Abstract
Premature ovarian failure (POF) syndrome, an early decline of ovarian function in women, is frequently associated with X chromosome abnormalities ranging from various Xq deletions to complete loss of one of the X chromosomes. However, the genetic locus responsible for the POF remains unknown, and no candidate gene has been identified. Using the Cre/LoxP system, we have disrupted the mouse X chromosome androgen receptor (Ar) gene. Female AR–/– mice appeared normal but developed the POF phenotype with aberrant ovarian gene expression. Eight-week-old female AR–/– mice are fertile, but they have lower follicle numbers and impaired mammary development, and they produce only half of the normal number of pups per litter. Forty-week-old AR–/– mice are infertile because of complete loss of follicles. Genome-wide microarray analysis of mRNA from AR–/– ovaries revealed that a number of major regulators of folliculogenesis were under transcriptional control by AR. Our findings suggest that AR function is required for normal female reproduction, particularly folliculogenesis, and that AR is a potential therapeutic target in POF syndrome.
Keywords: male hormone, nuclear receptor, female physiology, folliculogenesis, kit ligand
Premature ovarian failure (POF) is defined as an early decline of ovarian function after seemingly normal folliculogenesis (1). Genetic causes of POF have been frequently associated with X chromosome abnormalities (1, 2). Complete loss of one of the X chromosomes, as in Turner syndrome, and various Xq deletions are commonly identified as a cause of POF. However, responsible X-linked genes and their downstream targets have not been identified so far.
The androgen receptor (Ar) gene, which is the only sex hormone receptor gene on the X chromosome, is well known to be essential not only for the male reproductive system, but also for male physiology. In contrast, androgens are considered as male hormones; therefore, little is known about androgens' actions in female physiology, although AR expression in growing follicles has been described (3). However, because excessive androgen production in polycystic ovary syndrome causes infertility with abnormal menstrual cycles (4, 5), it is possible that AR-mediated androgen signaling also plays an important physiological role in the female reproductive system. Recently, using Cre/LoxP system, we generated an AR-null mutant mouse line (6) and demonstrated that inactivation of AR resulted in arrest of testicular development and spermatogenesis, impaired brain masculinization, high-turnover osteopenia, and late onset of obesity in males (7–9). At the same time, no overt physical or growth abnormalities were observed in female AR–/– mice. Therefore, to further examine potential role of AR in female physiology, we characterized female reproductive system in AR–/– females. Herein we show that female AR–/– mice develop the POF phenotype. At 3 weeks of age, AR–/– females had apparently normal ovaries with numbers of follicles similar to those in the wild-type females. However, thereafter the number of healthy follicles in the AR–/– ovary gradually declined, with a marked increase of atretic follicles, and by 40 weeks AR–/– mice became infertile, with no follicle detectable in the ovary. Reflecting this age-dependent progression in ovarian abnormality, several genes known to be involved in the oocyte–granulosa cell regulatory loop were identified by microarray analysis as AR downstream target genes. These findings clearly demonstrate that AR-mediated androgen signaling is indispensable for the maintenance of folliculogenesis and implicate impaired androgen signaling as a potential cause of the POF syndrome.
Materials and Methods
Generation of AR Knockout Mice. AR genomic clones were isolated from a TT2 embryonic stem cell genomic library by using human AR A/B domain cDNA as a probe (6). The targeting vector consisted of a 7.6-kb 5′ region containing exon 1, a 1.3-kb 3′ homologous region, a single loxP site, and a neo cassette with two loxP sites (10). Targeted clones (FB-18 and FC-61) were aggregated with single eight-cell embryos from CD-1 mice (11, 12). Floxed AR mice (C57BL/6) were then crossed with CMV-Cre transgenic mice (6). The two lines exhibited the same phenotypic abnormalities. The chromosomal sex of each pup was determined by genomic PCR amplification of the Y chromosome Sry gene (13).
Western Blot Analysis. To detect AR protein expression, ovarian cell lysates were separated by SDS/PAGE and transferred onto nitrocellulose membranes (14). Membranes were probed with polyclonal AR antibodies (N-20; Santa Cruz Biotechnology), and blots were visualized by using peroxidase-conjugated second antibody and an ECL detection kit (Amersham Pharmacia Biosciences).
Morphologic Classification of Growing Follicles. Sections were taken at intervals of 30 μm, and 6-μm paraffin-embedded sections were mounted on slides. Routine hematoxylin and eosin staining was performed for histologic examination by light microscopy. Follicle numbers in 12 sections per ovary were evaluated as primary follicles (oocyte surrounded by a single layer of cuboidal granulosa cells), preantral follicles (oocyte surrounded by two or more layers of granulosa cells with no antrum), or antral follicles (antrum within the granulosa cell layers enclosing the oocyte). Follicles were determined to be atretic if they displayed two or more of the following criteria within a single cross section: more than two pyknotic nuclei, granulosa cells within the antral cavity, granulosa cells pulling away from the basement membrane, or uneven granulosa cell layers (15).
Immunohistochemistry. Sections were subjected to a microwave antigen retrieval technique by boiling in 10 mM citrate buffer (pH 6.0) in a microwave oven for 30 min (16). The cooled sections were incubated in 1% H2O2 for 30 min to quench endogenous peroxidase and then incubated with 1% Triton X-100 in PBS for 10 min. To block nonspecific antibody binding, sections were incubated in normal goat serum for 1 h at 4°C. Sections were then incubated with anti-AR (1:100) or anti-cleaved caspase-3 (1:100) in 3% BSA overnight at 4°C. Negative controls were incubated in 3% BSA without primary antibody. The ABC method was used to visualize signals according to the manufacturer's instructions. Sections were incubated in biotinylated goat anti-rabbit IgG (1:200 dilution) for 2 h at room temperature, washed with PBS, and incubated in avidin–biotin–horseradish peroxidase for 1 h. After thorough washing in PBS, sections were developed with 3,3′-diaminobendizine tetrahydrochloride substrate, slightly counterstained with eosin, dehydrated through an ethanol series and xylene, and mounted.
Estrus Cycles and Fertility Test. To determine the stage of the estrus cycle (proestrus, estrus, and diestrus), vaginal smears were taken every morning and stained with Giemsa solution. For evaluation of female fertility for 15 weeks, an 8- or 24-week-old wild-type or AR–/– female was mated with a wild-type fertile male, replaced every 2 weeks with the other fertile male. Cages were monitored daily and for an additional 23 days, and the presence of seminal plugs and number of litters were recorded.
RNA Extraction and Quantitative Competitive RT-PCR. Total ovarian RNA was extracted by using TRIzol (Invitrogen) (16). Oligo-dT-primed cDNA was synthesized from 1 μg of ovarian RNA by using SuperScript reverse transcriptase (Gibco BRL, Gaithersburg, MD) in a 20-μl reaction volume, 1 μl of which was then diluted serially (2- to 128-fold) and used to PCR-amplify an internal control gene, cycA, to allow concentration estimation. Primers were designed from cDNA sequences of Kitl (M57647; nucleotides 1099–1751), Gdf9 (NM008110; nucleotides 720-1532), Bmp15 (NM009757; nucleotides 146–973), Ers2 (NM010157; nucleotides 1139–1921), Pgr (NM008829; nucleotides 1587–2425), Cyp11a1 (NM019779; nucleotides 761-1697), Cyp17al (M64863; nucleotides 522–932), Cyp19 (D00659; nucleotides 699-1049), Fshr (AF095642; nucleotides 625-1427), Lhr (M81310; nucleotides 592-1331), Ptgs2 (AF338730; nucleotides 3–605), and Ccnd2 (NM009829; nucleotides 150-1065) and chosen from different exons to avoid amplification from genomic DNA.
GeneChip Analysis. Ovaries were isolated and stabilized in RNA-later RNA Stabilization Reagent (Ambion, Austin, TX) before RNA purification (17). Total RNA was purified by using an RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. First-strand cDNA was synthesized from 5 μg of RNA by using 200 units of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), 100 pmol T7-(dT)24 primer [5′-GGCCAGTGA AT TGTA ATACGACTCACTATAGGGAGGCGG-(dT)24-3′], 1× first-strand buffer, and 0.5 mM dNTPs at 42°C for 1 h. Second-strand synthesis was performed by incubating first-strand cDNA with 10 units of Escherichia coli ligase (Invitrogen), 40 units of DNA polymerase I (Invitrogen), 2 units of RNase H (Invitrogen), 1× reaction buffer, and 0.2 mM dNTPs at 16°C for 2 h, followed by 10 units of T4 DNA polymerase (Invitrogen) and incubation for another 5 min at 16°C. Double-stranded cDNA was purified by using GeneChip Sample Cleanup Module (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions and labeled by in vitro transcription by using a BioArray HighYield RNA transcript labeling kit (Enzo Diagnostics, Farmingdale, NY). Briefly, dsDNA was mixed with 1× HY reaction buffer, 1× biotin-labeled ribonucleotides (NTPs with Bio-UTP and BioCTP), 1× DTT, 1× RNase inhibitor mix, and 1× T7 RNA polymerase and incubated at 37°C for 4 h. Labeled cRNA was then purified by using GeneChip Sample Cleanup Module and fragmented in 1× fragmentation buffer at 94°C for 35 min. For hybridization to the GeneChip Mouse Expression Array 430A or 430B or Mouse Genome 430 2.0 Array (Affymetrix), 15 μg of fragmented cRNA probe was incubated with 50 pM control oligonucleotide B2, 1× eukaryotic hybridization control, 0.1 mg/ml herring sperm DNA, 0.5 mg/ml acetylated BSA, and 1× hybridization buffer in a 45°C rotisserie oven for 16 h. Washing and staining were performed by using a GeneChip Fluidic Station (Affymetrix) according to the manufacturer's protocol. Phycoerythrin-stained arrays were scanned as digital image files and analyzed with genechip operating software (Affymetrix) (17).
Luciferase Assay. The Kitl promoter region (–2866 to –1 bp) was inserted into the pGL3-basic vector (Promega) for assay using the Luciferase Assay System (Promega) (14, 16). Cells at 40–50% confluence were transfected with a reference pRL-CMV plasmid (Promega) using Lipofectamine reagent (GIBCO/BRL, Grand Island, NY) to normalize transfection. Results shown are representative of five independent experiments.
Results and Discussion
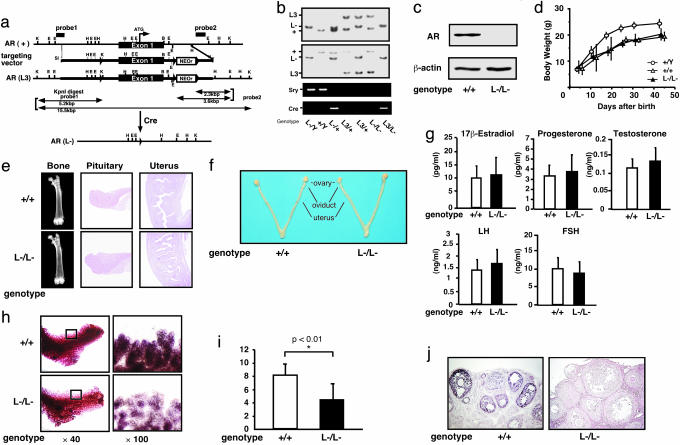
Subfertility of AR–/– Female Mice at 8 Weeks of Age. The Ar gene located on the X chromosome was disrupted in mice by using the Cre/Lox P system (6) (Fig. 1 a–c). Female AR–/– mice showed normal growth compared with the wild-type littermates (Fig. 1d), with no detectable bone loss (Fig. 1e) or obesity common for male AR–/Y mice (8, 9). Young (8-week-old) AR–/– females appeared indistinguishable from the wild-type littermates, displayed normal sexual behavior (7), and produced the first offspring of normal body size at the expected age. Macroscopic appearance of their reproductive organs, including uteri, oviducts, and ovaries, also appeared normal (Fig. 1f). Histological analysis showed no significant abnormality in the uterus or pituitary (Fig. 1e), whereas mammary ductal branching and elongation were substantially reduced, as revealed by whole-mount analysis (Fig. 1h). Serum levels of 17β-estradiol, progesterone, testosterone, luteinizing hormone, and follicle-stimulating hormone were also within normal range in 8-week-old mutant females at the proestrus stage (Fig. 1g), suggesting that the two-cell two-gonadotrophin system in female reproductive and endocrine organs (18) was intact in AR–/– mice at 8 weeks of age. The most obvious early sign of abnormal reproductive function in the AR–/– females was that their average numbers of pups per litter were only about half of those of the wild-type littermates, (AR+/+, 8.3 ± 0.4 pups per litter; AR–/–, 4.5 ± 0.5 pups per litter) (Fig. 1i).
Fig. 1.
Phenotypic characterization of AR knockout female mice. (a) Diagram of the wild-type Ar genomic locus (+), floxed AR L3 allele (L3), and AR allele (L-) obtained after Cre-mediated excision of exon 1. K, KpnI; E, EcoRI; H, HindIII; B, BamHI. LoxP sites are indicated by arrowheads. The targeting vector consisted of a 7.6-kb 5′ homologous region containing exon 1, a 1.3-kb 3′ homologous region, a single loxP site, and the neo cassette with two loxP sites. (b) Detection of the Y chromosome-specific Sry gene in AR–/Y mice by PCR. (c) Absence of AR protein in AR–/– mice ovaries by Western blot analysis using a specific C-terminal antibody. (d) Normal weight gain in AR–/– females. (e) Histology of pituitary, uterus, and bone tissues in AR+/+ and AR–/– females at 8 weeks of age. (f) Female reproductive organs were macroscopically normal in AR–/– mice. (g) Serum hormone levels at the proestrus stage in AR–/– mice were not significantly altered. Serum 17β-estradiol, progesterone, testosterone, luteinizing hormone (LH), and follicle-stimulating hormone (FSH) levels in AR+/+ (n = 13) and AR–/– (n = 10) females at 8–10 weeks of age are shown. (h) Lobuloalveolar development is impaired in AR–/– mammary glands. Whole mount of inguinal mammary glands (Left) and its higher magnification (Right) were prepared on day 3 of lactation. (i) Average number of pups per litter is markedly reduced in AR–/– mice at 8 weeks of age. Data are shown as mean ± SEM and analyzed by using Student's t test. (j) AR immunocytochemistry in AR+/+ and AR–/– ovaries. Sections were counterstained with eosin.
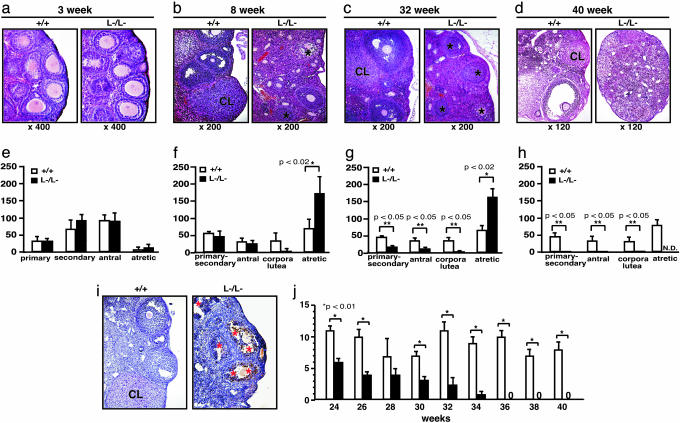
AR–/– Female Mice Developed POF Phenotypes. Histological analysis of 8-week-old AR–/– ovaries clearly showed that numbers of atretic follicles were significantly increased, with decreased numbers of corpora lutea (Fig. 2 b and f). This finding suggests that the reduced pup numbers were due to impaired folliculogenesis in AR-deficient ovaries. Indeed, AR protein expression was readily detectable in the wild-type 8-week-old ovaries (Fig. 1j), with AR expressed at the highest levels in growing follicle granulosa cells at all developmental stages and at relatively low levels in corpora lutea. Thus, AR appears to play a regulatory role in granulosa cells during their maturation to the luteal phase.
Fig. 2.
POF in AR–/– female mice. (a–d) Histology of AR+/+ and AR–/– ovaries at 3 weeks, 8 weeks, 32 weeks, and 40 weeks of age. All sections were stained with hematoxylin and eosin. An asterisk marks the atretic follicle. CL, corpus luteum. (e–h) Relative follicle counts at 3 weeks (e), 8 weeks (f), 32 weeks (g), and 40 weeks (h) of age. Numbers represent total counts of every fifth section from serially sectioned ovaries (n = 4 animals per genotype). (i) Immunohistochemical study for activated, cleaved caspase-3 revealed increased positive cells (apoptotic cells) in AR–/– ovaries. Sections were counterstained with hematoxylin. An asterisk marks the caspase-3-positive cell. CL, corpus luteum. (j) Age-dependent reduction in the number of pups per litter in AR–/– female mice. A continuous breeding assay was started at 24 weeks of age (n = 6–10 animals per genotype). For all panels, data are shown as mean ± SEM and were analyzed by using Student's t test.
To investigate this possibility, we examined the ovarian phenotype of female AR–/– mice at different ages. At 3 weeks, ovaries contain various stages of follicles, including primary, secondary, and antral follicles in wild-type animals (Fig. 2a) (19). In AR–/– ovaries at 3 weeks of age, the folliculogenesis appeared to be unaltered, with normal numbers and localization of primary and secondary follicles (Fig. 2 a and e). However, degenerated folliculogenesis became evident with further aging. Although follicles and corpora lutea at all developmental stages were still present, corpora lutea numbers were clearly reduced in 8-week-old AR–/– mutants (Fig. 2 b and f), similar to that observed in another mouse line (20). Expected apoptosis was seen in atretic follicles by activated caspase-3 immunohistochemistry assays (Fig. 2i). But, by 32 weeks of age, defects in folliculogenesis in AR–/– ovaries became profound, with fewer follicles observed and increased atretic follicles (Fig. 2 c and g), and >40% (5 of 12 mice) of the AR–/– females were already infertile. By 40 weeks, all AR–/– females became infertile, with no follicles remaining (Fig. 2 d and h); at the same age, AR+/+ females were fertile and had normal follicle numbers. Consistent with progressive deficiency in folliculogenesis, the pup number per litter steadily decreased in aging AR–/– females (Fig. 2i). These data indicate that AR plays an important physiological role at the preluteal phase of folliculogenesis.
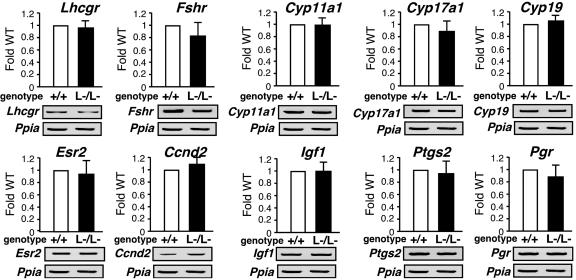
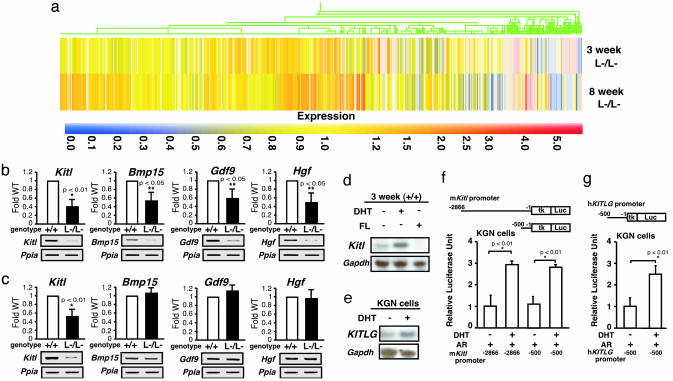
Alteration in Gene Expressions of Several Major Regulators Involved in the Oocyte–Granulosa Cell Regulatory Loop. To explore the molecular basis underlying the impaired folliculogenesis in AR–/– ovaries, we analyzed expression of several major known regulators and markers of folliculogenesis (21–23). Surprisingly, no significant alterations in mRNA levels of LH receptor (Lhr), FSH receptor (Fshr), p450 side chain cleavage enzyme (Cyp11a1), 17-α-hydroxylase (Cyp17a1), aromatase (Cyp19), estrogen receptor-β (Esr2), cyclin D2 (Ccnd2), or insulin-like growth factor 1 (Igf1) of 8-week-old AR–/– ovaries at the proestrus stage, and further cyclooxygenase 2 (Ptgs2) or progesterone receptor (Pgr) at the estrus stage, were detected by semiquantitative RT-PCR analysis (Fig. 3). Genome-wide microarray analysis (17) of RNA from 8-week-old AR–/– ovaries at the proestrus stage has been undertaken to identify AR-regulated genes. In comparison with AR+/+ ovaries, expressions of 772 genes were down-regulated, whereas 351 genes were up-regulated in AR–/– ovaries (Fig. 4a; see also Tables 1 and 2, which are published as supporting information on the PNAS web site). Several genes known to be involved in the oocyte–granulosa cell regulatory loop (24) were identified as candidate AR target genes, including KIT ligand (Kitl) (25), morphogenetic protein 15 (Bmp15) (26), growth differentiation factor-9 (Gdf9) (27), and hepatocyte growth factor (Hgf) (28). Impaired folliculogenesis had been reported in mice deficient in each of these three regulators (26, 27, 29). To validate the microarray data, we performed semiquantitative RT-PCR analysis of 8-week-old AR–/– ovary RNA and confirmed that expression of these factors was down-regulated (Fig. 4b). To identify a regulator downstream of the AR signaling at an earlier stage of folliculogenesis, 3-week-old AR–/– ovaries that, as pointed out earlier, display no apparent phenotypic abnormality were examined. Fewer genes had altered expression levels (519 genes up-regulated; 326 genes down-regulated) (Fig. 4a; see also Tables 3 and 4, which are published as supporting information on the PNAS web site), and, of the four regulators tested by RT-PCR, only Kitl was found to be down-regulated at this age (Fig. 4c). Because Kitl is a granulosa cell-derived factor and stimulates oocyte growth and maturation (29–31), down-regulation of the Kitl expression in 3-week-old or even younger AR–/– ovaries may trigger impairment in folliculogenesis at a later age. To test for possible Kitl gene regulation by AR, 3-week-old wild-type females were treated with 5α-dihydrotestosterone (DHT). At 4 h after hormone injection, a clear induction of Kitl expression was observed in the ovaries, whereas a known antiandrogen flutamide attenuated the induction by DHT (Fig. 4d). The induction of endogenous human kit ligand (KITLG) gene by DHT was also observed in human granulosa-like tumor cells (KGN) in culture (Fig. 4e). Furthermore, androgen-induced transactivation of mouse and human kit ligand promoters (32) was observed by a luciferase reporter assay (33) in KGN (Fig. 4 f and g), 293T, and HeLa (data not shown) cells. However, no response to DHT was detected in the similar assay using promoters of the Bmp15, Gdf9, and Hgf genes (data not shown). Thus, we have shown that, in a regulatory cascade controlling folliculogenesis, Kitl represents a direct downstream target of androgen signaling.
Fig. 3.
No significant alterations in mRNA levels of several major regulators in folliculogenesis. Shown is semiquantitative RT-PCR of LH receptor (Lhr), FSH receptor (Fshr), p450 side chain cleavage enzyme (Cyp11a1), 17-α-hydroxylase (Cyp17a1), Aromatase (Cyp19), estrogen receptor-β (Esr2), cyclin D2 (Ccnd2), insulin-like growth factor 1 (Igf1), cyclooxygenase 2 (Ptgs2), or progesterone receptor (Pgr) gene expression in AR+/+ and AR–/– ovaries. Results shown were representative (using one ovary per genotype in each experiment) of five independent experiments.
Fig. 4.
Genome-wide microarray analysis and semiquantitative RT-PCR revealed that expression of the oocyte–granulosa cell regulator loop was down-regulated in AR–/– ovaries. (a) Microarray analysis of AR–/– compared with AR+/+ ovaries at 3 and 8 weeks of age. Data obtained from microarray analysis as described in Materials and Methods were used to generate a cluster analysis. Each vertical line represents a single gene. The ratios of gene expression levels in AR–/– ovaries compared with wild type are presented. (b and c) Semiquantitative RT-PCR analysis of AR-regulated genes identified from the microarray study. Results shown are representative (using one ovary per genotype in each experiment) of five independent experiments. Data are shown as mean ± SEM and were analyzed by using Student's t test. (d) Comparison of Kitl gene expression by Northern blot analysis among placebo-, DHT-, and flutamide (FL)-treated AR+/+ mouse ovaries. (e) Induction of KITLG gene expression by DHT treatment in KGN cells. (f and g) Androgen responsiveness in the mouse and human kit ligand promoters by a luciferase assay performed by using KGN cells. Data are shown as mean ± SEM and were analyzed by using Student's t test.
As an upstream regulator, AR may also be indirectly involved in control of expression of other genes critical for folliculogenesis, because an age-dependent down-regulation of Bmp15, Gdf9, and Hgf gene expression was also observed in AR–/– ovaries. Bmp15 and Gdf9 are oocyte-derived factors that promote the development of surrounding granulosa cells in growing follicles (34, 35), whereas Hgf is secreted by theca cells and acts as a granulosa cell growth factor (36). Down-regulation of these factors, presumably due to decreased Kitl expression, may lead to impaired bidirectional communication between oocyte and granulosa cells (24) and, eventually, to early termination of folliculogenesis, as in POF syndrome.
Thus, we have identified AR as a novel regulator of folliculogenesis that apparently acts in the regulatory cascade upstream of the major factors controlling ovarian function, confirming the previous findings of the AR expression in granulose cells of growing follicles (3). Although not immediately relevant to the ovarian physiology, abnormal development of the mammary glands observed in our AR-deficient mice adds further strong evidence of an essential role of the AR not only in male, but also in female, reproductive function.
With increasing age of the first childbirth by women in the modern society, POF syndrome has become an important social and medical problem. Our findings suggest that POF syndrome may be caused by an impairment in androgen signaling and that X chromosomal mutations affecting the AR gene function may play a key role in hereditary POF. From clinical perspective, the present study provides evidence that AR can be a beneficial therapeutic target in treatment of POF syndrome patients.
Supplementary Material
Acknowledgments
We thank T. Iwamori and H. Tojo for expert advice on mammary gland anatomy, Y. Kanai for ovarian phenotypic analysis, members of the KO project team at the laboratory of Nuclear Signaling (Institute of Molecular and Cellular Biosciences) for their support, A. P. Kouzmenko for helpful suggestions, and H. Higuchi for manuscript preparation. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences and priority areas from the Ministry of Education, Culture, Sports, Science, and Technology (to S.K.).
Author contributions: J.K., H.Y., and S.K. designed research; H.S., T.M., T.S., K.I., J.M., S.T., M.S., I.T., and T.N. performed research; D.M. and P.C. contributed new reagents/analytic tools; H.S. analyzed data; and S.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AR, androgen receptor; DHT, 5α-dihydrotestosterone; POF, premature ovarian failure.
References
- 1.Laml, T., Preyer, O., Umek, W., Hengstschlager, M. & Hanzal, H. (2002) Hum. Reprod. Update 8, 483–491. [DOI] [PubMed] [Google Scholar]
- 2.Davison, R. M., Davis, C. J. & Conway, G. S. (1999) Clin. Endocrinol. (Oxford) 51, 673–679. [DOI] [PubMed] [Google Scholar]
- 3.Tetsuka, M., Whitelaw, P. F., Bremner, W. J., Millar, M. R., Smyth, C. D. & Hillier, S. G. (1995) J. Endocrinol. 145, 535–543. [DOI] [PubMed] [Google Scholar]
- 4.Ehrmann, D. A., Barnes, R. B. & Rosenfield, R. L. (1995) Endocr. Rev. 16, 322–353. [DOI] [PubMed] [Google Scholar]
- 5.Norman, R. J. (2002) Mol. Cell. Endocrinol. 191, 113–119. [DOI] [PubMed] [Google Scholar]
- 6.Kato, S. (2002) Clin. Pediatr. Endocrinol. 11, 1–7. [Google Scholar]
- 7.Sato, T., Matsumoto, T., Kawano, H., Watanabe, T., Uematsu, Y., Sekine, K., Fukuda, T., Aihara, K., Krust, A., Yamada, T., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato, T., Matsumoto, T., Yamada, T., Watanabe, T., Kawano, H. & Kato, S. (2003) Biochem. Biophys. Res. Commun. 300, 167–171. [DOI] [PubMed] [Google Scholar]
- 9.Kawano, H., Sato, T., Yamada, T., Matsumoto, T., Sekine, K., Watanabe, T., Nakamura, T., Fukuda, T., Yoshimura, K., Yoshizawa, T., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 9416–9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, M., Indra, A. K., Warot, X., Brocard, J., Messaddeq, N., Kato, S., Metzger, D. & Chambon, P. (2000) Nature 407, 633–636. [DOI] [PubMed] [Google Scholar]
- 11.Sekine, K., Ohuchi, H., Fujiwara, M., Yamasaki, M., Yoshizawa, T., Sato, T., Yagishita, N., Matsui, D., Koga, Y., Itoh, N. & Kato, S. (1999) Nat. Genet. 21, 138–141. [DOI] [PubMed] [Google Scholar]
- 12.Yoshizawa, T., Handa, Y., Uematsu, Y., Takeda, S., Sekine, K., Yoshihara, Y., Kawakami, T., Arioka, K., Sato, H., Uchiyama, Y., et al. (1997) Nat. Genet. 16, 391–396. [DOI] [PubMed] [Google Scholar]
- 13.Gubbay, J., Collignon, J., Koopman, P., Capel, B., Economou, A., Munsterberg, A., Vivian, N., Goodfellow, P. & Lovell-Badge, R. (1990) Nature 346, 245–250. [DOI] [PubMed] [Google Scholar]
- 14.Yanagisawa, J., Yanagi, Y., Masuhiro, Y., Suzawa, M., Watanabe, M., Kashiwagi, K., Toriyabe, T., Kawabata, M., Miyazono, K. & Kato, S. (1999) Science 283, 1317–1321. [DOI] [PubMed] [Google Scholar]
- 15.Britt, K. L., Drummond, A. E., Cox, V. A., Dyson, M., Wreford, N. G., Jones, M. E., Simpson, E. R. & Findlay, J. K. (2000) Endocrinology 141, 2614–2623. [DOI] [PubMed] [Google Scholar]
- 16.Ohtake, F., Takeyama, K., Matsumoto, T., Kitagawa, H., Yamamoto, Y., Nohara, K., Tohyama, C., Krust, A., Mimura, J., Chambon, P., et al. (2003) Nature 423, 545–550. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto, N., Igarashi, K., Kanno, J., Honda, H. & Inoue, T. (2004) J. Steroid Biochem. Mol. Biol. 91, 121–129. [DOI] [PubMed] [Google Scholar]
- 18.Couse, J. F. & Korach, K. S. (1999) Endocr. Rev. 20, 358–417. [DOI] [PubMed] [Google Scholar]
- 19.Elvin, J. A. & Matzuk, M. M. (1998) Rev. Reprod. 3, 183–195. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Y. C., Wang, P. H., Yeh, S., Wang, R. S., Xie, C., Xu, Q., Zhou, X., Chao, H. T., Tsai, M. Y. & Chang, C. (2004) Proc. Natl. Acad. Sci. USA 101, 11209–11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elvin, J. A., Yan, C., Wang, P., Nishimori, K. & Matzuk, M. M. (1999) Mol. Endocrinol. 13, 1018–1034. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, J., Kumar, T. R., Matzuk, M. M. & Bondy, C. (1997) Mol. Endocrinol. 11, 1924–1933. [DOI] [PubMed] [Google Scholar]
- 23.Burns, K. H., Yan, C., Kumar, T. R. & Matzuk, M. M. (2001) Endocrinology 142, 2742–2751. [DOI] [PubMed] [Google Scholar]
- 24.Matzuk, M. M., Burns, K. H., Viveiros, M. M. & Eppig, J. J. (2002) Science 296, 2178–2180. [DOI] [PubMed] [Google Scholar]
- 25.Joyce, I. M., Pendola, F. L., Wigglesworth, K. & Eppig, J. J. (1999) Dev. Biol. 214, 342–353. [DOI] [PubMed] [Google Scholar]
- 26.Yan, C., Wang, P., DeMayo, J., DeMayo, F. J., Elvin, J. A., Carino, C., Prasad, S. V., Skinner, S. S., Dunbar, B. S., Dube, J. L., et al. (2001) Mol. Endocrinol. 15, 854–866. [DOI] [PubMed] [Google Scholar]
- 27.Dong, J., Albertini, D. F., Nishimori, K., Kumar, T. R., Lu, N. & Matzuk, M. M. (1996) Nature 383, 531–535. [DOI] [PubMed] [Google Scholar]
- 28.Parrott, J. A., Vigne, J. L., Chu, B. Z. & Skinner, M. K. (1994) Endocrinology 135, 569–575. [DOI] [PubMed] [Google Scholar]
- 29.Driancourt, M. A., Reynaud, K., Cortvrindt, R. & Smitz, J. (2000) Rev. Reprod. 5, 143–152. [DOI] [PubMed] [Google Scholar]
- 30.Huang, E. J., Manova, K., Packer, A. I., Sanchez, S., Bachvarova, R. F. & Besmer, P. (1993) Dev. Biol. 157, 100–109. [DOI] [PubMed] [Google Scholar]
- 31.Packer, A. I., Hsu, Y. C., Besmer, P. & Bachvarova, R. F. (1994) Dev. Biol. 161, 194–205. [DOI] [PubMed] [Google Scholar]
- 32.Grimaldi, P., Capolunghi, F., Geremia, R. & Rossi, P. (2003) Biol. Reprod. 69, 1979–1988. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa, H., Fujiki, R., Yoshimura, K., Mezaki, Y., Uematsu, Y., Matsui, D., Ogawa, S., Unno, K., Okubo, M., Tokita, A., et al. (2003) Cell 113, 905–917. [DOI] [PubMed] [Google Scholar]
- 34.Otsuka, F. & Shimasaki, S. (2002) Proc. Natl. Acad. Sci. USA 99, 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce, I. M., Clark, A. T., Pendola, F. L. & Eppig, J. J. (2000) Biol. Reprod. 63, 1669–1675. [DOI] [PubMed] [Google Scholar]
- 36.Parrott, J. A. & Skinner, M. K. (1998) Endocrinology 139, 2240–2245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.