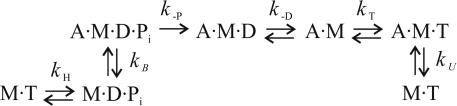Abstract
During skeletal muscle contraction, regular arrays of actin and myosin filaments slide past each other driven by the cyclic ATP-dependent interaction of the motor protein myosin II (the cross-bridge) with actin. The rate of the cross-bridge cycle and its load-dependence, defining shortening velocity and energy consumption at the molecular level, vary widely among different isoforms of myosin II. However, the underlying mechanisms remain poorly understood. We have addressed this question by applying a single-molecule approach to rapidly (≈300 μs) and precisely (≈0.1 nm) detect acto-myosin interactions of two myosin isoforms having large differences in shortening velocity. We show that skeletal myosin propels actin filaments, performing its conformational change (working stroke) in two steps. The first step (≈3.4–5.2 nm) occurs immediately after myosin binding and is followed by a smaller step (≈1.0–1.3 nm), which occurs much faster in the fast myosin isoform than in the slow one, independently of ATP concentration. On the other hand, the rate of the second phase of the working stroke, from development of the latter step to dissociation of the acto-myosin complex, is very similar in the two isoforms and depends linearly on ATP concentration. The finding of a second mechanical event in the working stroke of skeletal muscle myosin provides the molecular basis for a simple model of actomyosin interaction. This model can account for the variation, in different fiber types, of the rate of the cross-bridge cycle and provides a common scheme for the chemo-mechanical transduction within the myosin family.
Keywords: isoforms, optical tweezers, single molecule
It has been know for a long time that the rate of cross-bridge cycling, defining shortening velocity (V), is fundamental for the biological role (1, 2) and the energy consumption of skeletal muscle (3). The shortening velocity of skeletal muscle varies very widely in relation to the isoforms of myosin II (1, 2), conferring on muscles the remarkable capability to adapt their performance to the requirements of posture, movement, and locomotion, by a modulation of isoforms gene expression (1). Shortening velocity and, in turn, the rate of ATP splitting (4) by skeletal myosins strictly depend on the load applied (Fenn effect) (5). The load sensitivity is likely to play a significant role in coordinating the action of the multiple individual motors that in skeletal muscle work cooperatively in a complex ensemble.
It is widely accepted that, during its lifetime of attachment to actin (ton), myosin goes through a conformational change of amplitude δ (working stroke) coupled to the release of the products of ATP hydrolysis [inorganic phosphate (Pi) and ADP]. Actin filaments, therefore, slide at an average velocity V = δ/ton (6, 7). The rate of ADP release (k–D) has been suggested to define both the isoform dependence (8, 9) and the load sensitivity (10) of shortening velocity through a modulation of ton. However, no definitive evidence of such role has been provided for skeletal myosins so far.
In this respect, a deeper insight has been achieved in smooth and nonmuscle myosins based on the finding that those myosins produce their working stroke in two steps (11–13). The first step was shown to occur immediately after myosin binding to actin and was attributed to a conformational change generally related to Pi release (14). The second step (in the same direction but smaller than the first one) was attributed to a smaller conformational change in the myosin head, possibly following ADP release (11–13). The existence of the latter conformational change provided a possible simple explanation for a load-dependent ADP release in smooth and nonmuscle myosins. The load applied on the cross-bridge could, in fact, oppose the conformational change required for ADP release, increasing ton and decreasing V (13).
Unfortunately, evidence for a second step in the working stroke of skeletal muscle myosins has not been found (11), and structural studies are, so far, consistent with the lack of a conformational change upon ADP release in those myosins (15).
We have applied a single-molecule mechanical approach based on optical tweezers (16) to the study of the mechanisms underlying the modulation of sliding velocity in skeletal muscle. In particular, we have investigated two myosin II isoforms having very wide (4- to 5-fold) differences in shortening velocity (2): myosin 1 from rat (slow) and myosin 2B from mouse (fast).†† Pure isoforms of myosin subfragment 1 (S1) of skeletal myosin II were obtained in sufficient amounts for optical tweezers measurements by using a refined technique of myosin extraction from single muscle fibers of known myosin isoform content (9, 17). The low compliance of the single-molecule construct, in combination with a refined approach for event analysis, enabled us to rapidly (≈300 μs) and precisely (≈0.1 nm) detect acto-myosin interactions. The high resolution of the method allowed the first observation of a second step in the working stroke of skeletal myosins. This finding clarifies the kinetic mechanisms that modulate shortening velocity among skeletal myosin isoforms and provides a common scheme for the chemo-mechanical transduction within the myosin family.
Materials and Methods
Animals and Muscle Sampling. Fifteen adult male Wistar rats and 10 adult male C57 mice were used in this study. Mice were killed by cervical dislocation, and the gastrocnemius muscle was dissected. Rats were killed by ether anesthesia, and soleus muscle was dissected out. The experimental protocol for the study on animals was approved by the local animal ethics committee according to the procedures conformed to European Union directive 86/609/EEC. The mouse and rat muscles were immediately immersed in ice-cold skinning solution with the following composition: 150 mM potassium propionate/5 mM magnesium acetate/5 mM sodium ATP/5 mM EGTA/5 mM KH2PO4 (pH 7.0).
Isolation of Pure Myosin Isoforms. An approach based on myosin extraction from single fibers was used to obtain pure myosin isoforms (17). Single fibers mostly contain only a single type of MHC isoform; therefore, they are a convenient source of pure myosin isoforms. However, because of its short length, one fiber segment does not provide sufficient amount of myosin for the experiments. Therefore, (i) single fibers were dissected, (ii) SDS/PAGE was performed to identify MHC isoform content, (iii) 30–40 fibers shown to contain the same MHC isoform were pooled, and (iv) myosin was extracted from pooled fibers. Segments of fibers (at least 12 mm long) were manually isolated from muscle samples while immersed in skinning solution. The segments were chemically skinned (by immersion for 1 h in skinning solution containing 1% Triton X-100) and cut in two segments. The shorter segment (about 2 mm long) was used for the characterization of MHC isoform composition by SDS/PAGE. The longer segment (at least 10 mm long) was used for extraction of myosin according to the method described in detail in ref. 17. About 60–70 μg of pure myosin could be obtained from a pool of 30–40 fibers. Myosin heavy chains were separated on 8% SDS/PAGE as described by Pellegrino et al. (2). Densitometric analysis of gels was performed by a computer-assisted densitometer as described by Pellegrino et al. (2).
Flow Cell. The S1 subfragment from each pure myosin sample was prepared by papain digestion according to Harada et al. (18). G-actin was extracted from acetone powder and biotinylated as described by Ishijima et al. (19). Biotinylated G-actin was polymerized to form F-actin filaments and labeled with rhodamine phalloidin as described by Steffen et al. (20). Carboxylated fluorescent beads (1.1 μm diameter; L-4655, Sigma) were coated with neutravidin (#31000, Pierce), as described in Ishijima et al. (19). Silica beads (1.54 μm; SS04N/5303, Bangs Laboratories, Carmel, IN) were washed, resuspended in amylacetate with 0.1% nitrocellulose, and spread onto the surface of a coverslip. This was attached to a slide by using two pieces of double-sided tape (≈60 μm thick) to create a flow cell. Flow cells were incubated for 1 min with either ≈1 μg/ml S1 extracted from mice fibers type 2B or ≈5 μg/ml S1 extracted from rat fibers type 1, dissolved in AB buffer (25 mM KCl/4 mM MgCl2/25 mM Hepes/1 mM KH2PO4/0.02% NaN3, pH 7.2). The optimal concentration of S1 to be used was determined during optical trap experiments as the concentration that lead to detectable acto-myosin interactions every two to three silica beads investigated. Next, 1 mg/ml BSA was applied to the flow cell for 3 min. The flow cell was then washed with a reaction mix containing neutravidin-coated beads (≈0.002% solids), 1–2 nM biotinylated rhodamine-labeled F-actin, ATP at concentrations ranging from 1 to 20 μM, deoxygenating system (20 mM DTT/200 μg/ml glucose oxidase/50 μg/ml catalase/3 mg/ml glucose), and ATP regenerating system (2 mM creatine phosphate/100 μg/ml creatine phosphokinase) in AB buffer. Experiments were performed at room temperature (≈22°C).
Optical Trap Experiments. The experimental apparatus was developed at the European Laboratory for Non-linear Spectroscopy based on a custom optical microscope. A double tweezers setup (described in detail in ref. 21) was integrated within the optical microscope. A biotinylated actin filament was stretched between two trapped neutravidin-coated polystyrene beads (forming a structure named dumbbell) and presented to a silica bead attached to the coverslide and carrying, on average, less than one myosin molecule (Fig. 1a). Pretensioning in the range of 3–20 pN was applied to the actin filament. Corresponding stiffness of the two actin-bead links in series (kL/4) was measured from the slope of the force–extension curve of the dumbbell (modeling the actin filament as rigid) and ranged between 1 and 3 pN/nm, in agreement with previous measurements (22). A high-magnification (×2,000) image of the silica bead was projected onto a charge-coupled device camera, and its position along the three axes was measured to probe thermal drifts of the sample. A feedback system drove piezoelectric translators to correct the thermal drifts, thus stabilizing the position of the myosin molecule with respect to the actin filament within 1 nm (20, 21). The bandwidth of the position detector was ≈10 kHz, and position signals were recorded at 20-kHz sample rate. The detector response (nm/V) and the trap stiffness (kt/2) were calibrated from the power spectrum of the position signal of a bead trapped at half-depth within the sample chamber (23).
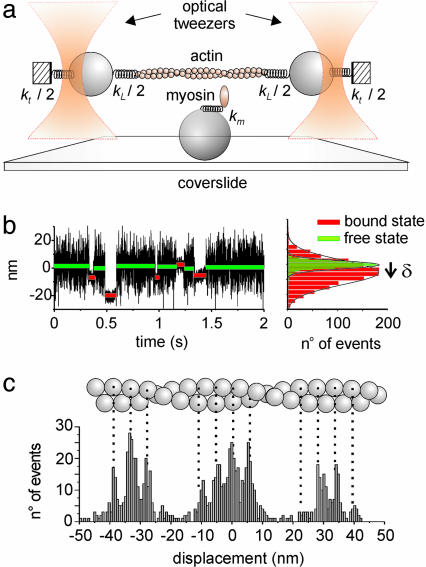
Fig. 1.
Experimental assay and events analysis. (a) In the single-molecule assay, an actin filament is connected to polystyrene beads through biotin–avidin links (stiffness kL/2). The beads are held by optical tweezers with stiffness kt/2. The myosin molecule (stiffness km) is carried on a third bead stuck to the coverslide. (b Left) Two seconds of a position recording of the dumbbell while myosin was interacting with actin. The red and green lines indicate the average position of bound and unbound events, respectively. (b Right) Distributions of the average position of bound and unbound events of a 100-s position recording containing several hundreds of interactions are shown. The working stroke (δ) was obtained from the displacement between the centers of the two distributions. (c) Distribution of the average position of bound events obtained while translating the myosin molecule by 72 nm at constant velocity along the actin filament. Displacement data were corrected for the constant drift in position of the myosin molecule to reveal the distribution modulated by the binding sites and the target zones along the actin filament.
Events Analysis. Acto-myosin interactions were detected from noise reduction of the bead position signal (24) (Fig. 1b). Recorded data were analyzed by fitting the signal variance with a two-state function, using a hidden Markov algorithm (22). This allowed for the separation of low-variance states (bound states) from high-variance states (unbound states). The window width (W) used to calculate the signal variance was chosen following Smith et al. (22), to result in zero or few false detected events, as evidenced by simulated data (see next section). The acto-myosin complex attachment rate (f) and detachment rate (g) were obtained as the inverse of the average durations of unbound (toff) and bound events (ton), respectively. The stiffness, km, of the myosin molecule in the bound state was obtained from the equipartition theorem, in the limit of rigid actin-bead links, as 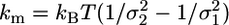 , where kB is the Boltzmann constant, T is the absolute temperature, and
, where kB is the Boltzmann constant, T is the absolute temperature, and 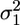 and
and 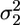 are the position variance of the unbound and bound state, respectively. km was obtained from experiments in which a combined trap stiffness (kt) in the range 0.05–0.1 pN/nm and pretensioning above 3 pN were used. The influence of compliant links is discussed in the results section. The myosin working stroke δ was evaluated from the displacement between the average position of bound and unbound events. This procedure provides a correct measurement of the working stroke only if the probability of myosin attachment is uniformly distributed along the actin filament. As already evidenced by Steffen et al. (20) and confirmed in our experiments (21), this is not the case (Fig. 1c): myosin binds stereo-specifically to the actin monomers, spaced ≈5.6 nm, and preferably to target zones spaced 36 nm (half of the actin helix period). When evaluating the working stroke, care was taken to average this effect to zero acquiring data as described by Steffen et al. (20).
are the position variance of the unbound and bound state, respectively. km was obtained from experiments in which a combined trap stiffness (kt) in the range 0.05–0.1 pN/nm and pretensioning above 3 pN were used. The influence of compliant links is discussed in the results section. The myosin working stroke δ was evaluated from the displacement between the average position of bound and unbound events. This procedure provides a correct measurement of the working stroke only if the probability of myosin attachment is uniformly distributed along the actin filament. As already evidenced by Steffen et al. (20) and confirmed in our experiments (21), this is not the case (Fig. 1c): myosin binds stereo-specifically to the actin monomers, spaced ≈5.6 nm, and preferably to target zones spaced 36 nm (half of the actin helix period). When evaluating the working stroke, care was taken to average this effect to zero acquiring data as described by Steffen et al. (20).
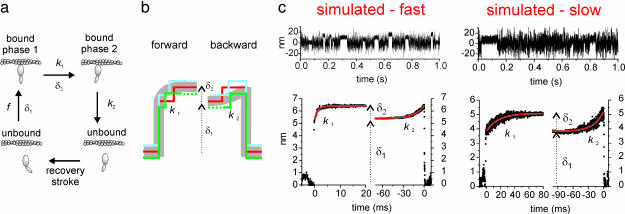
Two-Step Simulations and Analysis. Experimental data were analyzed by using an ensemble averaging method (discussed below) to detect possible conformational changes of the myosin molecule within the bound state. The averaging method was tested by using Monte Carlo simulations of the Langevin equation of motion of the dumbbell when myosin was interacting with the actin filament. The dumbbell was modeled as a rigid actin filament connected by compliant links (stiffness kL/2) to two microbeads held by optical tweezers with stiffness kt/2. The myosin molecule was modeled as an elastic element with stiffness km (Fig. 1a). The acto-myosin chemo-mechanical cycle was modeled as a three-state stochastic process (unbound state, phase 1 of bound state, and phase 2 of bound state, with average durations toff, ton1, and ton2, respectively). The transitions rates between states were defined as follows: attachment rate f = 1/toff, transition rate 1 (from phase 1 to phase 2) k1 = 1/ton1, and transition rate 2 (from phase 2 to the unbound state) k2 = 1/ton2 (Fig. 2a). The transition from the unbound state to phase 1 of the bound state and the transition from phase 1 to phase 2 were both supposed to be associated with tension development in the myosin molecule, with a relaxation leading to step δ1 and δ2, respectively. For simplicity, the probability of myosin binding to actin was supposed to be uniformly distributed along the actin filament. Integration of the Langevin equation of motion was performed according to the method of Smith (25). The dumbbell position was calculated at time intervals Δ = 10–6 – 10–7 s, and time series were then resampled at a 20-kHz sample rate. We considered two sets of simulated data, which approximate the two myosin isoforms investigated. Values km = 1.1 pN/nm and k1 = 103 s–1 were used for the “fast” simulated isoform, whereas km = 0.37 pN/nm and k1 = 43 s–1 were used for the “slow” isoform (see Results). The other parameters k2 = 5–100 s–1, kL/2 = 2–6 pN/nm, kt = 0.05 pN/nm, δ1 = 3.4–5.5 nm, δ2 = 1.0–1.5 nm, a = 0.55 μm (bead radius), η = 10–3 N·s/m2 (fluid viscosity), and T = 295 K were common for both sets of simulations.
Fig. 2.
Two-step simulations and analysis. (a) Model of the acto-myosin chemo-mechanical cycle used in simulated data. (b) Single events (continuous green, red, and blue lines) are aligned at their beginning (forward average) or end (backward average). In the forward average, the last position value of each event is replicated (dotted lines) to match the length of the longest event (red). Symmetrically, the first position value is replicated in the backward average. Ensemble averages (gray lines) are obtained by averaging the events point by point. (c Upper) Position traces of the dumbbell motion when simulated fast and slow myosin were interacting with the actin filament. (c Lower) Corresponding ensemble averages. The two steps δ1 and δ2 and the rate constants of the two phases k1 = 1/ton1 and k2 = 1/ton2 were obtained by fitting forward and backward ensemble averages with the exponential functions δfw = δ1 + δ2[1 – exp(–t/ton1)] and δbw = δ1 + δ2 exp(t/ton2), respectively.
The beginning and the end of each interaction (detected by the variance hidden Markov method) were obtained with high time resolution by using a threshold algorithm acting on the signal variance (threshold level 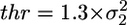 ), calculated with a running window (width W) proceeding point by point (50 μs) along the time series. Interactions detected by the hidden Markov method but characterized by a variance remaining above the threshold were discarded, resulting in zero false events for both sets of simulated data. Simulated data were used to calculate the mean (dt) and the standard deviation (σt) of time shifts between true and detected beginning of an event. The same procedure was followed with regard to the end of the events. σt represents the time resolution for detection of the event beginning or end. This resolution depended mostly on the ratio
), calculated with a running window (width W) proceeding point by point (50 μs) along the time series. Interactions detected by the hidden Markov method but characterized by a variance remaining above the threshold were discarded, resulting in zero false events for both sets of simulated data. Simulated data were used to calculate the mean (dt) and the standard deviation (σt) of time shifts between true and detected beginning of an event. The same procedure was followed with regard to the end of the events. σt represents the time resolution for detection of the event beginning or end. This resolution depended mostly on the ratio 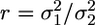 between free and bound states variance, which, in turn, depends on km and kL. For values r = 10 and r = 5 (which were typical for the fast and slow isoform, respectively), we found σt = 275 μs and σt = 860 μs, respectively. The values measured for dt (75 and 700 μs for r = 10 and r = 5, respectively) indicate a systematic phase shift between true and detected transitions. It is important to notice that such phase shift is systematic in all events. Therefore, the accuracy of the alignment of event beginnings or ends, as in the ensemble averages described below, does not depend on dt, but only on its standard deviation σt (see the supporting information, which is published on the PNAS web site).
between free and bound states variance, which, in turn, depends on km and kL. For values r = 10 and r = 5 (which were typical for the fast and slow isoform, respectively), we found σt = 275 μs and σt = 860 μs, respectively. The values measured for dt (75 and 700 μs for r = 10 and r = 5, respectively) indicate a systematic phase shift between true and detected transitions. It is important to notice that such phase shift is systematic in all events. Therefore, the accuracy of the alignment of event beginnings or ends, as in the ensemble averages described below, does not depend on dt, but only on its standard deviation σt (see the supporting information, which is published on the PNAS web site).
Ensemble averaging was performed similarly to the method of Veigel et al. (12) (Fig. 2b). Briefly, in the forward average procedure, the beginnings of bound events were aligned. Because the duration of each event is different, the last position value of each event was replicated to match the length of the longest event. The events were then averaged point by point. The backward average was obtained analogously, but in the backward time direction. Because the phase shift dt and its standard deviation σt on beginning and end positions are finite, event beginnings and ends were translated, respectively, by k × (dt + σt) and –k × (dt + σt) before averaging (k ≥ 1). This procedure assures that the prolonged position value really lies within the bound event. Moreover, the constant k should not exceed ton1/(dt + σt) or ton2/(dt + σt), to assure that the prolonged position value would really belong to the appropriate phase of the bound event.
Forward and backward averages were fitted with two exponentials to obtain ton1, ton2, δ1, and δ2 (Fig. 2c). Typical errors on the parameters obtained from fits were ≈0.5 ms for ton1 and ton2 and ≈0.02 nm for δ1 and δ2. However, we found that the uncertainty in the beginning and in the end of an event generally induced larger errors on ton1, ton2, δ1, and δ2. In fact, the parameters varied with higher standard deviations when k was varied between the lower (k ≥ 1) and upper [k ≤ min{ton1/(dt + σt); ton2/(dt + σt)}] limit. Typical deviations on ton1, ton2, δ1, and δ2 decreased with increasing ratios, r, between free and bound states variance and with increasing number of events, N. For fast isoform simulations (r ∼ 10 and N ∼ 1,500), we found Δton1 ≈ 0.1 ms, Δton2 ≈ 0.6 ms, and Δδ1 ≈Δδ2 ≈ 0.1 nm, whereas for slow isoform (r ∼ 5 and N ∼ 500), we found Δton1 ≈ Δton2 ≈ 5 ms and Δδ1 ≈ Δδ2 ≈ 0.2 nm.
The ensemble averaging method was finally tested by using simulations where only one step was present (δ2 = 0). In this case, analysis of simulated data gave a null second step within the error.
Results
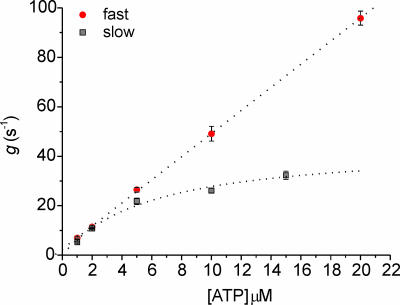
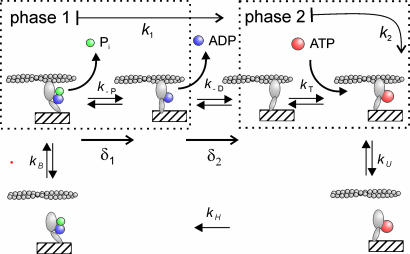
Kinetic Properties. Because shortening velocity of skeletal muscle fibers depends on the mechanical and kinetic properties of myosin (V = δ/ton), we investigated, at the single-molecule level, both aspects in the two myosin isoforms: slow (type 1 isoform from rat) and fast (type 2B isoform from mouse). Kinetics of the actomyosin interaction can be interpreted by following the Lymn–Taylor scheme (Scheme 1) (6). Here, M is myosin, A is actin, D is ADP, T is ATP, Pi is the phosphate group, and kH, kB, k-P, k–D, kT, and kU represent, respectively, the rate of ATP hydrolysis, myosin attachment to actin, phosphate release, ADP release, ATP binding to actomyosin, and myosin detachment from actin. According to this scheme, the detachment rate of the acto-myosin complex g = 1/ton can be limited either by the rate of ATP binding (kT) to the acto-myosin rigor complex (A·M) or by the preceding ADP release rate (k–D) [here, k–P and kU are supposed to be much faster than kT‡‡ and k–D, as suggested by experimental evidence (14)]. Because kT depends on [ATP], whereas k–D does not, we investigated which rate limits g by determining g of the slow and the fast isoforms at different ATP concentrations between 1 and 20 μM. In the fast isoform, g increased linearly with [ATP] through the 1–20 μM range [with a slope kT = (4.67 ± 0.03) × 106 M–1·s–1) and a correlation coefficient r = 0.99995 (see Fig. 3)]. On the contrary, in the slow isoform the relation was nonlinear and was well fitted (r = 0.993) by Michaelis–Menten function
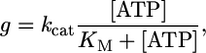 |
with kcat = 42.5 ± 2.6 s–1 and KM = 6.0 ± 0.9 μM.
Scheme 1.
Fig. 3.
The acto-myosin detachment rate g = 1/ton as a function of ATP concentration, for the two myosin isoforms investigated. Thirteen S1 molecules extracted from rat type 1 fibers (slow) and 10 extracted from mouse type 2B fibers (fast) were analyzed. Each point in the plot, depicted together with its standard error, is obtained from the average lifetime (ton) of some thousands of bound events.
In the fast isoform, the linear relation between g and [ATP] in the range 1–20 μM indicates that, in this experimental condition, g is limited only by kT. Therefore, k–D must be faster than the highest g that we were able to determine with this approach (≈100 s–1). In contrast, in the slow isoform the value of g increased much more slowly with [ATP] above 5 μM, and at saturating [ATP], it had to be defined by an ATP-independent rate, i.e., by k–D. The latter rate could be defined in the slow isoform as it corresponded to kcat (42.5 ± 2.6 s–1). In the range 1–5 μM [ATP] the relations g vs. [ATP] were linear and almost superimposed for both isoforms, suggesting very similar values of kT for the fast and slow isoform. The relation between g and [ATP] suggests that the differences in shortening velocity depend on k–D that is much higher in the fast than in the slow isoform, and not on kT that is very similar in the two isoforms.
Mechanical Properties. Because V of the slow and fast isoform could be modulated not only by their kinetic properties (ton) but also by the amplitude of the working stroke (δ), we determined the latter parameter in the two isoforms. The working stroke showed a large variation within the same isoform, ranging from 2 to 7 nm in the fast isoform and from 2 to 9 nm in the slow isoform. Such large spread might be attributed to the random attachment and orientation of the molecule on the glass surface, which modulate the amplitude of the working stroke, as reported by Tanaka et al. (26). The average size of the working stroke was smaller in the fast isoform (δ = 4.3 ± 0.6 nm) than in the slow isoform (δ = 6.0 ± 0.8 nm). A more pronounced difference in the mechanical properties of the two isoforms was found by measuring the stiffness of the myosin molecules km. In the limit of rigid actin-bead links, average km = 0.8 ± 0.1 pN/nm and km = 0.32 ± 0.06 pN/nm were found for the fast and slow isoform, respectively. These average values are underestimations of the real stiffness because of the presence of the compliant actin-bead links. Taking into account the stiffness of the actin-bead links, we estimate an average value for km in the range 0.9–1.4 pN/nm and 0.35–0.40 pN/nm for the fast and slow isoform, respectively. These results indicate that fast isoforms of skeletal muscle myosin are stiffer and produce a shorter stroke with respect to the slow isoforms. The higher stiffness of the fast isoform might be responsible for the smaller average stroke observed in our experiments. In fact, a higher stiffness restricts the possibility for the molecule to optimally orient itself when performing the working stroke. Because V = δ/ton, a smaller δ of the fast isoform goes in the opposite direction with respect to the higher shortening velocities of fast muscle fibers, indicating that only the kinetic properties (ton) of skeletal myosin isoforms are responsible for the differences in shortening velocities among skeletal muscle fibers.
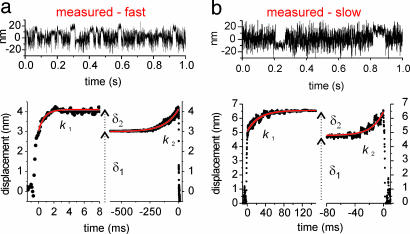
Two Steps in the Interaction. The relation of g vs. [ATP] provided several clues on the diversity among isoforms and suggested a major role for k–D in defining shortening velocity. However, a dramatic step forward was made possible when a careful analysis of the mechanical events surprisingly showed that the acto-myosin interaction was made of two steps. Experimental data were analyzed by using the ensemble averaging method discussed in Materials and Methods to reveal possible conformational changes of the myosin molecule when attached to actin. Such conformational changes could be detected with high temporal (≈300 μs for the fast isoform, and ≈1 ms for the slow one) and spatial (≈0.1 nm for the fast isoform, and ≈0.2 nm for the slow one) resolution, as demonstrated by Monte Carlo simulations of the Langevin equation of motion. The higher temporal resolution obtained with fast myosin was mainly due to the higher stiffness of the molecule, leading to a larger noise reduction during attachment with actin (see Materials and Methods and Discussion). This analysis evidenced that a first step δ1 (3.4 ± 0.5 nm and 5.2 ± 0.8 nm for the fast myosin isoform and slow myosin isoform, respectively) occurred very rapidly after myosin binding and was followed by a smaller step δ2 (1.0 ± 0.1 nm and 1.3 ± 0.1 nm for the fast myosin isoform and slow myosin isoform, respectively) in the same direction. δ1 and δ2 were found in all molecules investigated and in both isoforms. Such a finding indicates that, contrary to what believed so far, skeletal myosin II goes through two conformational changes during its interaction with actin. Thus, the bound state could be divided into two phases (with durations ton1 and ton2, respectively) corresponding to the two mechanical conformations of the myosin molecule when attached to actin.
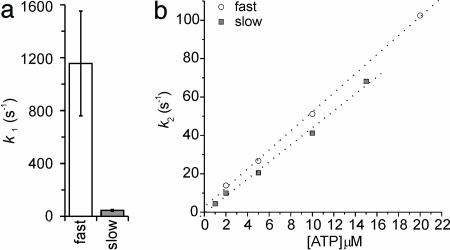
The durations of the two phases were found to be exponentially distributed, with apparently independent rate constants (Fig. 4). The duration of the first phase of the bound state (ton1) did not show a dependence on ATP concentration, whereas it was remarkably different between the two isoforms. The slow isoform showed an average rate constant k1 = 1/ton1 (42.8 ± 5.9 s–1) ≈20–30 times smaller than the fast isoform (1,156 ± 396 s–1) (Fig. 5a). The rate constant of the second phase of the bound state k2 = 1/ton2 showed, on the opposite, a linear dependence on ATP concentration (Fig. 5b) very similar in the two isoforms. Linear regression on experimental data gave a slope k2 = (5.0 ± 0.1) × 106 M–1·s–1 (r = 0.9996) and k2 = (4.4 ± 0.2) × 106 M–1·s–1 (r = 0.997) for the fast isoform and slow isoform, respectively.
Fig. 4.
The double step of skeletal muscle myosin is revealed by analysis of ensemble averages of bound events. In a Upper and b Upper, position traces of the dumbbell motion when myosin was interacting with the actin filament were obtained by using fast and slow myosin in the presence of 20 and 15 μM [ATP], respectively. The lower part shows forward and backward ensemble averages obtained by averaging 670 and 453 events using fast and slow myosin in the presence of 2 and 15 μM [ATP], respectively. Different time scales are used for forward and backward averages. Fitting parameters are k1 = 1,370 s–1, k2 = 8.7 s–1, δ1 = 3.0 nm, and δ2 = 1.1 nm for the fast isoform, and k1 = 40 s–1, k2 = 63 s–1, δ1 = 4.9 nm, and δ2 = 1.6 nm for the slow isoform.
Fig. 5.
Kinetics of the two phases of bound state. (a) The average rate of the first phase of the bound state (k1) was 1,156 ± 396 s–1 and 42.8 ± 5.9 s–1 for the fast isoform and slow isoform, respectively. (b) The rate of the second phase of bound state (k2) was linearly dependent on ATP concentration for both isoforms.
Discussion
Our data show two mechanical steps in the interaction of skeletal muscle myosin with actin. The data also indicate that the duration of the first phase of the bound state is related to the rate of ADP release from acto-myosin (k–D), whereas the duration of the second phase is related to the rate of ATP binding to acto-myosin (kT). In fact, the lack of ATP dependence for the first phase indicates that it ends before the formation of the rigor state (A·M) and, thus, that it must be determined by the transitions between A·M·D·Pi → A·M·D → A·M states. A significant effect of Pi release from acto-myosin in defining ton1 is very unlikely; in fact, Pi release is expected to occur very rapidly after acto-myosin binding and it is probably associated with development of the first step δ1 (14) (which is faster than our time resolution). Biochemical studies indicate that k–D defines the shortening velocity of skeletal muscle fibers by limiting the rate of actomyosin dissociation (g) (8, 9). Consistently, in the slow isoform, the rate of the first phase of the bound state, k1 (42.8 ± 5.9 s–1), is almost identical to kcat (42.5 ± 2.6 s–1, as determined from data in Fig. 3), namely the maximum g at saturating ATP concentrations. This finding is in full agreement with the conclusion that the first phase of the bound state is the rate-limiting step for the velocity of actomyosin dissociation and is defined by k–D.
The rate constant k1 has also the right order of magnitude to limit velocity of sliding of actin filaments on myosin (Vf) as measured in in vitro motility assays at saturating 2 mM ATP concentration (2). The detachment rate of the acto-myosin complex (1/ton) can be determined from Vf = δ/ton and compared with k1. We can use the δ values determined in the present work (4.3 and 6.0 nm for fast isoform and slow isoform, respectively) and the Vf values (2,011 and 312 nm·s–1 for fast isoforms and slow isoforms, respectively) previously determined for myosin at the same temperature and ionic strength (2) and estimated in this work for myosin S1 (27). The calculated values of 1/ton = Vf/δ result comparable (52 s–1 for the slow isoform, and 467 s–1 for the fast isoform) with the values of k1 for the same isoforms.
For the above reasons, the second step δ2, which marks the transition between the two phases, is very likely to be a conformational change associated with ADP release.
The rate of the second phase of bound state k2 is found to be dependent on ATP concentration and must, therefore, include the reactions A·M → A·M·ATP → A+M·ATP. In experiments of single-molecule mechanics the ATP concentration is low, and k2 is mainly limited by kT (14). This fact is confirmed by the observation that the relation between k2 and [ATP] is linear. The values of k2 reported here are fully consistent with previous determinations of the rate of ATP-induced acto-myosin dissociation by single-molecule mechanics in chicken myosin (7.6 × 106 M–1·s–1) (28) and in rabbit cardiac myosin (5.1 × 106 M–1·s–1 and 6.1 × 106 M–1·s–1 for V3 and V1, respectively) (29).
Previous attempts to detect a second step within the interaction cycle of skeletal muscle myosin failed, probably because of limited temporal and spatial resolution (11). A substantial limit in temporal resolution is set by the accuracy of detecting transitions between bound and unbound states, which scales with the overall stiffness of the dumbbell with bound myosin (see Materials and Methods and the supporting information). Previous attempts were made by using compliant actin-bead links [0.26 pN/nm reported by Veigel et al. (11) using NEM-myosin links, compared with 2–6 pN/nm of biotin-avidin links used in our experiments], resulting in lower temporal resolution. Detection of the second step of skeletal myosin also requires high spatial resolution, which depends primarily on the number of averaged events (N). Spatial resolution scales roughly as 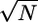 : the high attachment rate of the fast myosin isoform used in our experiments easily allows the collection of thousands of interactions and the attainment of a spatial resolution of ≈0.1 nm. Finally, an accurate data analysis assuring near-zero false events and a proper determination of the beginning and end of acto-myosin interactions is essential to prevent the introduction of additional sources of error.
: the high attachment rate of the fast myosin isoform used in our experiments easily allows the collection of thousands of interactions and the attainment of a spatial resolution of ≈0.1 nm. Finally, an accurate data analysis assuring near-zero false events and a proper determination of the beginning and end of acto-myosin interactions is essential to prevent the introduction of additional sources of error.
Structural studies have not shown a different conformation of skeletal muscle myosin with and without ADP bound, supporting the lack of a conformational change in the molecule upon ADP release (15, 30). However, structural studies might not be able to asses the ADP-bound state actually involved in the ADP-dependent conformational change. In fact, it has been suggested that two ADP-bound states exist, because slow myosin isomerization follows strong actin binding and precedes ADP release (A·M*·D → A·M·D → A·M+D) (31). For structural analysis, the ADP-bound state must be obtained by adding ADP to the system. The A·M*·D state cannot be obtained by reversal of the chemo-mechanical cycle from rigor state (A·M), but only through the hydrolysis of ATP by acto-myosin. Therefore, structural studies can assess only the A·M·D state and not the preceding A·M*·D state.
Overall, our findings can be easily summarized in the sequence of biochemical and mechanical events within the actomyosin interaction cycle shown in Fig. 6 consistent with what has been suggested by Veigel et al. (11) for myosin I, myosin V (12), and smooth muscle myosin (13). The large differences among the members of the myosin family appear to be solely due to variations in the rates and equilibrium constants of the acto-myosin interaction cycle and not to the existence of different molecular mechanisms. The double-step mechanism provides the molecular and kinetic basis for the wide variation in shortening velocity among skeletal isoforms, suggesting that the rate of ADP release defines V. Finally, the presence of a second mechanical event in the working stroke of skeletal muscle myosin provides a potential molecular basis for the strain dependence of the kinetics of acto-myosin interaction, as it has been demonstrated for smooth muscle myosin (13) and hypothesized in a comprehensive model of acto-myosin interaction (32).
Fig. 6.
The proposed mechanism of chemo-mechanical transduction for skeletal muscle myosin II. The mechanical states within the acto-myosin cycle and the corresponding transition rates are represented. Separation of the two phases of the bound state is defined by development of the second step δ2. A possible correspondence between mechanical and chemical states is also depicted. This correspondence enables us to link mechanical and chemical transition rates.
Supplementary Material
Acknowledgments
We thank Francesco Vanzi for stimulating discussion and suggestions on the paper and Daniela Romano for contributions in the preliminary phase of the work. This work was performed under European Union Contract RII3-CT-2003-506350 and supported by the Italian Ministry of University and Research (PRIN 2002 and 2004) and European Union Contract QLK6-CT2001-00323.
Author contributions: M. Capitanio, F.S.P., and R.B. designed research; M. Capitanio, M. Canepari, P.C., R.C., and M.M. performed research; M. Capitanio analyzed data; and M. Capitanio, V.L., and R.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Footnotes
Roman numerals indicate the myosin class, whereas Arabic numerals indicate the myosin II heavy chain isoform.
At the low ATP concentration used in the experiments (1–20 μM).
References
- 1.Schiaffino, S. & Reggiani, C. (1996) Physiol. Rev. 76, 371–423. [DOI] [PubMed] [Google Scholar]
- 2.Pellegrino, M. A., Canepari, M., Rossi, R., D'Antona, G., Reggiani, C. & Bottinelli, R. (2003) J. Physiol. 546, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill, A. (1938) Proc. R. Soc. London Ser. B 126, 136–195. [Google Scholar]
- 4.Kushmerick, M. J. & Davies, R. E. (1969) Proc. R. Soc. London Ser. B 174, 315–353. [DOI] [PubMed] [Google Scholar]
- 5.Fenn, W. O. (1923) J. Physiol. (London) 58, 175–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lymn, R. W. & Taylor, E. W. (1971) Biochemistry 10, 4617–4624. [DOI] [PubMed] [Google Scholar]
- 7.Huxley, H. E. (1990) J. Biol. Chem. 265, 8347–8350. [PubMed] [Google Scholar]
- 8.Siemankowski, R. F., Wiseman, M. O. & White, H. D. (1985) Proc. Natl. Acad. Sci. USA 82, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss, S., Rossi, R., Pellegrino, M. A., Bottinelli, R. & Geeves, M. A. (2001) J. Biol. Chem. 276, 45902–45908. [DOI] [PubMed] [Google Scholar]
- 10.Nyitrai, M. & Geeves, M. A. (2004) Philos. Trans. R. Soc. London B 359, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veigel, C., Coluccio, L. M., Jontes, J. D., Sparrow, J. C., Milligan, R. A. & Molloy, J. E. (1999) Nature 398, 530–533. [DOI] [PubMed] [Google Scholar]
- 12.Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. (2002) Nat. Cell Biol. 4, 59–65. [DOI] [PubMed] [Google Scholar]
- 13.Veigel, C., Molloy, J. E., Schmitz, S. & Kendrick-Jones, J. (2003) Nat. Cell Biol. 5, 980–986. [DOI] [PubMed] [Google Scholar]
- 14.Howard, J. (2001) Mechanics of Motor Proteins and the Cytoskeleton (Sinauer, Sunderland, MA).
- 15.Gulick, A. M., Bauer, C. B., Thoden, J. B. & Rayment, I. (1997) Biochemistry 36, 11619–11628. [DOI] [PubMed] [Google Scholar]
- 16.Finer, J. T., Simmons, R. M. & Spudich, J. A. (1994) Nature 368, 113–119. [DOI] [PubMed] [Google Scholar]
- 17.Canepari, M., Rossi, R., Pellegrino, M. A., Bottinelli, R., Schiaffino, S. & Reggiani, C. (2000) J. Muscle Res. Cell Motil. 21, 375–382. [DOI] [PubMed] [Google Scholar]
- 18.Harada, Y., Noguchi, A., Kishino, A. & Yanagida, T. (1987) Nature 326, 805–808. [DOI] [PubMed] [Google Scholar]
- 19.Ishijima, A., Kojima, H., Funatsu, T., Tokunaga, M., Higuchi, H., Tanaka, H. & Yanagida, T. (1998) Cell 92, 161–171. [DOI] [PubMed] [Google Scholar]
- 20.Steffen, W., Smith, D., Simmons, R. & Sleep, J. (2001) Proc. Natl. Acad. Sci. USA 98, 14949–14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capitanio, M., Cicchi, R. & Pavone, F. S. (2005) Eur. Phys. J. B 46, 1–8. [Google Scholar]
- 22.Smith, D. A., Steffen, W., Simmons, R. M. & Sleep, J. (2001) Biophys. J. 81, 2795–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capitanio, M., Romano, G., Ballerini, R., Giuntini, M. & Pavone, F. S. (2002) Rev. Sci. Instrum. 73, 1687–1696. [Google Scholar]
- 24.Molloy, J. E., Burns, J. E., Kendrick-Jones, J., Tregear, R. T. & White, D. C. S. (1995) Nature 378, 209–212. [DOI] [PubMed] [Google Scholar]
- 25.Smith, D. A. (1998) Biophys. J. 75, 2996–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka, H., Ishijima, A., Honda, M., Saito, K. & Yanagida, T. (1998) Biophys. J. 75, 1886–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyoshima, Y. Y., Kron, S. J., McNally, E. M., Niebling, K. R., Toyoshima, C. & Spudich, J. A. (1987) Nature 328, 536–539. [DOI] [PubMed] [Google Scholar]
- 28.Baker, J. E., Brosseau, C., Joel, P. B. & Warshaw, D. M. (2002) Biophys. J. 82, 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmiter, K. A., Tyska, M. J., Dupuis, D. E., Alpert, N. R. & Warshaw, D. M. (1999) J. Physiol. 519, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenfeld, S. S., Xing, J., Whitaker, M., Cheung, H. C., Brown, F., Wells, A., Milligan, R. A. & Sweeney, H. L. (2000) J. Biol. Chem. 275, 25418–25426. [DOI] [PubMed] [Google Scholar]
- 31.Sleep, J. A. & Hutton, R. L. (1980) Biochemistry 19, 1276–1283. [DOI] [PubMed] [Google Scholar]
- 32.Smith, D. A. & Geeves, M. A. (1995) Biophys. J. 69, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.