Abstract
Phytobilins (light harvesting and photoreceptor pigments in higher plants, algae, and cyanobacteria) are synthesized from biliverdin IXα (BV) by ferredoxin-dependent bilin reductases (FDBRs). Phycocyanobilin:ferredoxin oxidoreductase (PcyA), one such FDBR, is a new class of radical enzymes that require neither cofactors nor metals and serially reduces the vinyl group of the D-ring and A-ring of BV using four electrons from ferredoxin to produce phycocyanobilin, one of the phytobilins. We have determined the crystal structure of PcyA from Synechocystis sp. PCC 6803 in complex with BV, revealing the first tertiary structure of an FDBR family member. PcyA is folded in a three-layer α/β/α sandwich structure, in which BV in a cyclic conformation is positioned between the β-sheet and C-terminal α-helices. The basic patch on the PcyA surface near the BV molecule may provide a binding site for acidic ferredoxin, allowing direct transfer of electrons to BV. The orientation of BV is definitely fixed in PcyA by several hydrophilic interactions and the shape of the BV binding pocket of PcyA. We propose the mechanism by which the sequential reduction of the D- and A-rings is controlled, where Asp-105, located between the two reduction sites, would play the central role by changing its conformation during the reaction. Homology modeling of other FDBRs based on the PcyA structure fits well with previous genetic and biochemical data, thereby providing a structural basis for the reaction mechanism of FDBRs.
Keywords: phytobilin, redox, FDBR family, phycobilin, phycobilisome
Phytobilins are linear tetrapyrrole compounds used as chromophores for light harvesting and photoreceptor proteins in higher plants, algae, and cyanobacteria (1, 2). In higher plants, phytochromobilin (PΦB), one of the phytobilins, is the chromophore of phytochrome, a red-light-sensitive photoreceptor involved in photoperiodic induction of flowering, chloroplast development, leaf senescence, and leaf abscission (3). In red algae, cryptophytes, and cyanobacteria, phycobilins, a major group of phytobilins, are used as chromophores of phycobiliproteins. Phycobiliproteins are assembled on the outer surface of the thylakoid membrane as a large light-harvesting complex called phycobilisome (4). The characteristic blue–green and red colors of cyanobacteria and red algae reflect the presence of phycobilisomes.
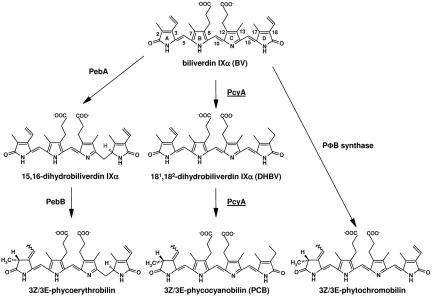
Biosynthesis of such phytobilins begins with the cleavage of the porphyrin ring of heme catalyzed by heme oxygenase (5). Biliverdin IXα (BV), a product of heme oxygenase, is further reduced by ferredoxin (Fd)-dependent bilin reductases (FDBRs) to phytobilins (1) (Fig. 1). FDBRs are found throughout oxygenic photosynthetic organisms and show weak sequence homology to each other (6). FDBRs are distinct from the NADPH-dependent biliverdin reductase found in mammals (7) and cyanobacteria (8) not only in their preference of Fd as a reductant but also in their amino acid sequences. Phycocyanobilin (PCB):Fd oxidoreductase (PcyA), a member of the FDBR family, is unique in that it catalyzes the reduction of BV by two sequential steps to produce 3Z/3E-PCB, one of major pigments in phycobilisomes. The first step is the reduction of the vinyl group of the BV D-ring to produce 181,182-dihydrobiliverdin IXα (181,182-DHBV), and the second step is the reduction of the A-ring of 181,182-DHBV to produce 3Z/3E-PCB (9) (Fig. 1). Each reduction step uses two electrons supplied by Fd. To achieve these sequential reductions, PcyA must possess a molecular structure that allows discrimination between the A- and D-rings of BV and control of the reaction sequence. Furthermore, low-temperature EPR spectroscopy has suggested that an organic radical is produced during the PcyA reaction (10). Many enzymes producing organic radical intermediates during catalysis possess metal and/or organic cofactors that mediate one-electron transfers from the reductant to the substrate (11). PcyA shows no such dependence on metals or organic cofactors, indicating that PcyA belongs to a novel class of radical enzymes (9, 10).
Fig. 1.
Biosynthetic pathway of bilin pigments by FDBRs. In higher plants, algae, and cyanobacteria, BV is reduced by FDBRs to produce phytobilins. PcyA reduces BV, with 181, 182-DHBV occurring as an intermediate.
In this study, we constructed an overexpression system for cyanobacterial PcyA and determined its crystal structure in complex with BV at 1.51-Å resolution. This crystallographic analysis revealed the first tertiary structure of an FDBR family member, suggesting how an FDBR recognizes its bilin substrate and Fd. On the basis of this structure, we propose a mechanism by which the sequential two-step reductions of BV might occur in PcyA.
Methods
Construction of PcyA Expression Plasmid. Sequence data for pcyA from Synechocystis sp. PCC 6803 was retrieved from CyanoBase (12). Two oligonucleotides were used for the PCR amplification of pcyA from Synechocystis sp. PCC 6803 chromosomal DNA by using ExTaq polymerase (Takara Biochemicals). The forward and reverse primers were, respectively, 5′-CATATGGCCGTCACTGATTTAAG-3′ and 5′-GGATCCGAGCTCATTGGATAACATCAAATAAGAC-3′ (the italics indicate the NdeI and BamHI sites). The PCR product was ligated into the pCR2.1-TOPO vector by using a TA cloning kit (Invitrogen), creating the plasmid pCR-pcyA. Sequence analysis verified that the construct was free of errors. The pcyA gene was excised by digestion with the restriction enzymes NdeI and BamHI (New England Biolabs) and ligated into the corresponding sites of the expression vector pET-21a (Novagen), creating the plasmid pET-21a-pcyA.
Expression and Purification of PcyA. Escherichia coli C41(DE3) (13) was transformed with pET-21a-pcyA, and the transformant was grown in 5 liters of TB medium with ampicillin (50 μg/ml) at 28°C. Induction with 500 μM isopropyl-β-d-thiogalactopyranoside was followed by growth for 16 h. The cells harvested by centrifugation were stocked at –30°C. All of the following steps were performed at 4°C or on ice. Frozen cells (14 g) were thawed and suspended in 50 ml of lysis buffer (100 mM NaCl/20 mM Tris-HCl, pH 8.0) and sonicated. The membrane fraction was removed by centrifugation at 27,000 × g for 30 min. The supernatant was subjected to the ammonium sulfate fractionation at 40% saturation and centrifuged at 27,000 × g for 20 min. The supernatant was loaded onto a HiPrep 16/10 Butyl FF column (Amersham Pharmacia Bioscience) equilibrated with 40% ammonium sulfate in 50 mM Tris-HCl, pH 8.0 (buffer A). The column was washed with buffer A, after which the protein was eluted with a decreasing linear gradient of 40% to 0% ammonium sulfate. Fractions containing PcyA were collected and dialyzed against 50 mM Tris-HCl, pH 8.0 (buffer B). The dialyzed sample was loaded onto a HiLoad 26/10 Q Sepharose HP column (Amersham Pharmacia Bioscience) equilibrated with buffer B. The column was washed with buffer B, and the protein was eluted with an increasing linear gradient of 0–400 mM NaCl. Fractions containing PcyA were concentrated, loaded onto a HiPrep 16/60 Sephacryl S-200 HR column (Amersham Pharmacia Bioscience) equilibrated with 100 mM NaCl/50 mM Tris-HCl, pH 8.0, and eluted with the same buffer. Fractions containing PcyA were monitored by SDS/PAGE analysis. Purified protein was concentrated up to 20 mg/ml by using Vivaspin (Viva Science) for crystallization.
Crystallization of PcyA–BV Complex. PcyA–BV complex was prepared by incubating equimolar PcyA and BV for 1 h on ice. Crystallization conditions for the PcyA–BV complex were screened by the hanging-drop vapor-diffusion method by using a WIZARD II kit (Jena Bioscience) at 293 K in the dark to avoid photochemical reactions. Solution containing PcyA–BV complex (11.5 mg/ml) was mixed with an equal volume of each reservoir solution and equilibrated. Plate crystals of PcyA–BV complex were obtained with a reservoir solution containing 2.0 M ammonium sulfate, 0.1 M sodium cacodylate (pH 7.0), and 0.2 M NaCl (condition A). Isomorphous crystals were also obtained with a reservoir solution containing 1.0 M sodium citrate, 0.1 M Tris-HCl (pH 7.0), and 0.2 M NaCl (condition B).
Data Collection and Structure Determination. PcyA–BV crystals obtained under condition A were soaked in crystallization solution containing glycerol up to 15% (vol/vol) as a cryoprotectant and flash-cooled with a nitrogen gas stream at 100 K. Heavy-atom derivatives were prepared by soaking PcyA–BV crystals obtained in condition B overnight in crystallization solution containing 10 mM KAu(CN)3 and Hg(CN)2, respectively. These derivative crystals were flash-cooled in the same way as for the native crystal. Diffraction data of the native crystal were collected at 100 K using synchrotron radiation (λ = 1.0000 Å) from the BL41XU beamline at SPring-8 and a Quantum 315 detector (Area Detector Systems). The hatch was kept dark during the data collection. Diffraction data for the two derivative crystals were collected at 100 K by using synchrotron radiation (λ = 0.9000 Å) from the BL44B2 beamline at SPring-8 and a Mar CCD detector (Mar USA, Evanston, IL). All diffraction data were processed, merged, and scaled with HKL2000 (14). Crystallographic statistics are summarized in Table 1.
Table 1. Data collection and phasing and refinement statistics.
| Native | Au derivative | Hg derivative | |
|---|---|---|---|
| Crystallographic data | |||
| Space group | P21212 | ||
| Unit cell dimensions, Å | |||
| a | 70.83 | 70.84 | 70.78 |
| b | 95.00 | 95.34 | 95.19 |
| c | 42.68 | 42.69 | 42.70 |
| Diffraction statistics | |||
| Resolution, Å | 1.51 | 2.0 | 1.9 |
| No. of observations | 357,569 | 147,360 | 164,266 |
| No. of unique reflections | 45,715 | 20,213 | 23,408 |
| Completeness,* % | 99.6 (98.0) | 99.8 (100.0) | 99.6 (99.4) |
| Mean Io/σ(I) | 10.9 | 10.5 | 12.3 |
| Rsym,*† % | 7.8 (33.5) | 8.9 (23.5) | 8.1 (28.5) |
| Phasing statistics | |||
| No. of heavy metal positions | — | 4 | 5 |
| Phasing power‡ (centric/acentric/anomalous) | —/—/— | 0.81/0.76/1.31 | 0.67/0.70/0.76 |
| Au—Hg phase combination | |||
| Mean figure of merit, final§ | 0.528 | ||
| Refinement statistics | |||
| Rcryst/Rfree,¶ % | 15.7/18.3 | ||
| No. of protein/biliverdin atoms | 2,025/43 | ||
| No. of solvent molecules (Na+/H2O) | 1/363 | ||
| rms deviations from ideal values | |||
| Bond lengths, Å | 0.017 | ||
| Bond angles, ° | 1.70 | ||
| Ramachandran plot | |||
| Most favored, % | 93.8 | ||
| Additionally allowed, % | 5.7 | ||
| Generously allowed, % | 0.5 |
Values in parentheses are for the outermost shells (1.56 to 1.51 Å for native, 2.07 to 2.00 Å for the Au derivative, and 1.97 to 1.90 Å for the Hg derivative).
Rsym = ΣhklΣi|Ii(hkl) - 〈I(hkl) 〉|/ΣhklΣi|Ii(hkl), 〈I(hkl) 〉 is the mean intensity for multiple recorded reflections.
Phasing power = rms (〈FH 〉/E), where FH is the calculated structure factor of the heavy atoms and E is the residual lack of closure. Isomorphous contributions for centric reflections (centric), acentric reflections (acentric), and anomalous contributions (anomalous) are separately shown.
Figure of merit = |F(hkl)best|/|F(hkl)| with F(hkl)best = ΣαP(α)Fhkl(α)/Σα|P (α).
Rcryst = Σ|Fobs(hkl) - Fcalc(hkl)|/Σ|Fobs(hkl)|. Rfree is the Rcryst calculate d for the 5% of the data set not included in the refinement.
Three gold binding sites were determined by the isomorphous and Bijvoet difference Patterson maps. Positions and occupancies of the three gold atoms were refined by using cns (15). The phases calculated by the SIRAS method yielded the Bijvoet difference Fourier maps, in which one additional gold and five mercury binding sites were found. Finally, phases were calculated by the MIRAS method by using the binding sites for the four gold and five mercury atoms. The resultant phases at 1.9 Å were improved and extended to 1.51 Å by density modification by using cns. The resultant electron density map was clear enough to define PcyA and BV. The model of PcyA was traced automatically by using arp/warp (16) and manually adjusted by using xtalview (17). BV, sodium ion, water molecules, and several multiple conformers of amino acid side chains were included in the model. The model was refined with cns and refmac5 (18) in the ccp4 package (19). Orientations of the amide side chains of Gln and Asn and the imidazole ring of His were carefully assigned on the basis of the temperature factors and hydrogen-bonding geometries. Phasing and refinement statistics are summarized in Table 1.
Results and Discussion
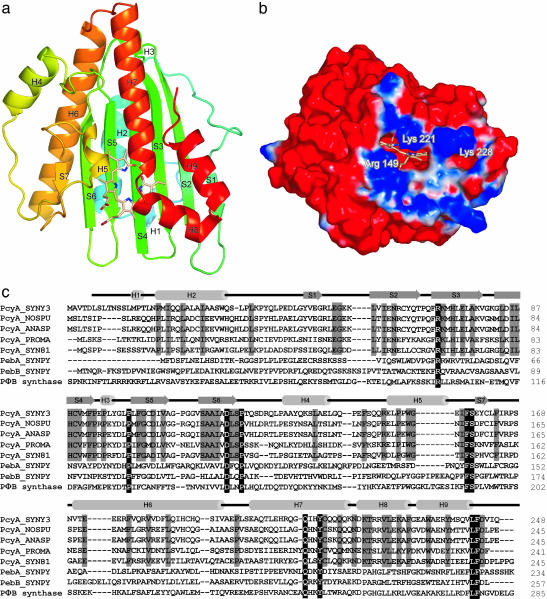
Overall Structure. The structure of PcyA–BV complex was determined by the MIRAS method and refined by using 1.51-Å resolution data to an R-factor of 0.157 and a free R-factor of 0.183 (Table 1). Except for five N-terminal residues, all residues of PcyA and BV in the complex were clearly visible in the electron density map. PcyA is folded in a three-layer α/β/α sandwich structure; four N-terminal α-helices (H1/H2/H4/H6), an anti-parallel β-sheet consisting of seven strands (S1-S7), and five C-terminal α-helices (H3/H5/H7/H8/H9) (Fig. 2a). BV is positioned in a cyclic conformation between the β-sheet and C-terminal α-helices. The five central β-strands (S2–S6) are longer than the edge strands (S1 and S7), such that this central region of the β-sheet forms a flattened and elongated BV binding pocket. A dali (20) search found that O2-dependent coproporphyrinogen III oxidases from yeast (21) and from the pathogenic protozoa Leishmania major (Protein Data Bank ID code 1VJU) have foldings similar to PcyA folding (rms deviations are 3.5 Å for 211 aa and 3.3 Å for 204 aa), even though their amino acid sequences share no significant similarity with that of PcyA. O2-dependent coproporphyrinogen oxidase is involved in porphyrin synthesis and catalyzes decarboxylation of coproporphyrinogen III to form the vinyl groups of protoporphyrinogen IX (21). The predicted substrate-binding pocket of this enzyme is located at a similar position to the BV binding site of PcyA, although the amino acid residues that make up their respective binding pockets are completely different.
Fig. 2.
Overall structure and electrostatic potential of the PcyA–BV complex. (a) Overall structure of the PcyA–BV complex. The ribbon model is displayed with the N-terminal side in blue and the C-terminal side in red. The BV molecule is shown as a stick model with carbon, oxygen, and nitrogen atoms in pale red, red, and blue, respectively. (b) Positive and negative surfaces are shown in blue and red, respectively. BV is shown as a stick model. Electrostatic potential was calculated with pymol and apbs (26), where solvent dielectric contribution (dielectric constant of solvent is 4) was taken into account. Basic residues conserved in PcyA are labeled on the surface. (c) Multiple sequence alignment of PcyA orthologs and other FDBRs. N- and C-terminal sequences of FDBRs extending beyond PcyA_SYNY3 are not shown. Completely conserved residues in the aligned sequences are depicted in dark shading with white letters. Residues conserved among the PcyA orthologs are depicted in gray shading with black letters. Secondary structures of PcyA are shown above the sequence alignment. SYNY3, Synechocystis sp. PCC 6803; NOSPU, Nostoc punctiforme; ANASP, Anabaena sp. PCC 7120; PROMA; Prochlorococcus marinus; SYN81, Synechococcus sp. WH 8102; SYNPY, Synechococcus sp. WH 8020. Database accession numbers are as follows: Protein Information Resource accession no. S76709 for PcyA_SYNY3; GenBank accession no. AF339057 for PcyA_NOSPU; GenBank accession no. AF339056 for PcyA_ANASP; National Center for Biotechnology Information (NCBI) accession no. AAK38599 for PcyA_PROMA; NCBI accession no. NP_897177 for PcyA_SYN81; NCBI accession no. AAK77915 for PebA_SYNPY; NCBI accession no. AAK77916 for PebB_SYNPY; NCBI accession no. Q9SR43 for PΦB synthase.
The electrostatic potential on the molecular surface is clearly divided into acidic and basic regions (Fig. 2b). A basic patch comprising Arg-52, Lys-79, Lys-82, Arg-149, Arg-166, Lys-221, Arg-224, and Lys-228 is located near to the two propionate groups of BV. Among these residues, Arg-149, Lys-221, and Lys-228 are completely conserved in PcyA orthologs (Fig. 2c). The acidic electron-transfer protein Fd has a negatively charged surface adjacent to the electron carrier site (22), and it is most likely that Fd binds to this basic patch. Two propionate groups of BV are exposed to the molecular surface, suggesting that Fd directly transfers electrons to BV bound to PcyA through the propionate groups.
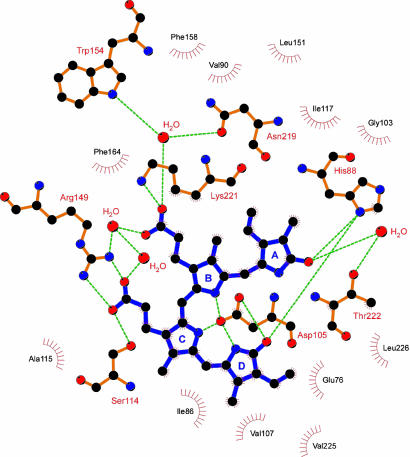
Substrate Recognition. The electron density of BV was so clear that its orientation and conformation were explicitly determined. There is neither a multiple conformation nor a multiple binding mode in BV. The D- and A-rings of BV are buried inside PcyA, whereas the two propionate groups bonded to the B- and C-rings are exposed to solution. Extensive hydrophobic contacts between PcyA and BV stabilize BV binding. In addition, all hydrophilic functional groups of BV are hydrogen-bonded and/or salt-bridged to PcyA (Fig. 3), resulting in tight and specific binding to PcyA. Specifically, the propionate group of the B-ring is salt-bridged to Lys-221 and is hydrogen-bonded through water molecules to Arg-149, Trp-154, and Asn-219, whereas the propionate group of the C-ring is salt-bridged to Arg-149 and is hydrogen-bonded to Ser-114. These residues fix the propionate groups of BV. Nδ of His-88 forms bifurcated hydrogen bonds with two lactam oxygen atoms of BV. The lactam oxygen atom of the A-ring is also hydrogen-bonded through a water molecule to Thr-222. The side chain of Asp-105 adopts two conformations. The major one forms two hydrogen bonds with the lactam group of D-ring and weakly hydrogen-bonds with the amide groups of the B- and C-rings (Fig. 4a). These interactions fix exactly the position and orientation of each pyrrole ring of BV.
Fig. 3.
Environment of the BV binding site. A schematic diagram of interactions between PcyA and BV is shown. Dashed lines indicate hydrogen bonds and salt bridges. The residues involved in van der Waals interaction with BV are also shown. For clarity, only the major conformation of Asp-105 is shown. Carbon, nitrogen, and oxygen atoms are colored in black, blue, and red, respectively. BV is colored in purple. This figure was prepared with ligplot (27).
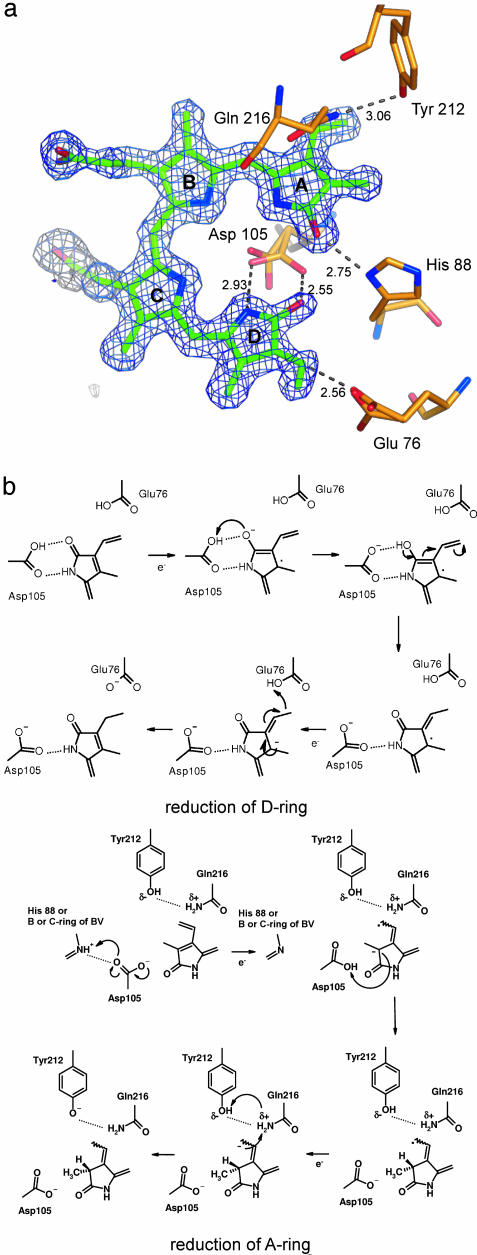
Fig. 4.
Proposed reaction mechanism of PcyA. (a) Close-up view of BV. Omit electron density map for BV (contoured at 2.5 σ) is superimposed on the stick model. The distances between the carboxyl group of Asp-105 in minor conformation and the lactam oxygen and nitrogen atoms of D-ring are 3.67 and 2.95 Å, respectively. (b) Proposed reaction mechanism catalyzed by PcyA. Reduction schemes at the D-ring (Upper) and at the A-ring (Lower) are shown.
Recognition of the face of BV by PcyA, which is essential for the sequential reductions of BV, appears to be achieved by detection of subtle asymmetry of side chain bonding to the pyrrole rings in the A- and D-rings; the positions of the methyl and vinyl groups of the A-ring are switched in the D-ring. The methyl group of the A-ring contacts with the side chains of His-88, Val-90, Tyr-212, and Phe-244, leaving little space for the vinyl group, whereas the space remains near the methyl group of the D-ring. These four residues are completely conserved in PcyA orthologs (Fig. 2c). Binding of BV in incorrect orientation of BV may be destabilized by the steric hindrance at the A-ring binding site (see Fig. 5, which is published as supporting information on the PNAS web site). In addition, van der Waals interactions of the methyl group of the D-ring in the inverted orientation with Phe-158 and Gly-103 appear to be weaker than those of the vinyl group of the A-ring. The incorrect BV binding to PcyA may also be destabilized by the less attractive interactions.
Reaction Mechanism. PcyA catalyzes the two sequential reductions of the D-ring and A-ring, each of which uses two electrons supplied by Fd (9). Lagarias and colleagues (10) detected radical intermediates in the course of this reaction and proposed a mechanism by which BV might be processed by PcyA. In the D-ring reduction step, after an electron is transferred from Fd to BV to form a radical on BV, the C182 or O19 atom of BV accepts the first proton. After the second electron transfer from Fd, the C181 atom of BV accepts the second proton to produce 181,182-DHBV. Subsequently, the A-ring is reduced in a similar way for the D-ring reduction; after an electron is transferred from Fd to 181,182-DHBV to form a radical on it, the C2 or O1 atom accepts a third proton, then the C32 atom accepts the fourth electron and proton to produce 3Z/3E-PCB. However, the previous studies did not address the specific contribution of PcyA in this reaction because of the lack of structural information.
The structure of the PcyA–BV complex elucidated by the present analysis reveals that Glu-76, His-88, Asp-105, Gln-216, and Tyr-212 are located near the A- and D-rings of BV (Fig. 4a); all of these residues are completely conserved among PcyA orthologs (Fig. 2c). In the context of the reaction scheme described above, we propose here how PcyA achieves the sequential two-step reduction of BV (Fig. 4b). Asp-105 and Glu-76 are potential candidates for the active residues in the D-ring reduction. The carboxyl group of Asp-105 in the major conformation is hydrogen-bonded to both the lactam oxygen and nitrogen atoms of the D-ring, whereas, in the minor conformation, this group is hydrogen-bonded to only the nitrogen atom (Fig. 4a). These structural features suggest that the carboxyl group of Asp-105 is protonated in the major conformation but deprotonated in the minor conformation, suggesting the primary role of this residue as a proton donor. Glu-76 is unusually close to the vinyl group of the D-ring and may donate the second proton in the D-ring reduction. The deprotonated Glu-76 in this reaction would be readily protonated by a proton transfer pathway composed of Tyr-238 and Asn-62 (see Fig. 6, which is published as supporting information on the PNAS web site). Multiple conformations in the carboxyl group of Asp-105 also indicates that the loss of the hydrogen bond between Asp-105 and the lactam oxygen of the D-ring during D-ring reduction may induce conformation change of this residue.
His-88, Gln-216, and Tyr-212 are located nearby the A-ring of BV (Fig. 4a) and appear to be involved in A-ring reduction. However, referring to the chirality at the C2 atom of the final product of PcyA, 3Z/3E-PCB (23), which is in the R-configuration, none of these residues is likely to act as the initial proton donor for the A-ring; although this first proton must be donated to 181,182-DHBV from the β-sheet side of PcyA, all of these are located on the opposite side. This discrepancy suggests that PcyA changes its local conformation around the A-ring with the D-ring reduction. Assuming that the flexibility of Asp-105 is increased upon D-ring reduction, its carboxyl groups may be rotated at the χl angle so as to situate it between the A-ring of 181,182-DHBV and the β-sheet of PcyA. This position is appropriate for donation of the first proton to the C2 atom of 181,182-DHBV with the correct chirality. A possible proton source for the deprotonated Asp-105 is His-88 or the amide group of B- or C-ring. The B- and C-rings are thought to be fully protonated based on the previous spectroscopic analysis of the PcyA–BV complex (10). Tyr-212 and Glu-216 may donate the second proton for A-ring reduction. It should be noted that the scheme proposed here also implies a control mechanism for the sequential reduction steps; reduction of the D-ring and the concomitant deprotonation of Asp-105 alter the conformation of this residue, which then leads to the state capable of reducing the A-ring.
Implications for Other FDBRs. Sequence and functional similarities among FDBRs suggest that the overall folding of PcyA is similar to that of other FDBR family enzymes, 15,16-DHBV:Fd oxidoreductase (PebA), phycoerythrobilin:Fd oxidoreductase (PebB), and PΦB synthase. Models of PebA, PebB, and PΦB synthase were constructed based on the PcyA structure (see Fig. 7, which is published as supporting information on the PNAS web site). In these models, the surfaces of PebA, PebB, and PΦB synthase are almost acidic, whereas surfaces nearby putative substrate binding pockets are basic, as occurs in PcyA. Fd would bind to basic patch of FDBRs and directly transfer electrons to substrate. Glu-76 in PcyA is substituted by Ala in PebB, resulting in a wider substrate binding pocket in PebB than in PcyA. This finding is consistent with the shape of the substrate of PebB, 15,16-DHBV; reduction at the C15⋕C16 double bond of BV loses the planarity. So far two point mutations that lack PΦB synthase activity were found; R252Q mutant in Arabidopsis thaliana and N140Y mutant in tomato (24, 25), which correspond to Gln-216 and Asp-105 in PcyA, respectively. These residues are candidates for active residues of PcyA as described above, implying that these residues in PΦB synthase may also be involved in substrate binding and/or reduction. Thus, homology models of FDBRs based on the PcyA structure are consistent with the genetic and biochemical characteristics of these molecules and their mutant forms.
Acknowledgments
We thank Drs. Masahide Kawamoto and Nobutaka Shimizu (Japan Synchrotron Radiation Research Institute) and Drs. Hisashi Naitow and Taiji Matsu (RIKEN Harima Institute) for their aid with data collection using the synchrotron radiation at SPring-8 (Proposal 2005A0214-NL1-np), Drs. Takakazu Kaneko and Satoshi Tabata (Kazusa DNA Research Institute) for their gift of cyanobacterial genomic DNA, and Prof. Masato Noguchi (Kurume University School of Medicine) for advice and encouragement. This work was supported in part by grants-in-aid for scientific research (Grants 16570095 and 17053014 to K.F.), by a grant-in-aid for Research Fellows of the Japan Society for the Promotion of Science (Grant 17·5063 to M.S.), and by a grant of the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions: Y.H., M.S., Y.T., and K.F. designed research; Y.H. and M.S. performed research; Y.H. and M.S. analyzed data; and Y.H., M.S., Y.T., and K.F. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BV, biliverdin IXα; DHBV, dihydrobiliverdin IXα; Fd, ferredoxin; FDBR, Fd-dependent bilin reductase; PCB, phycocyanobilin; PcyA, PCB:Fd oxidoreductase; PΦB, phytochromobilin; PebA, 15,16-DHBV:Fd oxidoreductase; PebB, phycoerythrobilin:Fd oxidoreductase.
Data deposition: Coordinates and structure factors of the PcyA–BV complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2D1E).
References
- 1.Beale, S. I. (1993) Chem. Rev. 93, 785–802. [Google Scholar]
- 2.Hughes, J. & Lamparter, T. (1999) Plant Physiol. 121, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schafer, E. & Bowle, C. (2002) EMBO Rep. 3, 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossman, A. R., Schaefer, M. R., Chiang, G. G. & Collier, J. L. (1993) Microbiol. Rev. 57, 725–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornejo, J., Willows, R. D. & Beale, S. I. (1998) Plant J. 15, 99–107. [DOI] [PubMed] [Google Scholar]
- 6.Frankenberg, N., Mukougawa, K., Kohchi, T. & Lagarias, J. C. (2001) Plant Cell 13, 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noguchi, M., Yoshida, T. & Kikuchi, G. (1979) J. Biochem. (Tokyo) 86, 833–848. [DOI] [PubMed] [Google Scholar]
- 8.Schluchter, W. M. & Glazer, A. N. (1997) J. Biol. Chem. 272, 13562–13569. [DOI] [PubMed] [Google Scholar]
- 9.Frankenberg, N. & Lagarias, J. C. (2003) J. Biol. Chem. 278, 9219–9226. [DOI] [PubMed] [Google Scholar]
- 10.Tu, S. L., Gunn, A., Toney, M. D., Britt, R. D. & Lagarias, J. C. (2004) J. Am. Chem. Soc. 126, 8682–8693. [DOI] [PubMed] [Google Scholar]
- 11.Stubbe, J. & van der Donk, W. A. (1998) Chem. Rev. 98, 705–762. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura, Y., Kaneko, T., Hirosawa, M., Miyajima, N. & Tabata, S. (1998) Nucleic Acids Res. 26, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miroux, B. & Walker, J. E. (1996) J. Mol. Biol. 260, 289–298. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 16.Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003) Methods Enzymol. 374, 229–244. [DOI] [PubMed] [Google Scholar]
- 17.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 18.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 20.Holm, L. & Sander, C. (1997) Nucleic Acids Res. 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips, J. D., Whitby, F. G., Warby, C. A., Labbe, P., Yang, C., Pflugrath, J. W., Ferrara, J. D., Robinson, H., Kushner, J. P. & Hill, C. P. (2004) J. Biol. Chem. 279, 38960–38968. [DOI] [PubMed] [Google Scholar]
- 22.Fukuyama, K. (2004) Photosynth. Res. 81, 289–301. [DOI] [PubMed] [Google Scholar]
- 23.Tu, S. L., Gunn, A., Toney, M. D., Britt, R. D. & Lagarias, J. C. (2005) J. Am. Chem. Soc. 127, 3230–3231. [DOI] [PubMed] [Google Scholar]
- 24.Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A. & Lagarias, J. C. (2001) Plant Cell 13, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramoto, T., Kami, C., Kataoka, H., Iwata, N., Linley, P. J., Mukougawa, K., Yokota, A. & Kohchi, T. (2005) Plant Cell Physiol. 46, 661–665. [DOI] [PubMed] [Google Scholar]
- 26.Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995) Protein Eng. 8, 127–134. [DOI] [PubMed] [Google Scholar]