Abstract
The formylglycine (FGly)-generating enzyme (FGE) uses molecular oxygen to oxidize a conserved cysteine residue in all eukaryotic sulfatases to the catalytically active FGly. Sulfatases degrade and remodel sulfate esters, and inactivity of FGE results in multiple sulfatase deficiency, a fatal disease. The previously determined FGE crystal structure revealed two crucial cysteine residues in the active site, one of which was thought to be implicated in substrate binding. The other cysteine residue partakes in a novel oxygenase mechanism that does not rely on any cofactors. Here, we present crystal structures of the individual FGE cysteine mutants and employ chemical probing of wild-type FGE, which defined the cysteines to differ strongly in their reactivity. This striking difference in reactivity is explained by the distinct roles of these cysteine residues in the catalytic mechanism. Hitherto, an enzyme–substrate complex as an essential cornerstone for the structural evaluation of the FGly formation mechanism has remained elusive. We also present two FGE–substrate complexes with pentamer and heptamer peptides that mimic sulfatases. The peptides isolate a small cavity that is a likely binding site for molecular oxygen and could host reactive oxygen intermediates during cysteine oxidation. Importantly, these FGE–peptide complexes directly unveil the molecular bases of FGE substrate binding and specificity. Because of the conserved nature of FGE sequences in other organisms, this binding mechanism is of general validity. Furthermore, several disease-causing mutations in both FGE and sulfatases are explained by this binding mechanism.
Keywords: posttranslational modification, oxygenase, enzyme mechanism
Sulfatases catalyze the hydrolysis of sulfate esters such as glycosaminoglycans, sulfolipids, and steroid sulfates in eukaryotic cells. The key catalytic residue in sulfatases is a unique formylglycine (FGly), which is generated from a cysteine precursor (Fig. 1a) and functions as a nucleophilic aldehyde hydrate in the initial addition reaction of sulfate ester hydrolysis (1). Inactivity of individual sulfatases in humans may lead to severe diseases such as mucopolysaccharidoses, metachromatic leukodystrophy, X-linked ichthyosis, and chondrodysplasia punctata. However, a severe reduction or complete lack of all sulfatase activities, termed multiple sulfatase deficiency, originates from mutations in the FGly-generating enzyme (FGE) (2, 3).
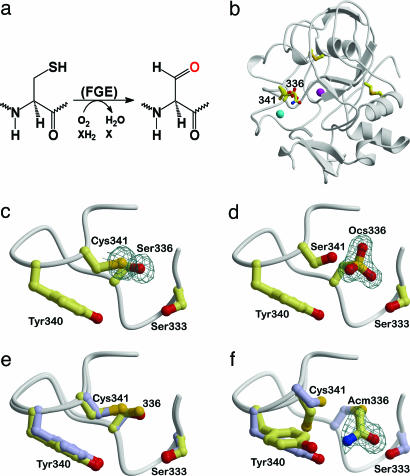
Fig. 1.
Overall fold of FGE and comparison of wild-type FGE (PDB entry 1Y1I) with the active site mutants C336S and C341S, and the IAM-modified wild-type FGE. (a) Scheme of the reaction catalyzed by FGE. (b) Ribbon representation of FGE with the cysteine residues drawn as stick models and structural Ca2+ ions displayed as magenta and cyan spheres. Cys-336 and Cys-341 are part of the active site. (c–f) Close-ups of the region around Cys-336 and Cys-341. (c) Cys336Ser mutant. (d) Cys341Ser mutant with Cys-336 oxidized to the sulfonic acid (Ocs). (e) Superposition of reduced wild-type FGE (yellow) with the Cys336Ser mutant (blue) showing the minor effect of the mutation on the structure. (f) Incubation of wild-type FGE with IAM leads exclusively to carboxamidomethylation of Cys-336 (Acm), whereas Cys-341 remains unaffected. The region Tyr-340–Cys-341 shows two alternate conformations, which are shown in yellow and blue. All σA-weighted mFo-DFc omit electron density maps, including Fig. 2c, are contoured at 3σ.
FGE is localized in the endoplasmic reticulum (ER) and modifies the unfolded form of newly synthesized sulfatases (4, 5). The generation of FGly from a cysteine residue is a multistep redox process that involves disulfide bond formation and requires a reducing agent (6) and molecular oxygen (J. Peng, B. Schmidt, A. Preusser-Kunze, M. Mariappan, K. von Figura, and T. Dierks, personal communication) but does not require any cofactors or metal ions. Peptides that contain the minimal motif C-[TSAC]-PSR with flanking sequences according to human sulfatases are FGE substrates and are converted to their FGly-containing counterparts with efficiencies that depend on the nature of the flanking sequences (7). This minimal motif is conserved in all human sulfatases, suggesting a general binding mechanism of substrate sulfatases by FGE.
The details of how O2-dependent cysteine oxidation is mediated by FGE are unknown. As a first step toward the elucidation of the molecular mechanism of FGly formation, we have previously determined crystal structures of FGE in various oxidation states (8). FGE adopts a novel fold with surprisingly little regular secondary structure and contains two structural Ca2+ ions and two permanent disulfide bonds (Fig. 1b). A third cysteine pair (Cys-336/Cys-341) was revealed by these apo-structures to exist in different oxidation states, being reduced, disulfide-bonded, or chemically modified at Cys-336, clearly establishing the involvement of Cys-336 and Cys-341 in catalysis (8). Cys-336 and Cys-341 border a groove on the surface of FGE, which we speculated to host the substrate binding site. The catalytic importance of the cysteine residues was further demonstrated by the inactivity of the respective point mutants (8). However, it remained unknown what structural consequences these mutations imposed on FGE and whether substitution of one cysteine residue would affect the redox activity of the other. In addition, a FGE–peptide complex crystal structure to define the molecular determinants for substrate recognition was lacking.
We describe here crystal structures of the Cys336Ser and Cys341Ser FGE mutants and a structure of wild-type FGE that has been covalently modified with the SH-reactive agent iodoacetamide (IAM). The structural integrity of the cysteine mutants is warranted, and they reveal strongly different redox activities, with Cys-336 being more reactive than Cys-341. More importantly, we present two complex crystal structures of the FGE Cys336Ser mutant covalently bound to pentamer and heptamer peptides derived from arylsulfatase A, which mimic reaction intermediates in the catalytic cycle of FGE. These structures reveal the general binding mechanism of sulfatases by FGE.
Materials and Methods
FGE was produced from HT1080 fibrosarcoma cells and crystallized as described in refs. 7 and 9. Binding of IAM to Cys-341 of FGE was achieved by incubating crystals of FGE in mother liquor (20–25% PEG 4000/0.1 M Tris·HCl, pH 8.0–9.0/0.2–0.3 M CaCl2) with 1 mM IAM for 1 d. For the FGE–CTPSR and FGE–LCTPSRA complexes, 181 μM FGE was preincubated with a 5-fold molar excess of peptide for 1 h at 4°C before setting up for crystallization. Crystals were cryo-cooled in mother liquor without additional cryoprotectant. All data were collected in-house at 100 K on a mar345dtb image plate detector (MAR-Research, Hamburg) mounted on either a MicroMax-007 or RU-H3R generator (Rigaku, Tokyo) and reduced with the hkl programs (HKL Research, Charlottesville, VA). This data-collection protocol eliminated radiation damage to the active-site cysteine residues. All structures were determined by molecular replacement with the same starting model (based on PDB entry 1Y1E) devoid of water molecules, alternate conformations, and mutated residues converted to alanine. Refinement was performed with refmac5 (10) with the same set of 5% of reflections reserved for Rfree cross-validation (11). Water oxygen atoms were assigned with arp/warp (10), visually inspected, and retained if they returned >1σ σA-weighted 2mFo-DFc electron density after refinement. The data collection and refinement statistics are summarized in Table 1. Possible hydrogen bonds, salt bridges, and van der Waals contacts were detected with hbplus (12) and contacsym (13) using default parameters. Buried surface areas and surface complementarity coefficients were calculated with ms (14) and sc (10), respectively. Electrostatic potentials were calculated with apbs (15) and displayed with pymol (www.pymol.org). Structure figures were created with bobscript (16) and rendered with raster3d (17).
Table 1. Data collection and refinement statistics.
| Data Set | 2AFT-C336S | 2AFY-C341S | 2AII-IAM | 2AIJ-CTPSR | 2AIK-LCTPSRA |
|---|---|---|---|---|---|
| Data collection | 30.0-1.66 | 30.0-1.48 | 50.0-1.54 | 30.0-1.55 | 30.0-1.73 |
| Resolution range, Å* | (1.72-1.66) | (1.53-1.48) | (1.60-1.54) | (1.61-1.55) | (1.79-1.73) |
| Measured reflections | 117,697 (3,763) | 167,117 (1,955) | 290,416 (14,208) | 182,700 (12,102) | 317,364 (8,425) |
| Unique reflections | 34,103 (2,228) | 45,114 (1,277) | 44,155 (3,759) | 42,603 (3,825) | 31,093 (2,688) |
| Completeness, % | 95.6 (63.7) | 89.8 (25.9) | 98.5 (85.5) | 97.5 (89.3) | 97.8 (87.0) |
| Mosaicity, ° | 0.45 | 0.63 | 0.28 | 0.60 | 0.85 |
| Rsym, %† | 6.3 (45.0) | 5.4 (23.7) | 4.5 (22.9) | 3.3 (18.1) | 7.1 (37.3) |
| Average I/σ(I) | 17.5 (1.5) | 23.3 (2.4) | 41.5 (4.9) | 41.2 (5.6) | 29.9 (1.8) |
| Refinement | 25.17-1.66 | 25.46-1.49 | 43.48-1.54 | 24.5-1.55 | 29.7-1.73 |
| Resolution range, Å | (1.71-1.66) | (1.53-1.49) | (1.58-1.54) | (1.59-1.55) | (1.78-1.73) |
| Rcryst, %‡ | 14.9 (32.5) | 13.9 (25.7) | 15.0 (27.1) | 14.4 (20.3) | 14.1 (0.20) |
| Rfree, %‡ | 18.7 (40.1) | 17.1 (46.9) | 17.6 (32.3) | 17.8 (25.5) | 17.4 (25.9) |
| # of residues/waters | 271/498 | 272/569 | 267/529 | 278/531 | 279/495 |
| Coordinate error, ŧ | 0.062 | 0.045 | 0.043 | 0.047 | 0.059 |
| rms bonds, Å/Angles, ° | 0.012/1.37 | 0.011/1.33 | 0.010/1.31 | 0.012/1.41 | 0.012/1.34 |
| Ramachandran plot, %¶ | 87.9/11.2/0/0.9 | 87.9/11.2/0/0.9 | 89.3/9.8/0/0.9 | 86.7/12.4/0/0.9 | 87.2/11.9/0/0.9 |
| Average B values, Å2 | 25.5 ± 9.8 | 24.2 ± 10.5 | 16.0 ± 10.0 | 24.2 ± 10.6 | 28.9 ± 10.5 |
Values in parenthesis correspond to the highest-resolution shell.
Rsym = 100·ΣhΣi|Ii(h) - 〈I(h) 〉|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h and 〈I(h) 〉 is the average value of the reflection intensity.
Rcryst = Σ|Fo| - |Fc|/Σ|Fo|, where Fo and Fc are the structure factor amplitudes from the data and the model. Rfree is Rcryst with 5% of test set structure factors.
Based on maximum likelihood.
Numbers reflect the percentage amino acid residues in the core, allowed, generous allowed, and disallowed regions, respectively.
Results
Comparison of Wild-Type and Mutant FGE Structures: Increased Reactivity of Cys-336. The importance of Cys-341 and Cys-336 for catalytic activity of FGE has been demonstrated in vitro by generation of the respective serine mutants, which are inactive for FGly formation (8). To assess whether these mutations influence the structure of the active site, crystal structures were determined and compared with the reduced wild-type FGE (PDB entry 1Y1I). Superposition of the structures revealed no gross structural changes except in the immediate vicinity of the active site. Small backbone and side-chain adjustments on the order of 0.2 Å compensate for the less voluminous serine compared with the cysteine side chain in the Cys336Ser mutant (Fig. 1 c and e). Interestingly, three independently determined structures of the Cys341Ser mutant all revealed the Cys-336 side chain to be fully oxidized to the sulfonic acid (Fig. 1d, Table 1, and data not shown). Because both mutants were crystallized under identical conditions, this result points to an increased redox activity (more negative potential) of Cys-336 compared with Cys-341.
In an attempt to directly distinguish the relative reactivity of the two cysteines, a crystal structure of wild-type FGE was determined from an IAM-modified crystal (Table 1). IAM is a strong electrophile that can covalently and irreversibly modify cysteine residues. Clear unbiased electron density for the carboxamidomethyl group was visible at Cys-336 but not at Cys-341 (Fig. 1f). The bulky carboxamidomethyl group leads to partial rearrangement of the Tyr-340 and Cys-341 side chains, but similar to the cysteine mutants, no gross structural differences with respect to wild-type FGE are apparent.
The facile oxidation of Cys-336 in the Cys341Ser mutant and the exclusive chemical modification of Cys-336 in wild-type FGE allow the conclusion that Cys-336 is more reactive than Cys-341. This result resonates with the observation that Cys-341 binds the substrate (see below) as it leaves the highly reactive Cys-336 free for the reaction with molecular oxygen as part of the substrate oxidation to FGly.
Covalent Substrate Binding. Previous apo–FGE crystal structures (8) and biochemical data (7) have helped in assigning a putative substrate binding site. A surface representation of FGE shows that the catalytically active Cys-336 and Cys-341 residues are located next to an oval-shaped groove of 20 Å length, 10 Å depth, and 12 Å width (8) (Fig. 2a). In addition, a photoreactive substrate peptide cross-linked to FGE residue Pro-182 (7), which also is close to this groove (Fig. 2a). However, further insight into substrate binding by FGE required crystallization of a FGE–peptide complex, where the peptide mimics an unfolded part of the natural sulfatase substrate. Initial efforts focused on cocrystallization of wild-type FGE with arylsulfatase-derived peptides of 5 to 13 residues in length. However, in all cases where crystals were obtained, the resulting structures did not contain bound peptide. Soaking of the FGE crystals with short peptides was also unsuccessful. Either the peptides did not bind to FGE or they were turned over before crystallization of FGE. Consequently, the inactive cysteine mutants offered an avenue to generate a stable FGE–substrate intermediate that would delineate the substrate binding mode and reveal the determinants of FGE substrate specificity.
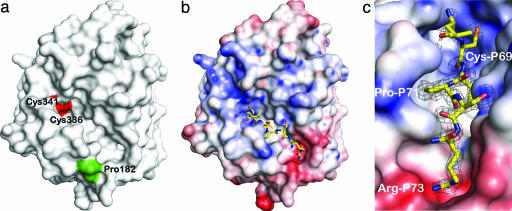
Fig. 2.
Substrate binding to FGE. (a) The surface representation of FGE shows a groove with the redox-active cysteine pair Cys-336/Cys-341 (red surface) at one end. Pro-182 (green surface) marks the site of a cross-link with a photoreactive substrate peptide (7) and hence is also close to the substrate binding site (8). (b) FGE–peptide complex. The peptide LCTPSRA binds to Cys-341 via an intermolecular disulfide bond. The FGE surface is colored according to electrostatic potential (±10 kT), showing a negative patch close to the C terminus of the peptide, which is neutralized by Arg-P73. (c) Close-up of b rotated 45° clockwise showing the exquisite surface complementarity of the peptide with FGE.
The cocrystallization trials were repeated by using the FGE Cys336Ser and Cys341Ser mutants and peptides CTPSR and LCTPSRA, comprising arylsulfatase A residues 69–73 and 68–74, respectively. Only in case of the Cys336Ser mutant was electron density visible that emerged from the Sγ atom of Cys-341 and stretched into the groove of FGE (Fig. 2c). Thus, the earlier (8) assignment of the substrate binding site to the groove bordering the Cys-336/Cys-341 pair proved to be correct. No density corresponding to a peptide was visible in three independently determined Cys341Ser structures (Fig. 1d and data not shown), establishing unambiguously that Cys-341, and not Cys-336, is responsible for substrate binding. The structure obtained from cocrystallization of FGE Cys336Ser with the LCTPSRA peptide includes all salient features of the CTPSR-bound structure and will be described further. The peptide binds at the surface of FGE (Fig. 2b) in an extended conformation that does not follow any regular secondary structure element. Five residues, CTPSR, are placed into the binding groove and engage in numerous interactions that are described in detail below.
Conformational Changes Associated with Substrate Binding. Superposition of the peptide-containing FGE Cys336Ser structures with four apo–FGE structures (PDB entries 1Y1E, 1Y1F, 1Y1H, and 1Y1I) revealed rms deviations of ≈0.2 Å over 272 common Cα atoms. Larger conformational changes are limited to the immediate vicinity of the substrate binding site with regions Ser-336–Tyr-340 and Phe-265–Pro-266 undergoing rigid body backbone shifts of 0.5 Å and 0.4 Å toward the peptide, respectively (Fig. 4, which is published as supporting information on the PNAS web site). As the Ser-336–Tyr-340 region moves closer toward the binding site due to its tethering to the substrate via a disulfide bond to Cys-341, the Tyr-340 side chain rotates away from the peptide (80° around χ1), resulting in a 6.4 Å distance between the apexes of the side chains in apo–FGE and the complex (Fig. 3c). This change in the side-chain rotamer is the largest difference between the apo– and peptide–FGE structures. The presence of the peptide is also sensed by residues Phe-156, Trp-179, Gln-351, and Asn-352, whose side chains move slightly outwards to accommodate the substrate (Fig. 4). The magnitude of these side-chain adjustments is on the order of <0.5 Å, indicating that only minor adjustments are necessary to fit the peptide. When the spheres of hydration in the four apo–FGE structures (see above) are compared, 14 water molecules are present in at least two of these structures, and 8 water molecules are structurally conserved. The 14 water molecules are displaced upon substrate binding, and the peptide satisfies the hydrogen bonding potentials of the FGE side chains lining the groove that were previously held by solvent molecules (Fig. 3a). Other solvent molecules are rearranged in the complex and form an integral part of the substrate binding pocket (see below). Overall, the small conformational changes in FGE that accompany substrate binding indicate that the substrate binding site is already preformed in apo–FGE and defines a rigid scaffold for the conserved CTPSR motif in sulfatases.
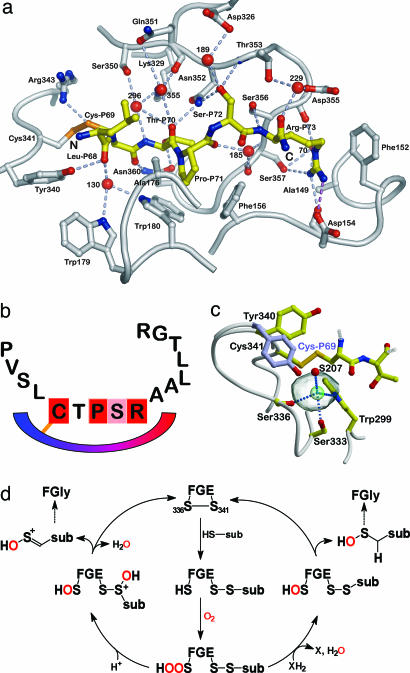
Fig. 3.
Substrate binding and mechanistic details. (a) Hydrogen bonds are shown as dashed lines, and water molecules are drawn as red spheres. (b) General binding mechanism of FGE to all human sulfatases. The schematic drawing generalizes the binding of unfolded sulfatases to FGE as the first step in FGly formation. (c) Magnification of the region adjacent to the intermolecular disulfide bond. The orientation of the Tyr-340 side chain in the apo– and peptide–FGE structures differs by 6.4 Å (compare with Fig. 4). Only residues Cys-P69 and Thr-P70 of the peptide are drawn. The solvent-inaccessible volume between the disulfide bond and serine residues 333 and 336 (transparent gray sphere) is occupied by Cl– (green) in the complex structure. (d) Possible mechanisms after the activation of molecular oxygen by FGE. Atoms from O2 are indicated in red. A novel hydroperoxide intermediate is formulated from which two alternative avenues for FGly formation are conceivable. Currently, no distinction between these two pathways is possible.
Sequence Specificity of FGE Substrates. The substrate peptide buries 80%, or 498 Å2, of its total surface area, which is below the range of 1,600 ± 400 Å2 typical of protein–protein recognition sites (18), and resembles more that of strong peptide–MHC interactions (19). Yet, the apparent affinity of polypeptides for FGE has been estimated to be high with KM values in the 13 nM range (6). A similarly small area of 680 Å2 that mediates a high-affinity interaction with a Kd value of close to 10 nM has been found in the Rab escort protein 1–geranylgeranyltransferase complex (20), indicating that small contact areas need not correlate with low affinity. Another hallmark for high-affinity interactions is extensive surface complementarity (21). The surface of the substrate to FGE has a high Sc value of 0.64, where a value of 1 would denote perfect complementarity. This value increases to 0.75 when water molecules are included in the calculation, showing that solvent strongly contributes to the good fit of substrate and enzyme and, thus, constitutes an important part of the substrate recognition by FGE.
The high surface complementarity between substrate and FGE results in a total of 50 van der Waals contacts and 24 hydrogen bonds, half of which are water-mediated (Fig. 3a and Table 3, which is published as supporting information on the PNAS web site). Thus, the main roles of water in the FGE–substrate complex are shaping of the binding site and providing additional binding energy for the substrate. This importance of water is in contrast to the paucity of peptide main-chain interactions with FGE: Of the 12 direct hydrogen bonds, only 4 are formed by peptide main-chain atoms, stressing the importance of the side chains for determining substrate specificity (Table 3). In cases where substrate-contacting residues are not conserved, they are substituted by residues of similar size and chemical properties in homologous FGEs. For instance, Asp-154 is an asparagine in sea urchin FGE, and Ala-176 and Gln-351 are serine and glutamic acid, respectively, in FGE from tunicates.
Apart from forming the disulfide bond with Cys-341, Cys-P69 (P denotes peptide substrate residues, the numbering is according to arylsulfatase A) also entertains a hydrogen bond with the side chain of Asn-360. The next residue in the substrate, Thr-P70, can also be Ala, Cys, or Ser in sulfatases. This degeneracy is surprising because the Thr-P70 side chain points down into the groove, which would anticipate specific interactions between this side chain and FGE. However, there are no van der Waals interactions of the Thr-P70 side chain and only a single hydrogen bond with Asn-360. The paucity of Thr-P70 interactions thus explains the observed sequence variation at this position. Most important for substrate specificity is Pro-P71, which binds in a pocket formed by the conserved FGE residues Phe-156 and Trp-180 (Fig. 2c), and also to Ala-176, resulting in 25 van der Waals contacts. The side chain of Ser-P72 points upwards from the binding site and hydrogen bonds with Asn-352 and Thr-353. Interestingly, there is no sequence variation among sulfatases in this position, although space and hydrogen bonding requirements would allow other small side chains as in the case of Thr-P70. Similarly, in the folded sulfatase Ser-P72 forms only one hydrogen bond so its strict conservation remains unexplained. The guanidinium group of Arg-P73 is fixed by a strong charged hydrogen bond to Asp-154 and three additional hydrogen bonds to the carbonyl groups of Asp-154 and Asp-355, and the Oγ atom of Ser-357 (Fig. 3a). The use of the full hydrogen bonding potential of Arg-P73 discriminates against the other positively charged residues histidine and lysine, which are not observed in sulfatases at this position (Table 2).
Table 2. Minimal FGE binding motifs and currently known natural motif mutations in human sulfatases.
| Sulfatase | Motif | Mutation | Associated FGE residues | Ref. |
|---|---|---|---|---|
| ARS B | CTPSR | T92M | N360 | 26 |
| R95Q | A149, F152, D154, D355, S356, S357 | 29 | ||
| SGSH | CSPSR | R74C/H | See ARS B R95Q | 31, — |
| GALNS | CSPSR | S80L | N360 | — |
| IDS | CAPSR | A85T | N360 | 27 |
| P86L/Q/R | F156, A176, W180 | 28, 29, 30 | ||
| S87N | F156, N352, T353 | 36 | ||
| R88C/H/L/P | see ARS B R95Q | 27, 28, 29, 28 |
Mutated residues in the modification motifs are underlined. The residue numbering follows the respective sulfatase. ARS B, arylsulfatase B; SGSH, sulfoglucosamine sulfamidase; GALNS, N-acetylgalactosamine-6-sulfatase; IDS, iduronate 2-sulfatase.
In summary, two major strategies for conferring substrate specificity are apparent from the FGE–substrate complexes: Pro-P71 binds with high surface complementarity into a hydrophobic pocket provided by Phe-156 and Trp-180, and Arg-P73 displays electrostatic complementarity by neutralizing the charge of Asp-154. These few but strong side-chain interactions provide the structural basis for conservation of the minimal modification sequence among the sulfatases.
A General Sulfatase Binding Mode. Inspection of the unbiased electron density (Fig. 2c) for the peptide reveals that only the central CTPSR sequence has defined electron density, whereas the N-terminal Leu-P68 and the C-terminal Ala-P74 residues are poorly defined, probably because of increased mobility. Residue Ala-P74 was excluded from the final model, but the electron density was clear enough to assign the amide nitrogen atom for this residue (Fig. 3a). Both terminal residues point up and away from the body of the FGE molecule. Their disposition and increased flexibility are congruent with the limitation of FGE recognition to a short sequence motif while the rest of the sulfatase sequence is irrelevant for FGly formation (Fig. 3b) but rather influences the turnover rate of the substrate (7). The elongated binding mode is further evidence that sulfatase modification in the ER occurs before folding (1). The in vivo complex between FGE and authentic sulfatase should therefore include properties reminiscent of chaperones, prolyl isomerases, or protein disulfide isomerases bound to their unfolded substrates.
Mechanistic Implications. The peptide connects to FGE via a disulfide bond between FGE Cys-341 and the FGly precursor Cys-P69. Formation of this disulfide bond generates a small cavity of 32 Å3 that is buried between Ser-336 (catalytically active Cys-336 in wild-type FGE) and the disulfide bond (Fig. 3c). In previous FGE crystal structures, this space was occupied by a putative sulfenic acid or a peroxide moiety at Cys-336 (8). The size and location of this cavity fit the requirement for the essential oxygen molecule during catalysis (J. Peng, B. Schmidt, A. Preusser-Kunze, M. Mariappan, K. von Figura, and T. Dierks, personal communication), which is then close to all three cysteine residues of FGE and the substrate and is also shielded from bulk solvent. In both FGE–peptide complexes, this cavity is occupied by a Cl– ion, which is defined by a3.5σ peak in an anomalous difference map (data not shown) and which has not been observed in any apo–FGE structure. The Cl– ion is hydrogen-bonded to the side chains of Trp-299, Ser-333, Ser-336, and water molecule S207 (Fig. 3c). Ca2+ as alternative ion at this position was excluded based on coordination geometry and electrostatic potential considerations in apo–FGE, which displays a positive patch at the location of the catalytically active cysteine pair (Fig. 2 b and c), in line with binding of a negatively charged Cl–. Halide ions display some degree of hydrophobicity, which is also true for molecular oxygen. The small cavity may represent the catalytic volume that is closed off by the disulfide bond and which will contain the reactive oxygen species that are likely formed from molecular oxygen by FGE (8). The question as to the activation of molecular oxygen by FGE in the absence of cofactors may be answered by the conserved Trp-299 residue, which could act as a molecular oxygen sensitizer to produce a Cys-336-hydroperoxide from O2 and water molecule 207. Oxygen sensitization by tryptophan residues has also been implicated in the antibody-catalyzed reduction of O2 by water to produce H2O2 (22). Several other cofactor-free oxygenases such as the quinone-forming monooxygenases and dioxygenases involved in the degradation of quinolines (23) conserve histidine residues in their active sites and might similarly rely on this mode of O2 activation.
Discussion
FGE executes the limiting step in the activation of all sulfatases. The mechanism of FGly formation by this unique enzyme is still ill understood, most notably because of the absence of redox-active cofactors and metal ions, which implies a novel oxygenase mechanism. Our findings establish that the catalytically active cysteine residues display strongly different redox activity. Both cysteines are involved in redox reactions. Whereas Cys-341 covalently binds to the substrate via a disulfide bond, Cys-336 must react with molecular oxygen as a first step in the introduction of an oxygen atom into the substrate cysteine. The reaction with molecular oxygen may require the observed increase in activity of Cys-336, whereas occlusion of a small reaction volume by the newly formed disulfide bond (Fig. 3c) prevents overoxidation, and thus inactivation, of Cys-336.
Most importantly, the FGE–peptide complexes described here delineate the general substrate binding mode used by FGE for all human sulfatases during their maturation. The binding of the central CTPSR sequence into pockets of high surface and electrostatic complementarity has elements of the mode of peptide binding by class I MHC molecules (24). Because of the high sequence conservation of the modification motifs also in prokaryotic sulfatases, FGEs from all kingdoms of life will bind their substrates similarly, and the mechanism of FGE-mediated FGly formation is universal. The FGE substrate binding site displays exquisite sensitivity to alterations in the sulfatase sequence as seen by scanning mutagenesis (25) and by natural mutations in human sulfatases (Table 2) that lead to mucopolysaccharidoses. As the mutated sulfatases most likely fail to bind to FGE and are therefore not modified, these diseases further cement the notion that the five residues of the C-[TSAC]-PSR motif are necessary and sufficient for specific FGE–sulfatase interaction. For instance, the second residue in this motif has a small side chain and mutation to the larger side chains methionine in arylsulfatase B (26) or leucine in N-acetylgalactosamine-6-sulfatase leads to Maroteaux–Lamy syndrome or Morquio A syndrome, respectively. Apparently, a larger side chain at the position after the modified cysteine residue is only tolerated in iduronate sulfatase, where the A85T mutation (27) leads to only a mild form of Hunter syndrome. The importance of the shape complementarity between the pocket formed by FGE residues Phe-156 and Trp-180 and the conserved proline residue in the C-[TSAC]-PSR motif is brought out by three mutations in iduronate sulfatase, where this residue is mutated to leucine (28), glutamine (29), or arginine (30) with intermediate to severe forms of Hunter syndrome. The conserved arginine residue of the C-[TSAC]-PSR motif has been found mutated to glutamine in arylsulfatase B (29), cysteine (31) or histidine in sulfamidase, or cysteine (27), histidine (28), leucine (29), or proline (28) in iduronate sulfatase. All mutations lead to intermediate or severe forms of mucopolysaccharidoses, stressing the importance of this arginine for FGE recognition.
Because FGly formation is a reaction before protein folding, any protein sequence complying with the C-[TSAC]-PSR motif that enters the ER should be a substrate for FGE. A database search using this motif yielded 15 non-sulfatase entries, of which 9 were functionally annotated. Among the annotated entries were cytoplasmic proteins such as kinesin-like protein KIF14, CDK5 regulatory subunit-associated protein 2, centrosome-localized kendrin, the phosphotyrosine-interacting protein APBA2, the RNA binding protein regulator of differentiation 1 (Rod1), and the Ca2+-dependent activator protein for secretion (CADPS). Another class of molecules containing the FGly determination sequence comprises the mitochondrial thioredoxin reductase, oxidoreductase EALL419, and the selenoprotein Zf2. Given their cytoplasmic or mitochondrial localization, none of these proteins is likely to traverse the ER, rendering a possible FGly residue unlikely. However, little is known about these proteins, and it cannot be excluded that another function for FGly than in sulfatases may exist.
Similar to FGE, its paralogue pFGE is located in the ER and possibly involved in sulfatase maturation (32). pFGE shares 50.6% sequence identity with FGE and adopts a very similar structure (33). However, because of a lack of the catalytic cysteine residues, pFGE has no FGly generating activity, which raises the question as to its biological function. Recently, FGE/pFGE homo- and heterodimerization has been observed by coimmunoprecipitation of the proteins from transfected Cos7 cells (34). In addition, this study reported a trimolecular complex of FGE, pFGE, and sulfatase. Superposition of pFGE and the FGE–peptide complex structures revealed that pFGE could bind sulfatases in a similar mode to FGE because there are no steric conflicts between the FGE-bound peptide and pFGE. Most residues contacting the peptide in the FGE complexes are conserved in pFGE. Notable exceptions are tryptophan at FGE position Asp-154 and alanine at position Ser-357. An appealing mode of dimerization for both FGE and pFGE was inferred by the presence of a dimer in the crystal structure of pFGE (33). However, in this dimer the peptide binding groove is occluded by a face-to-face binding of pFGE. A lysine residue of pFGE is in the same position as Arg-P73 in the FGE-LCTPSRA complex, excluding a ternary complex with this pFGE/FGE geometry. If such a ternary complex exists, it is predicted to have a different pFGE/FGE disposition than that of the pFGE dimer. Further structural and biochemical studies are required to fully assess the potential complex formation of FGE, pFGE, and sulfatases.
Multiple sulfatase deficiency is a rare disease with, so far, only 18 different described missense mutations that are distributed over 32 patients. An analysis of the potential effect of these mutations on the FGE structure was done previously but left two mutants, Ala177Pro (35) and Trp179Ser (8), largely uncharacterized because these residues are located in a loop region close to the substrate binding site. The FGE–peptide complex structures confirm that positions 177 and 179 are indeed close to the bound peptide (Figs. 3a and 4). Trp-179 is engaged in a water-mediated hydrogen bond that would be destroyed by mutation to serine. Likewise, although Ala-177 is not in direct contact with the substrate peptide, Ala-176 does form six van der Waals contacts with Pro-P71 (Table 3). The Ala177Pro mutation could alter the conformation of the neighboring residues and abrogate the Ala-176 contacts with the substrate. Although no data exist on the Trp179Ser mutant, the phenotype of the Ala177Pro mutant is mild (35), indicating that proper contacting of the substrate by adjoining loop regions is important albeit not indispensable for FGly formation.
In summary, the trapped FGE–substrate intermediate analogue together with the structures of the inactive cysteine mutants described here define important cornerstones in the structural delimitation of the catalytic cycle of FGE. The putative reaction volume isolated by formation of the disulfide bond between FGE and substrate opens the possibility of a binding site for molecular oxygen, which would poise it for reaction with Cys-336. High sequence conservation and natural mutations in the sulfatase modification motif leading to mucopolysaccharidoses emphasize the conserved binding mode of FGE and sulfatases. This conservation is also underscored by natural FGE mutants near the substrate binding site that lead to multiple sulfatase deficiency. Based on the results presented here, structural analysis of FGE mutants and reaction intermediates should lead to a full description of this novel oxygenase mechanism.
Supplementary Material
Acknowledgments
M.G.R. thanks Dagmar Klostermeier for key discussions and helpful comments on the manuscript and Ralf Ficner for continuous support. We thank a reviewer for suggesting an alternative mechanism for FGly formation. This work was supported by grants from the Deutsche Forschungsgemeinschaft (separate grants to M.G.R. and T.D.) and Fonds der Chemischen Industrie (to K.v.F.).
Author contributions: K.v.F. and M.G.R. designed research; D.R., K.G., J.G.W., and M.G.R. performed research; A.P.-K. and B.S. contributed new reagents/analytic tools; D.R., T.D., K.v.F., and M.G.R. analyzed data; and D.R., T.D., and M.G.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; FGly, formylglycine; FGE, formylglycine-generating enzyme; IAM, iodoacetamide.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2AFT, 2AFY, 2AII, 2AIJ, and 2AIK).
References
- 1.von Figura, K., Schmidt, B., Selmer, T. & Dierks, T. (1998) BioEssays 20, 505–510. [DOI] [PubMed] [Google Scholar]
- 2.Dierks, T., Schmidt, B., Borissenko, L. V., Peng, J., Preusser, A., Mariappan, M. & von Figura, K. (2003) Cell 113, 435–444. [DOI] [PubMed] [Google Scholar]
- 3.Cosma, M. P., Pepe, S., Annunziata, I., Newbold, R. F., Grompe, M., Parenti, G. & Ballabio, A. (2003) Cell 113, 445–456. [DOI] [PubMed] [Google Scholar]
- 4.Dierks, T., Schmidt, B. & von Figura, K. (1997) Proc. Natl. Acad. Sci. USA 94, 11963–11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dierks, T., Lecca, M. R., Schmidt, B. & von Figura, K. (1998) FEBS Lett. 423, 61–65. [DOI] [PubMed] [Google Scholar]
- 6.Fey, J., Balleininger, M., Borissenko, L. V., Schmidt, B., von Figura, K. & Dierks, T. (2001) J. Biol. Chem. 276, 47021–47028. [DOI] [PubMed] [Google Scholar]
- 7.Preusser-Kunze, A., Mariappan, M., Schmidt, B., Gande, S. L., Mutenda, K., Wenzel, D., von Figura, K. & Dierks, T. (2005) J. Biol. Chem. 280, 14900–14910. [DOI] [PubMed] [Google Scholar]
- 8.Dierks, T., Dickmanns, A., Preusser-Kunze, A., Schmidt, B., Mariappan, M., von Figura, K., Ficner, R. & Rudolph, M. G. (2005) Cell 121, 541–552. [DOI] [PubMed] [Google Scholar]
- 9.Roeser, D., Dickmanns, A., Gasow, K. & Rudolph, M. G. (2005) Acta Crystallogr. D 61, 1057–1066. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computing Project, Number 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 11.Brünger, A. T. (1992) Nature 355, 472–475. [DOI] [PubMed] [Google Scholar]
- 12.McDonald, I. K. & Thornton, J. M. (1994) J. Mol. Biol. 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 13.Sheriff, S., Hendrickson, W. A. & Smith, J. L. (1987) J. Mol. Biol. 197, 273–296. [DOI] [PubMed] [Google Scholar]
- 14.Connolly, M. L. (1993) J. Mol. Graphics 11, 139–141. [DOI] [PubMed] [Google Scholar]
- 15.Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnouf, R. M. (1997) J. Mol. Graphics 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 17.Merritt, E. A. & Murphy, M. E. P. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 18.Lo Conte, L., Chothia, C. & Janin, J. (1999) J. Mol. Biol. 285, 2177–2198. [DOI] [PubMed] [Google Scholar]
- 19.Rudolph, M. G., Stevens, J., Speir, J. A., Trowsdale, J., Butcher, G. W., Joly, E. & Wilson, I. A. (2002) J. Mol. Biol. 324, 975–990. [DOI] [PubMed] [Google Scholar]
- 20.Pylypenko, O., Rak, A., Reents, R., Niculae, A., Sidorovitch, V., Cioaca, M. D., Bessolitsyna, E., Thoma, N. H., Waldmann, H., Schlichting, I., et al. (2003) Mol. Cell 11, 483–494. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, M. C. & Colman, P. M. (1993) J. Mol. Biol. 234, 946–950. [DOI] [PubMed] [Google Scholar]
- 22.Wentworth, P., Jr., Jones, L. H., Wentworth, A. D., Zhu, X., Larsen, N. A., Wilson, I. A., Xu, X., Goddard, W. A., III, Janda, K. D., Eschenmoser, A. & Lerner, R. A. (2001) Science 293, 1806–1811. [DOI] [PubMed] [Google Scholar]
- 23.Fetzner, S. (2002) Appl. Microbiol. Biotechnol. 60, 243–257. [DOI] [PubMed] [Google Scholar]
- 24.Rudolph, M. G., Luz, J. G. & Wilson, I. A. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 121–149. [DOI] [PubMed] [Google Scholar]
- 25.Dierks, T., Lecca, M. R., Schlotterhose, P., Schmidt, B. & von Figura, K. (1999) EMBO J. 18, 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litjens, T., Brooks, D. A., Peters, C., Gibson, G. J. & Hopwood, J. J. (1996) Am. J. Hum. Genet. 58, 1127–1134. [PMC free article] [PubMed] [Google Scholar]
- 27.Li, P., Bellows, A. B. & Thompson, J. N. (1999) J. Med. Genet. 36, 21–27. [PMC free article] [PubMed] [Google Scholar]
- 28.Balzano, N., Villani, G. R., Grosso, M., Izzo, P. & Di Natale, P. (1998) Hum. Mutat. 11, 333. [DOI] [PubMed] [Google Scholar]
- 29.Hopwood, J. J., Bunge, S., Morris, C. P., Wilson, P. J., Steglich, C., Beck, M., Schwinger, E. & Gal, A. (1993) Hum. Mutat. 2, 435–442. [DOI] [PubMed] [Google Scholar]
- 30.Bunge, S., Steglich, C., Zuther, C., Beck, M., Morris, C. P., Schwinger, E., Schinzel, A., Hopwood, J. J. & Gal, A. (1993) Hum. Mol. Genet. 2, 1871–1875. [DOI] [PubMed] [Google Scholar]
- 31.Bunge, S., Ince, H., Steglich, C., Kleijer, W. J., Beck, M., Zaremba, J., van Diggelen, O. P., Weber, B., Hopwood, J. J. & Gal, A. (1997) Hum. Mutat. 10, 479–485. [DOI] [PubMed] [Google Scholar]
- 32.Mariappan, M., Preusser-Kunze, A., Balleininger, M., Eiselt, N., Schmidt, B., Gande, S. L., Wenzel, D., Dierks, T. & von Figura, K. (2005) J. Biol. Chem. 280, 15173–15179. [DOI] [PubMed] [Google Scholar]
- 33.Dickmanns, A., Schmidt, B., Rudolph, M. G., Mariappan, M., Dierks, T., von Figura, K. & Ficner, R. (2005) J. Biol. Chem. 280, 15180–15187. [DOI] [PubMed] [Google Scholar]
- 34.Zito, E., Fraldi, A., Pepe, S., Annunziata, I., Kobinger, G., Di Natale, P., Ballabio, A. & Cosma, M. P. (2005) EMBO Rep. 6, 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosma, M. P., Pepe, S., Parenti, G., Settembre, C., Annunziata, I., Wade-Martins, R., Di Domenico, C., Di Natale, P., Mankad, A., Cox, B., et al. (2004) Hum. Mutat. 23, 576–581. [DOI] [PubMed] [Google Scholar]
- 36.Popowska, E., Rathmann, M., Tylki-Szymanska, A., Bunge, S., Steglich, C., Schwinger, E. & Gal, A. (1995) Hum. Mutat. 5, 97–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.