Abstract
Microorganisms are spectacularly diverse phylogenetically, but available estimates of their species richness are vague and problematic. For example, for comparable environments, the estimated numbers of species range from a few dozen or hundreds to tens of thousands and even half a million. Such estimates provide no baseline information on either local or global microbial species richness. We argue that this uncertainty is due in large part to the way statistical tools are used, if not indeed misused, in biodiversity research. Here we develop a powerful synthetic statistical approach to quantify biodiversity. It provides statistically sound estimates of microbial richness at any level of taxonomic hierarchy. We apply this approach to a large original 16S rRNA dataset on marine bacterial diversity and show that the number of bacterial species in a sample from marine sediments is (2.4 ± 0.5 SE) × 103. We argue that our methodology provides estimates of microbial richness that are reliable and general, have biologically meaningful SEs, and meet other fundamental statistical standards. This approach can be an essential tool in biodiversity research, and the estimates of microbial richness presented here can serve as a baseline in microbial diversity studies.
Keywords: global biodiversity, microorganisms, number of species
The number of microbial species in nature may be in the millions (1), but most have never been observed or otherwise detected; the existence of these species is predicted. Regrettably, the available predictions are essentially guesswork, conjectured from high (but equally uncertain) estimates of local microbial species richness. The latter cannot be measured directly, for the same reason that the global diversity has yet to be quantified; it's simply too large for the methodology available. [There are several measures or indices of diversity in a population (2), but here we focus on the number of species or species richness.] The best tool for microbial detection is the rRNA approach (3), but even large 16S rRNA clone libraries seem to capture only a small fraction of the original richness. Thus, the number of species in all but the simplest communities can only be estimated statistically, typically on the basis of a small subset of species (or their rRNA sequences) observed directly. Remarkably, even for samples obtained from similar environments (e.g., soils), such estimates vary widely: from a few dozen and hundreds (4, 5) to tens of thousands (6) to half a million (7). Clearly, the validity of such estimates is questionable. The quality of microbial richness predictions is however an important issue as they serve as a basis for all of the paradigms of biodiversity, its role, function, and meaning. It is therefore of principal interest to know the true extent of microbial diversity, starting from that in a single environmental sample. The question therefore is: what is the total number of microbial species in a sample, habitat, and biosphere?
Two approaches have been developed and used to answer this question. Historically first was the idea to use parametric distributions to approximate the frequency distribution of captured species, and to project the given distribution so as to estimate how many more species must be present in the community to account for the empirically collected data (8). This powerful tool has often not been used to its potential. While there is an infinite number of candidate parametric distributions, only one, the lognormal, has been commonly used. This choice was apparently based largely on theoretical considerations (6), but it has been frequently challenged (9). Furthermore, microbial data are frequencies of specific PCR products and clones in rRNA gene libraries, which may have little to do with the frequency distribution of real species in nature, even if the latter is indeed lognormal. In short, there is no convincing a priori reason to rely exclusively on the lognormal distribution to predict the number of microbial species. Perhaps more importantly, the applications of the lognormal and a handful of other distributions tested on occasion [the Poisson, negative binomial, and inverse Gaussian-mixed Poisson (10)] have often been statistically incorrect. To the best of our knowledge, previous applications in this area did not use maximum likelihood (ML) estimation of model parameters, reliable goodness-of-fit assessment, or correct ML SEs. In addition, previous literature has sometimes failed to take into account relevant existing and current research in mathematical statistics, and the inconclusive nature of theoretical justifications of the choice of parametric model. The published applications of parametric distributions to estimate species richness have erred in other important ways as well, which we discuss later in this paper.
The second group of species richness estimation methods uses coverage-based nonparametric estimators, such as Chao's estimators ACE and ACE1 (11). This approach dominates the landscape of microbial diversity research (5) but in many cases may be inappropriate for the purpose. To perform well, coverage-based nonparametric estimators require a large empirical database that covers the total diversity well (10). In case of microbial communities, this condition is often not met because, save for a few exceptions, even the largest rRNA gene libraries capture only a small fraction of all of the species. As a result, coverage-based nonparametric estimates of microbial richness are likely to underestimate the true diversity.
In the biological literature, rarefaction analysis is often used to address this issue (12). Such analysis is not counterintuitive and may have heuristic value (but see ref. 13 for an early critique). However, the theoretical foundations of resampling methods such as rarefaction, the jackknife, bootstrap, etc., in the problem of species richness estimation are not yet established. Indeed the problem may violate certain technical regularity conditions for the validity (asymptotic convergence) of resampling methods, at least without careful modifications of these methods (see, e.g., ref. 14); this is an open problem in mathematical statistics. Fortunately, we do not require resampling methods for variance estimation (SEs), because classical asymptotic theory provides direct formulas for SEs for both parametric and coverage-based nonparametric estimators. Furthermore, our model selection and choice of right truncation point (maximum frequency to be analyzed) are based on goodness-of-fit statistics rather than resampling. A new methodology, nonparametric ML estimation (which we have not used here) does appear to require resampling for variance estimation (15); this paper also includes a careful application of the bootstrap, and further theoretical development in mathematical statistics will be needed to justify and/or modify resampling methods for this application.
In the end, microbial diversity research has generated a substantial pool of rRNA richness data but has not gathered sufficient statistical resources to use these data to quantify the total microbial species richness. We think that this is the principal reason why the available estimates of microbial richness differ so dramatically as to provide essentially no baseline information. Here we develop a comparative approach that employs several parametric and nonparametric tools, including all those used to date and also two that are new to biodiversity research (the Pareto-mixed and mixture-of-exponentials-mixed Poisson distributions). We analyze the performance of all parametric models and nonparametric estimators on all possible right-truncated subsets of the data, and we choose the best performer on the basis of (i) two goodness-of-fit tests, (ii) ability to produce a biologically meaningful SE, and (iii) use of the maximum amount of the empirical species frequency data (largest right truncation point). [Another very useful approach to model selection is based on information-theoretic assessments such as the Akaike information criterion (16). However, in our view these methods have not yet achieved the flexibility needed to address the multifaceted model-selection problem faced here.] To carry out the ML computations, we constructed a new, accelerated expectation-maximization algorithm (15), with locally adaptive approximations to the distribution functions (particularly the lognormal- and Pareto-mixed Poisson). This algorithm allows us to obtain correct ML estimates of the parameters and to compute (asymptotically) correct SEs, simultaneously for all parametric models. We use the newly available software spade (17) for the coverage-based nonparametric estimates. We apply this strategy to a large original 16S rRNA survey of bacteria in a marine sediment sample, and we analyze the amount of “missing” diversity at different phylogenetic levels, from operational taxonomic units (OTUs) combining very similar organisms to OTUs representing large clades (99–60% sequence identity as cut-off values). For each identity cut-off value, we find that there is at least one model with acceptable goodness-of-fit and SE, and we choose this model to estimate the sample's microbial richness. We argue that the resulting estimates are accurate and reliable, and can therefore be used as a baseline in microbial diversity research.
Methods
Microbial samples came from an intertidal sand flat in Massachusetts Bay, near the Marine Science Center of Northeastern University (Nahant, MA). We collected an undisturbed core of sediment 15 cm deep and 13 cm in diameter, thoroughly mixed it, and subsampled the mix. We extracted DNA from a 5-g subsample after ref. 18, PCR-amplified the 16S rRNA gene using 27F and 1492R primers (19), and cloned and sequenced the PCR products. After manual editing and elimination of potentially chimeric sequences (7% of total number), the 16S rRNA gene sequences were grouped into OTUs based on 99%, 98%, 97%, 96%, 95%, 90%, 80%, 70%, 60%, and 50% sequence similarity cut-off values. This grouping was achieved by first making all possible pair-wise sequence alignments by using clustalw at default settings, and calculating % sequence identities, followed by clustering the sequences into OTUs by using the unweighted pair group method with arithmetic mean as implemented in the oc clustering program (20). The OTU grouping was checked manually to verify that all OTUs were assembled at the cut-off level desired. The number of OTUs and their frequencies at each cut-off value became the subject of statistical analyses.
We applied two families of statistical procedures to these frequency data; for a summary of the statistical theory see ref. 10. In the first, we use six parametric models (the ordinary Poisson and the gamma, inverse Gaussian, lognormal, Pareto, and mixture-of-2-exponentials mixed Poisson), and we fit each to the frequency data by the method of ML. The ordinary (unmixed) Poisson assumes equal species abundances, and the gamma, inverse Gaussian, and lognormal are 2-parameter abundance distributions that have been applied in the literature (10) (although with approximate computations and without ML SEs). The latter two distributions are new to this problem. We selected a 2-parameter (shape + scale) Pareto for its ability to model extreme phenomena (such as very abundant or rare species). The mixture-of-2-exponentials attempts to represent the species abundances as a mixture (convex combination) of two groups or subpopulations, each represented by a different exponential abundance distribution; this is a 3-parameter model.
Typically no currently available parametric model will fit a complete dataset of this type, so we separate the observations into “rare” vs. “abundant” species, i.e., those with sample frequencies less than or equal to some right-truncation point, and those with frequencies above this point. We fit all models to all possible collections of “rare” frequencies (all possible right-truncation points), and calculated the ML SE, two χ-square goodness-of-fit statistics (one straightforward or “naïve,” and one with adjacent frequencies concatenated so as to achieve an expected frequency count of at least 5, and hence an asymptotically correct P value). All numeric computations used the same basic algorithm so as to yield directly comparable results. Finally, we selected the “best of the best” as the final parametric analysis, searching for the smallest SE, largest right-truncation point, and best goodness-of-fit.
The second family of procedures consists of the coverage-based nonparametric estimators (11, 21). The coverage of the sample is the fraction of the population represented by the species that have been discovered. These estimators start with a nonparametric coverage-based richness estimate, and further adapt nonparametrically to the degree of variability in the frequency counts; different estimators are recommended depending on the degree of variability observed (11). For the required computations, we used the software spade (17). We calculated these estimates and their SEs for the collections of rare frequencies corresponding to the right-truncation points resulting from the best parametric analyses. Because the coverage-based nonparametric estimators do not fit parametric models, goodness-of-fit does not apply. We selected the “best” coverage-based nonparametric estimator using recommendations based on findings in the research literature as summarized in the spade documentation.
Results
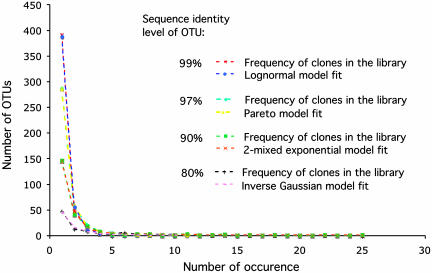
The bacterial assemblage recovered by the application of the rRNA approach appeared very diverse, as typical for the chemically diverse environment of marine tidal flats (Table 1). The 556 clones obtained grouped into 459, 405, 380, 351, 328, 233, 92, 16, and 1 OTUs, respectively, at the sequence identity cut-off values of 99%, 98%, 97%, 96%, 95%, 90%, 80%, 70%, and 60%. The frequency distributions of OTUs at selected levels of identity are given in Fig. 1.
Table 1. Microbial phyla detected among sequenced clones.
| Phylum | Representation, no. of clones | |
|---|---|---|
| 1 | Proteobacteria | |
| Alpha | 31 | |
| Beta | 1 | |
| Gamma | 139 | |
| Delta | 65 | |
| Epsilon | 11 | |
| Unclear affiliation | 26 | |
| 2 | Bacteriodetes | 133 |
| 3 | Chlorobi | 11 |
| 4 | Fibrobacteres | 2 |
| 5 | Gemmatimonadetes | 2 |
| 6 | Planctomycetes | 14 |
| 7 | Verrucomicrobia | 28 |
| 8 | Acidobacteria | 18 |
| 9 | Cyanobacteria | 26 |
| 10 | Spirochaetes | 2 |
| 11 | Fusobacteria | 1 |
| 12 | Firmicutes | 3 |
| 13 | Actinobacteria | 11 |
| 14 | Chloroflexi | 11 |
| 15 | Aquificae | 1 |
| 16 | OP3 | 1 |
| 17 | WS3 | 3 |
| 18 | TM6 | 1 |
| 19 | OD1 | 4 |
| 20 | OP1 | 1 |
| 21 | SR1 | 2 |
| 22 | Unclear affiliation | 8 |
Fig. 1.
Frequency distribution of OTUs in the clone library versus parametric models' fitted values. The fitted and empirical values are extremely close, illustrating quality fits.
Six parametric models were tested for their ability to describe the probability distribution of the OTUs' frequencies, with OTUs defined as clusters with varying rRNA identity (99% to 70% in 8 steps). There appeared to be no hands-down winner as no single model performed universally well at all OTU identity levels. Two models, the Poisson (equal species sizes) and the negative binomial (gamma-mixed Poisson), provided no realistic estimates of microbial richness with meaningful SEs, or exhibited unacceptable goodness-of-fit, or both (data not shown). The performance of the other four parametric models depended on the level of OTU grouping. The lognormal model appeared to be optimal when the number of OTUs was the highest (99% sequence identity cut-off value) and the frequency distribution had a long upward tail due to a large number of singletons (Fig. 1). As the frequency distribution gradually became long tailed along both the horizontal and vertical axes, the best parametric distribution to describe it became the Pareto at 98% and 97%, and 2-mixed exponential at 96%, 95%, and 90%. Finally, the probability distribution of OTUs at the 80% and 70% cut-off levels were best described by the inverse Gaussian model. Parametric maximum-likelihood-based estimates of the total richness and the corresponding SE, along with goodness-of-fit statistics, are given in Table 2. The estimates of the sample's richness based on two frequently used nonparametric estimators, ACE and ACE1, are also given in Table 2.
Table 2. Microbial richness of the sample.
| Estimate of the total sample's richness
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Parametric model
|
Nonparametric estimators
|
|||||||
| OTU boundary | Statistic | Sample's richness detected | Inverse Gaussian | Log normal | Pareto | 2-mixed exponential | ACE | ACE 1 |
| 99% | No. of OTU | 459 | 15,247,678 | 4,011 | 1,627 | 14,715 | 2,078 | 2,714 |
| SE | >1010 | 1,578 | NA | 64,376 | 278 | 530 | ||
| Asym. GOF | 0.41 | NA | 0.00 | 0.56 | NP | NP | ||
| Naïve GOF | 0.64 | 0.32 | 0.00 | 0.45 | NP | NP | ||
| TP | 6 | 6 | 6 | 6 | NP | NP | ||
| 98% | No. of OTU | 405 | 31,905 | 2,803 | 2,752 | 2,889 | 1,470 | 2,179 |
| SE | 194,114 | 1,017 | 589 | 1,078 | 197 | 494 | ||
| Asym. GOF | 0.96 | 0.55 | 0.51 | 0.60 | NP | NP | ||
| Naïve GOF | 0.07 | 0.05 | 0.08 | 0.03 | NP | NP | ||
| TP | 9 | 9 | 9 | 9 | NP | NP | ||
| 97% | No. of OTU | 380 | 3,351 | 2,565 | 2,434 | 2,181 | 1,385 | 2,296 |
| SE | 2,282 | 964 | 542 | 606 | 198 | 592 | ||
| Asym. GOF | 0.63 | 0.55 | 0.52 | 0.29 | NP | NP | ||
| Naïve GOF | 0.67 | 0.37 | 0.47 | 0.40 | NP | NP | ||
| TP | 8 | 12 | 12 | 10 | NP | NP | ||
| 96% | No. of OTU | 351 | 1,628 | 1,259 | 1,333 | 1,343 | 1,158 | 1,947 |
| SE | 575 | 289 | 195 | 145 | 155 | 785 | ||
| Asym. GOF | 0.22 | 0.07 | 0.06 | 0.14 | NP | NP | ||
| Naïve GOF | 0.48 | 0.31 | 0.26 | 0.00 | NP | NP | ||
| TP | 7 | 7 | 7 | 13 | NP | NP | ||
| 95% | No. of OTU | 328 | 14,831 | 2,028 | 1,683 | 1,522 | 1,081 | 1,818 |
| SE | 6,517 | 798 | 348 | 317 | 142 | 416 | ||
| Asym. GOF | 0.92 | 0.60 | 0.38 | 0.25 | NP | NP | ||
| Naïve GOF | 0.11 | 0.06 | 0.02 | 0.10 | NP | NP | ||
| TP | 12 | 12 | 10 | 12 | NP | NP | ||
| 90% | No. of OTU | 233 | 934 | 755 | 779 | 715 | 835 | 1,983 |
| SE | 212 | 206 | 111 | 94 | 124 | 1,228 | ||
| Asym. GOF | 0.91 | 0.69 | 0.60 | 0.36 | NP | NP | ||
| Naïve GOF | 0.07 | 0.04 | 0.02 | 0.22 | NP | NP | ||
| TP | 8 | 8 | 8 | 25 | NP | NP | ||
| 80% | No. of OTU | 92 | 282 | 241 | 240 | 286 | 176 | 236 |
| SE | 96 | 98 | 71 | 65 | 28 | 59 | ||
| Asym. GOF | 0.18 | 0.11 | 0.05 | 0.47 | NP | NP | ||
| Naïve GOF | 0.19 | 0.11 | 0.03 | 0.03 | NP | NP | ||
| TP | 10 | 10 | 25 | 26 | NP | NP | ||
| 70% | No. of OTU | 16 | 31 | 29 | 25 | 164,823 | 26 | 33 |
| SE | 16 | 17 | 6 | >109 | 9 | 17 | ||
| Asym. GOF | NA | NA | NA | NA | NP | NP | ||
| Naïve GOF | 0.03 | 0.03 | 0.04 | 0.05 | NP | NP | ||
| TP | 9 | 9 | 9 | 9 | NP | NP | ||
Boldface values represent the best estimates. Asymp. GOF, asymptotically correct goodness-of-fit; NA, not available; NP, estimation not possible.
Discussion
The main rationale for this research is that knowledge of microbial diversity is crucial for our understanding of the structure, function, and evolution of biological communities (1, 7, 22–25), but current estimates of numbers of microbial species are either vague or inaccurate. The coverage-based nonparametric estimators are widely used to estimate microbial richness (4, 5) because of their attractive simplicity. However, they likely underestimate this diversity, in accordance with theoretical expectations (10): the smaller and more diverse the empirical dataset is, the greater the underestimation. Our environmental clone library contains >500 clones and >400 hundred unique species and strains (defined as sequence clusters with 97–99% identity), and as such it is larger than the absolute majority of such libraries reported in literature (5). Even though we achieved a better coverage of the extant diversity than most previous studies, ACE and ACE1 estimate its richness at a 10–50% lower level than the parametric methods (Table 2). Typically, environmental clone libraries are a small percentage of the size of the one obtained here (5), and the use of coverage-based nonparametric estimators on such smaller datasets should lead to an even larger degree of underestimation of the “missing” diversity. This may well explain why microbial richness appears rather low whenever it is estimated with nonparametric tools (4, 5). Clearly, coverage-based nonparametric procedures are not the final word when the object is highly diverse microbial communities.
The applications of parametric distributions in microbial diversity research are few and not always well executed. In the Introduction, we pointed to the fact that these distributions have sometimes been used without optimal parameter estimation, goodness-of-fit assessment, or SE calculation. In some cases, parametric procedures have been used that are not well grounded in relevant statistical theory [although the stream of research in mathematical statistics on the problem dates from the 1940s and is now well developed (8)]. For example, the lognormal distribution is sometimes fitted by using suboptimal parameter estimates and without specifying an underlying stochastic sampling model that generates the observed frequency counts.
A notable parametric approach to estimate microbial richness is based on the reassociation kinetics of environmental DNA, pioneered in ref. 26. Recently, a further development of this approach produced a sensationally high estimate of almost 107 microbial species in a small sample of soil (27). We note that although this approach uses a very different (from rRNA surveys) kind of biological data, some of the statistical analyses are shared, and the requirement of statistical rigor stays the same. In particular, standard statistical practice requires that an estimate of a quantity such as species richness be accompanied by a SE that is derived from the underlying statistical model that generated the estimate, but the basis of the SE in this case is an approximation of unknown precision. (Analogous considerations hold for model choice and goodness-of-fit assessment.) In fact, we have observed that suboptimal (although not necessarily incorrect) methods can in some cases produce estimates of a given sample's diversity ranging from a few species to millions of species. Often, such disparate estimates have SEs that are orders of magnitude higher than the highest estimate itself. Dramatic estimates have high visibility, but such errors render the estimates unusable. Regrettably, statistically correct error calculation has not yet become standard practice in biodiversity research.
To advance the application of state-of-the-art statistical methods to practical species richness estimation, greater two-way communication is needed between the statistical community and biological scientists who require such methods. For example, there is a new, third family of procedures based on nonparametric estimation of abundance distributions, that has not yet been generally applied (15, 21, 28). These procedures are based on the same statistical model as we used here (the mixed Poisson), but the underlying species abundance (mixture) distribution is specified nonparametrically rather than parametrically. This approach has its own advantages and disadvantages, but it will ultimately yield another plausible set of analyses for comparison.
In this paper, we adopt an empirical approach of making no assumptions about the nature of a parametric distribution underlying microbial data. Instead, we systematically apply to our 16S rRNA dataset all of the models used to date, as well as two more that are new to the field of biodiversity. We then choose the one that fits the specific dataset the best, gives a reasonable SE, and uses as much of the clone data as possible. It is important to note that the choice of a “winner” is multidimensional, and in many cases there is no single choice. Identifying the “winner” thus involves a certain degree of subjectivity. Luckily, we face the dilemma of choice because we have too many well performing models, not because they all perform equally poorly. Therefore, we are truly looking for the best and not merely identifying the lesser evil. Our guiding principle is to first consider the goodness-of-fit, and if it is similarly good for more than one model, prefer the one giving the lowest SE. (For goodness-of-fit, we look at not only the χ2 P values but also at the actual fitted values at lower frequencies, especially frequency = 1). Following this logic, we suggest that at the level of bacterial strains (99% rRNA gene sequence similarity), our data can be best described by the lognormal distribution, which estimates the number of such strains in our sample at 4,011 ± 1,578 (SE). At the level of species (29), the Pareto model gives an overall better combination of the goodness-of-fit and SE. This model is well known in statistics (30), but it is used in biodiversity research here for the first time. This model predicts that our sediment sample contained at a minimum 2,434 ± 542 (SE) bacterial species.
Bacterial genera, families/classes, and phyla can be difficult to identify with a specific 16S sequence distance value, but as the first approximation 5%, 10%, and 20% have been proposed and used for the respective taxonomic groups above (24, 31, 32). At the 5% and 10% divergence levels as OTU criteria, the 2-mixed exponential model outperformed other parametric models used, and estimated the microbial richness at 1,522 ± 317 (95% cut-off value) and 715 ± 94 (90%). Interestingly, this parametric model also is new to biodiversity research. We also noted good performance of the inverse Gaussian model, especially at estimating microbial richness at the phylum level and above (80% and 70% sequence identity cut-off values; Table 2).
It is important to reemphasize that no single parametric model was of universal applicability, and specifics of the dataset dictated the nature of the model best describing the frequency distribution therein. Indiscriminate application of a single model to any dataset is likely to lead to erroneous results. We note that some of our estimates, notably those for the total number of bacterial species, are about an order of magnitude higher than typical nonparametric estimates (4, 5). The likely explanation is that the nonparametric procedures were not compared to other possible competitors, and the resulting values were underestimates. However, our estimate of microbial species richness is over an order of magnitude lower than the few parametric ones obtained earlier (6), but the latter came with no SEs, and this renders comparisons futile. We therefore argue that, on a per sample basis, the estimates of microbial richness provided here are the first statistically sound estimates of microbial diversity in general, and in marine sediments in particular.
Although a systematic evaluation of different parametric models seems to be necessary to correctly estimate the sample's microbial richness at the levels of strains, species, and genera, this requirement appears relaxed at the level of larger bacterial clades. It was interesting to see how the decrease in the cut-off values defining OTUs from 99% to 70%, and a corresponding increase in estimated sample coverage of the total diversity from ≈10% to 50%, led to progressively greater similarity in the estimates made by different methods. At what appears to be the phylum level (80% sequence identity), four parametric distributions (lognormal, Pareto, inverse Gaussian, and 2-mixed exponential) and both ACE and ACE1 estimators made essentially the same predictions (Table 2). This result shows that a clone library of about 500 clones captures enough of microbial diversity at the higher end of taxonomic hierarchy for the differences between individual models to become insignificant, and consequently many different approaches converge to predict the same microbial richness. If these estimates are correct, then even for OTUs defined as clusters with 70–80% sequence identity, our library did not reach saturation, and as much as 50% and more of the larger clades evaded our sequencing efforts. Because we detected representatives of approximately half of all of the bacterial phyla recognized or proposed to date (23, 24), it seems possible that the library might have in fact contained representatives of essentially all of the known phyla, albeit some at such low abundances that they went undetected. It would be particularly interesting to apply our approach to data from the existing literature and to compare the estimates of microbial richness.
We recognize that there are factors outside statistics that also limit our ability to fully estimate microbial richness. The holy grail of microbial biodiversity studies is to know microbial richness in nature, yet all of the available estimates, including ours, are those of rRNA genes in the clone library, and not the original sample. Obviously, before the latter can be achieved, biases associated with DNA extraction, PCR, and cloning have to be minimized. Until such time, the estimates of microbial richness such as provided here remain very conservative.
In conclusion, we developed a synthetic statistical approach to evaluate the amount of biological species on the basis of a small sample of all of the species. We also developed an algorithm for applying this approach to empirical frequency data. As a result, we estimate that between 2,000 and 3,000 bacterial species are present in a single sample of marine sediments. This number is conservative because of as-yet-unresolved biases of the rRNA approach.
Acknowledgments
We are grateful to Chesley Leslin and Valya Ilyin (Northeastern University) for their outstanding help in performing tens of thousands of pairwise sequence comparisons we needed to correctly cluster sequences into OTUs. We appreciate the assistance of Kathryn Barger with computing and critical comments. We thank two anonymous reviewers and members of the Editorial Board for valuable and constructive criticisms and suggestions that significantly improved the manuscript. This work was supported by National Science Foundation Grants OCE-0221267 and MCB-0348341 (to S.S.E.). S.-H.H. was supported by the postdoctoral fellowship program of the Korean Science and Engineering Foundation. This research was conducted with the use of Cornell Theory Center resources; the Center receives funding from Cornell University, New York State, federal agencies, foundations, and corporate partners. This is contribution no. 253 of the Marine Science Center of Northeastern University.
Author contributions: S.-H.H., J.B., and S.S.E. designed research; S.-H.H., J.B., S.-O.J., and S.S.E. performed research; S.-S.H., J.B., S.-O.J., and S.S.E. analyzed data; J.B. and S.S.E. contributed new reagents/analytic tools; and J.B. and S.S.E. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ML, maximum likelihood; ACE, abundance-based coverage estimator; OTU, operational taxonomic unit.
References
- 1.Tiedje, J. M. (1994) ASM News 60, 524–525. [Google Scholar]
- 2.Gove, J. H., Patil, G. P., Swindel, B. F. & Taillie, C. (1994) in Handbook of Statistics Volume 12: Environmental Statistics, eds. Patil, G. P., Rao, C. R. & Ross, N. P. (North–Holland/Elsevier, New York), pp. 409–462.
- 3.Olsen, G. J., Lane, D. J., Giovannoni, S. J., Pace, N. R. & Stahl, D. A. (1986) Annu. Rev. Microbiol. 40, 337–365. [DOI] [PubMed] [Google Scholar]
- 4.Hughes, J. B., Hellmann, J. J., Ricketts, T. H. & Bohannan, B. J. (2001) Appl. Environ. Microbiol. 67, 4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kemp, P. F. & Aller, J. Y. (2004) FEMS Microbiol. Ecol. 47, 161–177. [DOI] [PubMed] [Google Scholar]
- 6.Curtis, T. P., Sloan, W. T. & Scannell, J. W. (2002) Proc. Natl. Acad. Sci. USA 99, 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykhuizen, D. E. (1998) Antonie van Leeuwenhoek 73, 25–33. [DOI] [PubMed] [Google Scholar]
- 8.Bunge, J. & Fitzpatrick, M. (1993) J. Am. Stat. Assoc. 88, 364–373. [Google Scholar]
- 9.Williamson, M. & Gaston, K. J. (2005) J. Anim. Ecol. 74, 409–422. [Google Scholar]
- 10.Chao, A. & Bunge, J. (2002) Biometrics 58, 531–539. [DOI] [PubMed] [Google Scholar]
- 11.Chao, A. (2006) in Encyclopedia of Statistical Sciences, eds. Balakrishnan, C., Read, B. & Vidakovic, B. (Wiley, New York), in press.
- 12.Gotelli, N. J. & Colwell, R. K. (2001) Ecol. Lett. 4, 379–391. [Google Scholar]
- 13.Tipper, J. C. (1979) Paleobiology 5, 423–424. [Google Scholar]
- 14.Shao, J. & Tu, D. (1995) The Jackknife and Bootstrap (Springer, New York).
- 15.Bohning, D. & Schon, D. (2005) J. R. Stat. Soc. Ser. C 54, 721–738. [Google Scholar]
- 16.Burnham, K. P. & Anderson, D. R. (1998) Model Selection and Inference (Springer, New York).
- 17.Shen, T.-J., Chao, A. & Lin, C.-F. (2003) Ecology 84, 798–804. [Google Scholar]
- 18.Zhou, J., Bruns, M. A. & Tiedje, J. M. (1996) Appl. Environ. Microbiol. 62, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. J. (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds. Stackebrandt, E. & Goodfellow, M. (Wiley, Chichester, U.K.), pp. 115–175.
- 20.Siddiqui, A. S., Dengler, U. & Barton, G. J. (2001) Bioinformatics 17, 200–201. [DOI] [PubMed] [Google Scholar]
- 21.Mao, C. X. & Colwell, R. K. (2005) Ecology 86, 1143–1153. [Google Scholar]
- 22.Colwell, R. R. (1997) J. Ind. Microbiol. Biotechnol. 18, 302–307. [DOI] [PubMed] [Google Scholar]
- 23.Rappe, M. S. & Giovannoni, S. J. (2003) Annu. Rev. Microbiol. 57, 369–394. [DOI] [PubMed] [Google Scholar]
- 24.Schloss, P. D. & Handelsman, J. (2004) Microbiol. Mol. Biol. Rev. 68, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis, T. P. & Sloan, W. T. (2004) Curr. Opin. Microbiol. 7, 221–226. [DOI] [PubMed] [Google Scholar]
- 26.Torsvik, V., Goksoyr, J. & Daae, F. L. (1990) Appl. Environ. Microbiol. 56, 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gans, J., Wolinsky, M. & Dunbar, J. (2005) Science 309, 1387–1390. [DOI] [PubMed] [Google Scholar]
- 28.Mao, C. X. (2004) J. Am. Stat. Assoc. 99, 1108–1118. [Google Scholar]
- 29.Stackebrandt, E. & Goebel, B. M. (1994) Int. J. Syst. Bacteriol. 44, 846–849. [Google Scholar]
- 30.Perline, R. (2005) Stat. Sci. 20, 68–88. [Google Scholar]
- 31.Hugenholtz, P., Goebel, B. M. & Pace, N. R. (1998) J. Bacteriol. 180, 4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sait, M., Hugenholtz, P. & Janssen, P. H. (2002) Environ. Microbiol. 4, 654–666. [DOI] [PubMed] [Google Scholar]