during Botrytis infection, regardless of the presence or absence of a functional BIK1 (Figure 7E). These findings suggest that BIK1 functions independently of these components of the MAPK cascades and OXI1 kinase.
In addition, we examined the Botrytis-induced expression of potential target genes encoding transcription factors downstream of the MAPKs (Figure 7E). Recently, the transcription factors WRKY22 and WRKY29 were placed downstream of FLS2 and MPK3/MPK6 in a basal defense response pathway triggered by flagellin and also implicated in Botrytis resistance (Asai et al., 2002). WRKY6 regulates defense- and senescence-related genes in Arabidopsis (Robatzek and Somssich, 2002). The WRKY6 and WRKY29 genes showed increased transcripts in response to Botrytis infection in both wild-type and bik1 plants (Figure 7E). WRKY6 is strongly induced at 1 d after infection, whereas WRKY29 transcript increased at later stages of infection in the wild type, with an early induction in the mutant. Similar results were obtained with the BOS1 gene encoding an R2R3MYB transcription factor that is required to restrict Botrytis growth in inoculated plants (Mengiste et al., 2003). These data suggest that BIK1 may not be a direct upstream component of these transcription factors, either as a negative or a positive regulator.
BIK1 Is Required for Normal Plant Growth and Development
bik1 plants show defects in some aspects of plant growth and development. Mutant plants grown in tissue culture without disease or stress conditions produce shorter primary roots and longer and significantly more root hairs and lateral roots than wild-type plants (Figure 8A). Mutant plants also have leaves with serrated margins and wrinkled surfaces that display occasional curling (Figure 8B). Mature bik1 plants flower an average of 5 to 7 d earlier than wild-type plants and have a weaker stem strength, leading to reduced standing ability and lodging (Figure 8C, left). In addition, bik1 plants have reduced fertility and smaller siliques (Figure 8C, right). These data suggest that wild-type BIK1 is required for normal plant growth and development.
Figure 8.
bik1 Plants Show Altered Growth Traits.
(A) Root growth phenotypes of wild-type and bik1 plants. bik1 plantlets growing on Murashige and Skoog (MS) medium show decreased primary root elongation and increased root hair growth compared with wild-type plants. At right are close-ups of the roots to show the root hairs.
(B) Soil-grown plants revealing the wrinkled leaf surfaces in bik1 (left), and close-ups of the leaves that emerged at different stages showing serrated leaf margins (right).
(C) Wild-type and bik1 plants grown without stress conditions showing increased lodging in bik1 plants (left), and close-ups of the inflorescences with smaller siliques (right).
To confirm that the altered growth traits of bik1 are also attributable to the mutation in the BIK1 gene, bik1 plants expressing the BIK1 cDNA were analyzed. The altered growth characteristics of bik1, including the length of the primary root and the number and length of the root hairs, were fully restored to wild-type levels, confirming that BIK1 is required for normal plant growth and development and that the bik1 mutation resulted in the altered growth traits (see Supplemental Figure 6 online).
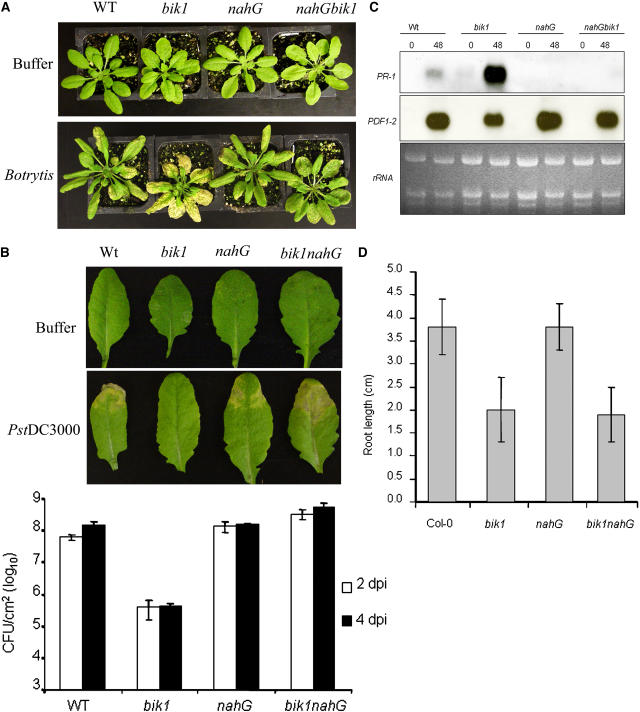
BIK1 Functions through SA-Dependent and SA-Independent Pathways
To investigate the role of SA in bik1 disease responses and growth-related phenotypes, we crossed bik1 to plants expressing the nahG gene encoding a bacterial salicylate hydroxylase that degrades SA to catechol (Delaney et al., 1994). F2 segregating plants were analyzed by PCR for the presence of the bik1 mutation and the nahG gene. Homozygous bik1 plants containing the nahG gene were identified, and their disease responses were studied (Figure 9). The bik1 nahG plants were comparable to wild-type and nahG plants in their resistance to Botrytis (Figure 9A). In the case of PstDC3000, bik1 nahG plants showed increased disease symptoms and supported significantly more bacterial growth than nahG or wild-type plants (Figure 9B) (Duncan's test; P = 0.05). These data suggest that the disease response of bik1 is SA-dependent and that BIK1 may act through the SA pathway. The increased susceptibility of bik1 nahG plants to PstDC3000 relative to nahG plants suggests that BIK1 may act through another pathway in addition to the SA pathway. The differences in bacterial growth between wild-type and nahG plants are consistent with previous reports (Delaney et al., 1994; Fouts et al., 2003; DebRoy et al., 2004). The patterns of SA levels in bik1 are consistent with the pattern of expression of PR-1 and the disease responses of bik1 nahG plants. As expected, expression of the PR-1 gene was eliminated in nahG and bik1 nahG plants, whereas PDF1.2 expression increased over that in bik1 plants (Figure 9C). This finding is consistent with the suppression of JA/ET-regulated genes by increased SA levels and the antagonistic interactions between the JA/ET and SA response pathways (Kunkel and Brooks, 2002; Spoel et al., 2003). By contrast, the growth phenotypes of bik1 were not affected by nahG. The primary root length and root hair phenotypes in bik1 nahG plants were comparable to those of wild-type bik1 plants, suggesting that the function of BIK1 in root growth is independent of SA (Figure 9D). Among 190 F2 seedlings of the cross between bik1 and nahG evaluated for root length and root hair patterns, 54 were similar to bik1 and 136 showed wild-type root growth patterns. This segregation is consistent with no effect from the nahG gene and a 3:1 segregation ratio for a single recessive mutation rather than 15:1 if nahG had an effect (χ2 = 1.011; P ≤ 0.01). The mean primary root length of bik1 plants was approximately twofold shorter than that of wild-type plants after 10 d on MS plates. nahG plants did not differ from wild-type plants in root growth characteristics (Figure 9D). The early flowering and reduced fertility of bik1 were also not restored to wild-type levels by nahG.
Figure 9.
The Role of SA in Disease and Growth Traits of bik1.
Comparisons of wild-type, bik1, nahG, and bik1 nahG are shown for the following features.
(A) Responses to mock (top row) or Botrytis (bottom row) inoculation.
(B) Responses to buffer infiltration (top row) and PstDC3000 (bottom row). The bar graph shows bacterial growth at 2 and 4 d after infiltration with PstDC3000 (OD600 = 0.001). Duncan's multiple range test for counts was performed to separate mean values to determine the significance of the differences. The mean values of bacterial counts in nahG and bik1 nahG were significantly different from each other at P = 0.05. Data on bacterial counts represent averages ± se from four experiments.
(C) Expression of PR-1 and PDF1.2 genes after Botrytis inoculation. Each lane contained 6 μg of total RNA.
(D) Primary root length. Root length was measured at 10 d of growth on MS plant growth medium. Each data point represents the average from measurements of 12 plants ± se. The experiments were repeated at least three times.
DISCUSSION
In this study, we identified BIK1 as a crucial component of host response signaling required to activate the resistance responses to Botrytis and A. brassicicola infection in Arabidopsis. BIK1 encodes a regulatory protein, specifically a protein kinase, predicted to be specific to Ser/Thr residues, that is similar to receptor-like cytoplasmic protein kinases and may act early in the disease response pathway. Additionally, the bik1 mutant phenotypes suggest that the BIK1 kinase function cannot be fully complemented by other endogenous kinases. The BIK1 gene transcript is strongly upregulated in response to challenge with Botrytis and a ROI-generating compound. The bik1 mutant is susceptible to Botrytis and A. brassicicola and shows attenuated expression of the plant defensin PDF1.2 gene, but it is competent for the activation of SAR. Race-specific resistance to Pst is not altered, based on plant resistance to a bacterial strain with the Avr protein. However, bik1 plants exhibit increased resistance to a virulent bacterial pathogen that is based on SA-dependent and SA-independent pathways and independent of cell death lesions. Interestingly, bik1 plants accumulate increased levels of SA before and after Botrytis infection, suggesting that BIK1 acts upstream of SA accumulation. These data implicate BIK1 as a negative regulator of SA accumulation and basal defense against virulent bacterial pathogens, because the bik1-null mutant supported less bacterial growth and a total absence of disease symptoms.
The function of BIK1 as a disease resistance factor may be to regulate normal levels of SA required for resistance to necrotrophs. Upon infection, BIK1 may be activated to trigger the Botrytis and A. brassicicola resistance response, including the regulation of optimal levels of SA synthesis. In the absence of BIK1, this regulation is relieved and SA level increases, which may lead to suppression of the mechanisms required for necrotrophic resistance, consistent with previous reports (Penninckx et al., 1996; Clarke et al., 1998; Gupta et al., 2000; Kunkel and Brooks, 2002; Spoel et al., 2003). Accordingly, bik1 contains increased SA levels, reduced PDF1.2 expression, and distinct disease resistance responses to biotrophic and necrotrophic pathogens. These distinct responses are further substantiated by data from bik1 nahG plants that show wild-type levels of resistance to Botrytis infection and expression of PDF1.2. The ET/JA functions independent of defense responses are normal, as bik1 plants exhibited wild-type sensitivity to plant hormones and lacked hormone-related phenotypes. The loss of resistance to necrotrophic pathogens but enhanced resistance to virulent pathogens suggests that BIK1 modulates the crosstalk between different defense signaling pathways.
The role of SA in regulating responses to Botrytis and other necrotrophs appears to be complex and may be multidimensional, with both positive and negative regulatory roles. Increased SA associated with lesion-mimic mutants correlated with susceptibility to Botrytis, and this may be an indirect effect rather than a defect in a bona fide resistance mechanism. In addition, SA synthesized via Phe ammonia lyase, and not via isochorismate synthase, was required for local resistance in Arabidopsis, whereas defects in SA signaling had no effect (Ferrari et al., 2003). Increased SA also suppresses the expression of JA-responsive genes (Spoel et al., 2003) normally required for full resistance to necrotrophic pathogens (Thomma et al., 1998, 1999). The function of BIK1 as a positive regulator of resistance to Botrytis and a negative regulator of resistance to Pst is dependent on normal levels of SA. When SA increases above a certain threshold level, it may trigger the suppression of mechanisms required for resistance to Botrytis and A. brassicicola while still promoting mechanisms for resistance to Pst. Alternatively, BIK1 may suppress SA accumulation and basal resistance to Pst, and when removed, it may lead to the activation of defense responses that some necrotrophs (Botrytis and A. brassicicola) exploit for their benefit. Consistent with this notion, the hypersensitive cell death, a particularly strong defense response, promotes susceptibility to Botrytis (Govrin and Levine, 2000). Our data and emerging data suggest no effect or limited effects linked to SA deficiency, whereas increased SA (and PR-1 expression) is positively correlated with susceptibility to Botrytis in Arabidopsis.
Localization of BIK1 to the plasma membrane suggests that BIK1 may act as an early component of the plant defense response, either directly in pathogen recognition or early in the signaling cascade. A protein with a similar structure to BIK1, the OXI1 protein kinase, is activated in response to active oxygen species and functions through the positive regulation of MPK3 and MPK6 (Rentel et al., 2004). The loss-of-function oxi1 mutant has reduced root hair growth and increased susceptibility to virulent strains of the oomycete pathogen Peronospora parasitica, which was attributed to altered ROI signaling (Rentel et al., 2004). The root hair phenotypes and disease responses of bik1 and oxi1 mutants suggest that the two proteins act antagonistically or that BIK1 functions upstream of OXI1, MPK3, and MPK6 as a negative regulator. In response to Botrytis infection, the kinase activity of MPK3 and MPK6 increased, but no consistent differences were observed between bik1 and wild-type plants. Similarly, tissue culture–grown 14-d-old seedlings of bik1 showed no significant difference from wild-type plants for MPK3, MPK4, MPK6, and OXI1 kinase activities. This, coupled with the lack of evidence for direct physical interaction between BIK1, MAPKs, or OXI1, implicates BIK1 as a negative regulator of root hair growth and basal resistance to pathogens independent of the MAPK pathway. We cannot exclude the possibility that BIK1 functions through other components of the MAPK pathway and that MAPK activity may be transient and tissue-specific. However, further and detailed examination of the relationship between BIK1 and this group of proteins and other MAPKs is warranted.
BIK1 may be activated by pathogen-derived signals or signals resulting from pathogen–plant interactions regardless of the strain. The increased transcript accumulation of BIK1 in response to Botrytis, Pst strains, and paraquat is consistent with this hypothesis. The kinase activity of BIK1 may be required to transduce pathogen-derived signals to downstream molecules that are targeted for negative or positive regulation depending on the pathogen. The reduced activation of the ROI-responsive PDF1.2 in response to paraquat suggests an altered ROI signaling in bik1 plants. Previously, the attenuated expression of PDF1.2 in response to paraquat was linked to altered ROI signaling and susceptibility to necrotrophic pathogens in Arabidopsis (Tierens et al., 2002). Paraquat treatment leads to the production of H2O2, O2−, and OH− in plant cells (Suntres, 2002). ROI production and signaling are required for plant defense responses and normal root hair growth in Arabidopsis (Levine et al., 1994; Lamb and Dixon, 1997; Foreman et al., 2003; Rentel et al., 2004). Reduced ROI levels resulted in reduced cell death and resistance to biotrophic pathogens (Torres et al., 2002). In the case of Botrytis, host susceptibility positively correlated with the levels of O2− or H2O2 produced (Govrin and Levine, 2000). Consistent with the disease responses of bik1, H2O2 levels in Botrytis-infected leaf tissue was slightly higher than that in wild-type plants, based on histochemical assays. The sensitivity to ROI-generating compounds was not different between bik1 and wild-type plants. This finding excludes the possibility that susceptibility to Botrytis is the result of plant sensitivity to ROIs.
Among defense responses associated with resistance to necrotrophs in Arabidopsis, in bik1, camalexin accumulation was not affected, whereas PR-1 was expressed more strongly and the induced expression of PDF1.2 was reduced. PR-1 expression positively correlates with resistance to biotrophic pathogens but negatively correlates with resistance to Botrytis in some mutants, including bos3 and ssi2, and provides a clear distinction between host responses to the two classes of pathogens (Kachroo et al., 2001; Veronese et al., 2004). In the bos mutants described previously, expression of PR-1 and camalexin accumulation varied, with no consistent correlations to enhanced Botrytis susceptibility (Mengiste et al., 2003; Veronese et al., 2004). This lack of consistent correlation in different mutants may indicate that the roles of the pathways represented by the defense markers are not clear and may vary in the different mutants, because there may be cross-regulation of the different pathways. Similarly, the bos mutants varied in terms of their disease responses to biotrophic pathogens, with no consistent antagonistic or coordinated genetic control of resistance (Mengiste et al., 2003; Veronese et al., 2004).
The Arabidopsis genome encodes many putative Ser/Thr kinases whose functions are not yet determined. The R genes PBS1 and PTO from Arabidopsis and tomato, respectively, encode Ser/Thr protein kinases that have similar structures to that of BIK1 and show significant sequence similarity. In contrast with BIK1, however, PBS1 and PTO are involved in race-specific resistance to Pst (Martin et al., 1993; Swiderski and Innes, 2001). It is possible that the BIK1 protein may be a cellular target for an effector protein(s) and may interact with and be guarded by an unknown R protein, consistent with the guard hypothesis (Dangl and Jones, 2001). In that case, inactivation of BIK1 could result in upregulated basal resistance similar to the responses of the rin4 mutant (Mackey et al., 2003; Belkhadir et al., 2004). In sum, the bik1 mutation represents a defect in a novel gene affecting disease responses and plant growth. Our data suggest that bik1 is affected in signaling required for the activation of cellular mechanisms involved in plant development and the pathogen response, and they provide a novel avenue for further dissection of the components of disease response pathways.
METHODS
Plant Cultivation
Procedures for growing plants in tissue culture and in soil have been described (Mengiste et al., 2003). Arabidopsis thaliana plants were grown in soil under fluorescent light (150 μE·m−2·s−1) at 22 ± 4°C with 60% RH and a 12-h-light/12-h-dark cycle. For axenic growth, seeds were sterilized and sown on a medium solidified with 1% agar that contained MS salts (PhytoTechnology Laboratories) and 2% (w/v) sucrose. Conditions for axenic growth were 12 h of light of 60 μE·m−2·s−1.
Fungal Culture and Disease Assays
The Botrytis cinerea strain BO5-10 used in all of our experiments was cultured on 2× V8 agar (36% V8 juice, 0.2% CaCO3, and 2% Bacto-agar) and incubated at 20 to 25°C. Collection of conidia and plant inoculation were as described (Mengiste et al., 2003). Conidia were resuspended in 1% Sabouraud maltose broth buffer (Difco) before inoculation. Alternaria brassicicola strain MUCL20297 was grown on potato dextrose agar, and spores were suspended in distilled water for plant inoculation. The procedures for inoculation and determination of susceptibility have been described (Mengiste et al., 2003; Veronese et al., 2004). All of the Botrytis disease assays described here were performed on whole plants by spraying a spore suspension (2 × 105 spores/mL) in Sabouraud maltose broth buffer. The disease assay for A. brassicicola was performed on detached leaves. A single 5-μL spore suspension (5 × 105 spores/mL) in water was deposited on each detached leaf. Inoculated plants were kept under a transparent cover to maintain high humidity. A. brassicicola spores in inoculated plants were counted as described with minor modifications (van Wees et al., 2003).
Bacterial Disease Assay
Leaves of 4-week-old plants were infiltrated with bacterial suspensions (OD600 = 0.001 in 10 mM MgCl2) as described (Mengiste et al., 2003). The bacterial strain Pseudomonas syringae pv tomato DC3000, strain DC3000 harboring a plasmid carrying the avrRpm1 gene, and the bean-infecting strain Pseudomonas syringae pv phaseolicola strain 1448A race 6 used in our assays were from J. Dangl (University of North Carolina, Chapel Hill). To determine bacterial growth, leaf discs from infected leaves were collected at 0, 2, and 4 d after inoculation. Each experiment for bacterial growth assay was performed in three replicates. At each time point, 10 leaves were collected from each wild-type plant and other genotypes tested for each replicate. Leaf discs of the same size were made using a hole puncher, and bacterial titer per leaf area was determined. Analysis of variance and Duncan's multiple range test for counts were performed on the bacterial count data in Figure 9 using the SAS program.
SA Analysis
Free SA levels in leaves were determined by grinding tissue (1 g) to a fine powder under liquid nitrogen and extracting with 100% methanol (2.5 mL) for 20 min with shaking. Extracts were then passed through a 0.45-μm filter (Nylon Acrodisc 13; Pall Corporation) and analyzed with an HPLC apparatus equipped with an analytical C-18 column (Econosphere, 150 mm × 4.6 mm; Alltech) and a fluorescence detector (Shimadzu Scientific Instruments). The HPLC program consisted of a linear gradient from 25% acetonitrile and 75% 0.1 M sodium phosphate buffer, pH 3.3 (v/v), to 75% acetonitrile and 25% 0.1 M sodium phosphate buffer, pH 3.3 (v/v), over the course of 14 min, with the detector set at 305-nm excitation and 407-nm emission. SA levels were quantified by comparing peak areas of samples with a standard curve generated from a SA standard (Sigma-Aldrich).
Chlorophyll Extraction and Assays
Chlorophyll was extracted essentially as described (Veronese et al., 2003). Briefly, shoot tissues were ground into powder in liquid nitrogen and weighed. Chlorophyll was extracted in 80% acetone, and the concentration per gram fresh weight was calculated as described by Lichtenthaler (1987). Chlorophyll was extracted from pools of six uninoculated and six inoculated plants for each genotype.
Induction Treatments
Induction treatments were performed by spraying 4-week-old soil-grown plants with 2 × 105 Botrytis spores/mL, 5 mM SA (Aldrich), 100 μM MeJA (Aldrich), 0.5 mM ACC (Sigma-Aldrich), or 100 μM paraquat (methyl viologen; Sigma-Aldrich). The chemical compounds were dissolved in 0.1% ethanol. The controls were sprayed with 0.1% ethanol.
Sensitivity to Plant Hormones
Surface-sterilized seeds were sown on 1.0% (w/v) agar medium containing MS salts, 2% (w/v) sucrose, and different concentrations of MeJA, ACC, 2,4-D, or IAA, pH 5.7. The plant hormones were added to the autoclaved medium from filter-sterilized stock solutions. Plates were incubated in a vertical position. The effects of MeJA and ACC on primary root growth and/or hypocotyl elongation were determined essentially as described (Ton et al., 2002). The primary root length was measured after 8 d of growth in culture. In each case, 25 randomly selected seedlings were measured. The sensitivity of root elongation to auxin (2,4-D or IAA) was done in a similar manner. IAA was tested at 0.01, 0.1, 1, 10, and 100 μM; 2,4-D was tested at 100, 200, 300, 400, and 500 nM; ACC was tested at 1, 3, and 6 μM; and MeJA was tested at 0.2, 0.4, 0.6, 0.8, and 1 μM. The experiments were repeated at least twice on different seed lots.
RNA Blots
RNA was extracted from harvested leaf tissue frozen in liquid nitrogen using phenol/chloroform followed by lithium chloride precipitation (Lagrimini et al., 1987). RNA was separated on formaldehyde agarose gels. The gels were then blotted onto Hybond N+ nylon membranes (Amersham Pharmacia Biotech). Probes were labeled with 32P by random priming using a commercial kit (Sigma-Aldrich). Hybridization of probe and subsequent washings were performed as described (Church and Gilbert, 1984).
Identification of bik1 nahG Plants
The bik1 nahG plants were identified using PCR. Plants carrying the nahG gene were identified using primers specific to nahG (5-ATGAAAAACAATAAACTTGGCTTG-3′ and 5′-CTGACCTTCCAGCACATGACTAC-3′). The primers from the BIK1 gene (5′-CAGGTCACTTGAATGCAAGAAGCG-3′ and 5′-GGGTATGGGACATGTAACCGGAAA-3′) and primers from the T-DNA insertion in bik1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) (Alonso et al., 2003) were used to identify plants carrying the bik1 mutation. The procedure for the extraction of plant genomic DNA for PCR was described previously (Veronese et al., 2004).
Histochemical Assays
The presence of H2O2 in leaf samples was determined using the DAB staining method (Thordal-Christensen et al., 1997). Leaves were detached and placed in 1 mg/mL DAB-HCl, pH 3.8 (D-8001; Sigma-Aldrich). In the presence of endogenous peroxidase activity, DAB generates a reddish-brown DAB polymer that can be detected at the site of H2O2 formation. After staining, leaves were cleared in 96% boiling ethanol and observed with a microscope. Trypan blue staining of leaves was performed as described (Koch and Slusarenko, 1990).
Subcellular Localization
For the analysis of the subcellular localization of BIK1 using the GFP reporter gene, the BIK1 coding sequence was cloned into a C-terminal GFP fusion vector kindly provided by Ron Coolbaugh (Purdue University). The vector was constructed by assembling the CaMV35S promoter from pBI121 (Clontech), the multicloning site from the vector pSP72 (Promega), and the GFP from the vector pUhGFPC3-N (Carlsberg Laboratory). BIK1 coding sequence was amplified using the BD Advantage-HF 2 PCR kit (BD Biosciences, Clontech) using the primers 5′-CGGGATCCCGATGGGTTCTTGCAG-3′ and 5′-GCCTAGGACACAAGGTGCCTGCCA-3′. The correct amplification was verified by sequencing.
In Vitro Kinases and Immunocomplex Kinase Assays
The open reading frame of BIK1 was cloned into GST fusion protein expression vector pGEX4T-1 (Pharmacia). Expression of the GST fusion protein and affinity purification were performed as described previously (Cardinale et al., 2002). The protein concentrations of the recombinant proteins were determined with the Bio-Rad detection system using BSA as a standard. Kinase reactions were performed in 15 μL of kinase buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 5 mM EGTA, and 1 mM DTT) containing 5 μg of GST fusion protein, 5 μg of myelin basic protein, 0.1 mM ATP, and 2 μCi of [γ-32P]ATP. The protein kinase reactions were performed at room temperature for 30 min, and the reactions were stopped by adding 4× SDS loading buffer. The phosphorylation of myelin basic protein was analyzed by autoradiography after separation by 12.5% SDS-PAGE. Immunocomplex kinases assays were performed as described (Cardinale et al., 2002).
Complementation Experiments
The full-length BIK1 cDNA was obtained from the ABRC (clone F17A14). The cDNA was reamplified using the BD Advantage-HF 2 PCR kit (BD Biosciences, Clontech), sequenced, and inserted after the CaMV35S promoter into a modified version of the binary vector pCAMBIA 1200 kindly provided by Huazhong Shi (Texas Tech University). Hygromycin-resistant homozygous T2 plants were identified from progeny of primary transformants and selfed to produce the T3 generation of bik1 plants expressing the BIK1 cDNA. Five independent transformants were examined for the various phenotypes of bik1, as described in Results.
Phylogenetic Analysis
For construction of the BIK1 phylogenetic tree (see Supplemental Figure 2 online), the full-length amino acid sequences were aligned using ClustalW (Thompson et al., 1994) (see Supplemental Figure 3 online) with default gap penalties, and the alignment was manually adjusted where necessary. Mean character distances were used to construct an unrooted neighbor-joining phylogeny (Saitou and Nei, 1987) from the PAUP version 4.0 package (Swofford, 2000). Statistical support of the branches was tested with 1000 bootstrap resamples.
Accession Numbers
Sequence data for the genes described in this study can be found in the GenBank/EMBL data libraries under the following accession numbers: BIK1 (At2g39660), NAK (At5g02290), PBS1 (AT5g13160), APK1B (At2g28930), APK1A (At1g07570), APK2B (At2g02800), MPLKe (BAD12263), PTO (A49332), putative protein kinase (At3g55450), Solanum demissum putative kinase (AAT40481), Oryza sativa putative protein kinases (NP_910058, XP_493889), and Solanum demissum putative protein kinase (AAT40481).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Deduced Amino Acid Sequence of the BIK1 Protein.
Supplemental Figure 2. Phylogenetic Tree Showing the Relationship between BIK1 and Related Plant Protein Kinases.
Supplemental Figure 3. Multiple Sequence Alignment Used for Phylogenetic Analysis.
Supplemental Figure 4. Expression of BIK1 in Arabidopsis Wild-Type, ein2, coi1, nahG, and pad2 Plants during Botrytis Infection.
Supplemental Figure 5. Immunocomplex Assays Showing the Activation of MPK3 and MPK6 in Wild-Type and bik1 Plants in Response to Botrytis Infection.
Supplemental Figure 6. The BIK1 cDNA Rescues the bik1 Altered Growth Traits to Wild-Type Levels.
Supplementary Material
Acknowledgments
We thank Charles Woloshuk for his help with the analysis of SA in plants. This research is funded by Grant IOB-0422702 to T.M. from the National Science Foundation. This is Purdue University Agricultural Program paper 2005-17656.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tesfaye Mengiste (mengiste@purdue.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035576.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, E.P., ten Have, A., van't Klooster, J.W., and van Kan, J.A.L. (1998). Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur. J. Plant Pathol. 104 207–220. [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Cardinale, F., Meskiene, I., Ouaked, F., and Hirt, H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14 703–711. [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares, A.J., Aleu, J., Duran-Patron, R., Collado, I.G., and Hernandez-Galan, R. (2002). The putative role of botrydial and related metabolites in the infection mechanism of Botrytis cinerea. J. Chem. Ecol. 28 997–1005. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- DebRoy, S., Thilmony, R., Kwack, Y.B., Nomura, K., and He, S.Y. (2004). A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA 101 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton, N., Muckenschnabel, I.I., Goodman, B.A., and Williamson, B. (1999). Lipid peroxidation and the oxidative burst associated with infection of Capsicum annuum by Botrytis cinerea. Plant J. 20 485–492. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., and Ryals, J. (1994). A central role of salicylic acid in plant diseases resistance. Science 266 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dewdney, J., Reuber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24 205–218. [DOI] [PubMed] [Google Scholar]
- Diaz, J., ten Have, A., and van Kan, J.A. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich, W., Lorenz, G., Lyr, H., Nega, E., and Pommer, E.H. (1989). New aspects of the infection mechanism of Botrytis cinerea. Neth. J. Plant Pathol. 95 53–56. [Google Scholar]
- Ferrari, S., Plotnikova, J.M., De Lorenzo, G., and Ausubel, F.M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35 193–205. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Fouts, D.E., Badel, J.L., Ramos, A.R., Rapp, R.A., and Collmer, A. (2003). A Pseudomonas syringae pv. tomato DC3000 Hrp (type III secretion) deletion mutant expressing the Hrp system of bean pathogen P. syringae pv. syringae 61 retains normal host specificity for tomato. Mol. Plant Microbe Interact. 16 43–52. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2002). Infection of Arabidopsis with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol. Biol. 48 267–276. [DOI] [PubMed] [Google Scholar]
- Gupta, V., Willits, M.G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant Microbe Interact. 13 503–511. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200 38–62. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 97–131. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., and Oka, A. (1992). Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol. Biol. 20 653–662. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N., Noble, M.E., and Owen, D.J. (1996). Active and inactive protein kinases: Structural basis for regulation. Cell 85 149–158. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 325–331. [DOI] [PubMed] [Google Scholar]
- Lagrimini, L.M., Burkhart, W., Moyer, M., and Rothstein, S. (1987). Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: Molecular analysis and tissue-specific expression. Proc. Natl. Acad. Sci. USA 84 7542–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 583–593. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. 2. Methods Enzymol. 18 350–382. [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 1432–1436. [DOI] [PubMed] [Google Scholar]
- Mengiste, T., Chen, X., Salmeron, J.M., and Dietrich, R.A. (2003). The BOS1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F.L., van Pelt, J.A., Pieterse, C.M., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D.O., and De Bondt, H.L. (1994). Protein kinase regulation: Insights from crystal structure analysis. Curr. Opin. Cell Biol. 6 239–246. [DOI] [PubMed] [Google Scholar]
- Muckenschnabel, I., Goodman, B.A., Williamson, B., Lyon, G.D., and Deighton, N. (2002). Infection of leaves of Arabidopsis thaliana by Botrytis cinerea: Changes in ascorbic acid, free radicals and lipid peroxidation products. J. Exp. Bot. 53 207–214. [DOI] [PubMed] [Google Scholar]
- Murase, K., Shiba, H., Iwano, M., Che, F.S., Watanabe, M., Isogai, A., and Takayama, S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303 1516–1519. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt, B.F., III, and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275 7521–7526. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Podell, S., and Gribskov, M. (2004). Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics 5 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins, T.W., Tudzynski, P., Tiedemann, A.V., Tudzynski, B., ten Have, A., Hansen, M.E., Tenberge, K., and van Kan, J.A.L. (2000). Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In Fungal Pathology, J.W. Kronstad, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 33–64.
- Rentel, M.C., Lecourieux, D., Ouaked, F., Usher, S.L., Petersen, L., Okamoto, H., Knight, H., Peck, S.C., Grierson, C.S., Hirt, H., and Knight, M.R. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 858–861. [DOI] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16 473–485. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. a). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. b). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001 RE22. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.M., and Walker, J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntres, Z.E. (2002). Role of antioxidants in paraquat toxicity. Toxicology 180 65–77. [DOI] [PubMed] [Google Scholar]
- Swiderski, M.R., and Innes, R.W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26 101–112. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2000). PAUP. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, G.A., Jr., and Okuyama, H. (2000). Lipid-linked proteins of plants. Prog. Lipid Res. 39 19–39. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Tiedemann, A.V. (1997). Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol. Mol. Plant Pathol. 50 151–166. [Google Scholar]
- Tierens, K.F., Thomma, B.P., Bari, R.P., Garmier, M., Eggermont, K., Brouwer, M., Penninckx, I.A., Broekaert, W.F., and Cammue, B.P. (2002). Esa1, an Arabidopsis mutant with enhanced susceptibility to a range of necrotrophic fungal pathogens, shows a distorted induction of defense responses by reactive oxygen generating compounds. Plant J. 29 131–140. [DOI] [PubMed] [Google Scholar]
- Ton, J., De Vos, M., Robben, C., Buchala, A., Metraux, J.P., Van Loon, L.C., and Pieterse, C.M. (2002). Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 29 11–21. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S., Chang, H.-S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., Uknes, S., Kessmann, H., and Ryals, J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese, P., Chen, X., Bluhm, B., Salmeron, J., Dietrich, R.A., and Mengiste, T. (2004). The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J. 40 558–574. [DOI] [PubMed] [Google Scholar]
- Veronese, P., Narasimhan, M.L., Stevenson, R.A., Zhu, J.K., Weller, S.C., Subbarao, K.V., and Bressan, R.A. (2003). Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 35 574–587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.