Abstract
The eukaryotic initiation factor 5A (eIF5A), a factor essential for eukaryotic cell proliferation, is the only cellular protein containing the polyamine-derived amino acid hypusine [Nε-(4-amino-2-hydroxybutyl)lysine]. Hypusine is formed in a posttranslational modification that involves two sequential enzymatic steps catalyzed by deoxyhypusine synthase and deoxyhypusine hydroxylase (DOHH). By screening a Saccharomyces cerevisiae GST-ORF library for expression of DOHH activity, we have cloned YJR070C as the gene encoding DOHH and identified the human homolog DOHH gene, HLRC1. Purified recombinant yeast and human DOHH enzymes effectively catalyzed hydroxylation of the deoxyhypusine residue in the eIF5A intermediate. Overexpression of human DOHH along with eIF5A precursor and deoxyhypusine synthase was required for overproduction of mature, hypusine-containing eIF5A in 293T and other mammalian cells. The Saccharomyces cerevisiae strain with deletion of YJR070C contained only deoxyhypusine but no hypusine, indicating that YJR070C was the single DOHH gene in this organism. One highly conserved DOHH homolog gene is found in a variety of eukaryotes from yeast to human. Sequence and structural analyses reveal that DOHH belongs to a family of HEAT-repeat-containing proteins, consisting of eight tandem repeats of an α-helical pair (HEAT motif) organized in a symmetrical dyad. The predicted structure is unrelated to the double-stranded β-helix type structures of the Fe(II)- and 2-oxoacid-dependent dioxygenases, such as collagen prolyl or lysyl hydroxylases. However, metal coordination sites composed of four strictly conserved histidine–glutamate sequences were identified, suggesting that DOHH enzymes have convergently evolved an iron-dependent hydroxylation mechanism.
Keywords: eukaryotic initiation factor 5A, hypusine, YJR070C
Deoxyhypusine hydroxylase (DOHH) is the second enzyme in the posttranslational modification of the precursor of eukaryotic initiation factor 5A (eIF5A). One specific lysine residue of the eIF5A precursor is modified to the unique amino acid, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] to form the mature, active eIF5A (for reviews, see refs. 1 and 2). In the first step, deoxyhypusine synthase (DHS) catalyzes the transfer of the 4-amino butyl moiety of the polyamine spermidine to the ε-amino group of one specific lysine residue (Lys-50 for the human protein) of the eIF5A precursor to form a deoxyhypusine residue (3). This intermediate is subsequently hydroxylated by DOHH (4) to form hypusine in mature eIF5A. Hypusine synthesis, which occurs only in eIF5A, represents one of the most specific protein modifications known to date and defines a specific function of polyamines in eukaryotic cell proliferation (1, 2, 5, 6).
eIF5A, a small acidic protein, is highly conserved throughout the eukaryotes. Its homolog is also found in archaea (7, 8). Sequence conservation is high, especially around the hypusine residue, underscoring the importance of this unusual modification throughout eukaryotic evolution. X-ray structures of archaeal eIF5A homologs (9, 10) reveal that this protein consists of two well-defined domains: the N-terminal domain, which contains the hypusine modification site in an exposed loop, and the C-terminal domain, which is similar to the oligonucleotide-binding domain found in several RNA-binding proteins. Furthermore, eIF5A is homologous to the translation elongation factor EF-P of eubacteria (11) in sequence and topology. The essential requirement for the eIF5A protein and its posttranslational modification was established from gene disruption studies in Saccharomyces cerevisiae. Disruption of the eIF5A genes (TIF51A and TIF51B) (12, 13) or of the DHS gene (14, 15) is lethal, indicating the importance of the deoxyhypusine modification in the viability of eukaryotic cells (12, 13). However, no genetic study directly addressing the role of the second step, deoxyhypusine hydroxylation, has been reported, because the DOHH gene has not been cloned or identified hitherto.
DHS, which catalyzes the first step in the activation of eIF5A, is a tetrameric enzyme comprising four identical subunits. The structure and mechanism of this enzyme has been extensively characterized (16–18). In contrast to DHS, the structural and catalytic properties of DOHH are not well understood. This enzyme appears to share some properties with the non-heme Fe(II)- and 2-oxoacid-dependent dioxygenases, such as collagen prolyl 4-hydroxylase and lysyl hydroxylases (19, 20). In common with these enzymes, DOHH of mammalian cells or tissues is inhibited by various metal chelators, including 2,2′-dipyridyl, mimosine, deferoxamine, 1,10-phenanthroline, deferiprone, and ciclopirox olamine (4, 21–23). However, partially purified DOHH from rat testis did not catalyze the release of CO2 from α-ketoglutarate (4). In the present study, we used a unique biochemical genomics approach to clone the DOHH gene from a S. cerevisiae GST-ORF library (24, 25). By screening this library, we have identified (i) YJR070C as the single gene for DOHH in S. cerevisiae and (ii) the human homolog HLRC1 (MGC4293) as a DOHH gene.
Sequence profile searches reveal that DOHH belongs to a family of HEAT-repeat proteins, which includes human Huntingtin, Elongation factor 3, a subunit of protein phosphatase 2A, and the Target of rapamycin, as well as importin α proteins, β-catenin, and clathrin-associated adaptor proteins. In a variety of bacterial and eukaryotic proteins, termed HEAT-repeat-containing proteins, the HEAT motif, an α-helical hairpin (a pair of α-helices) of ≈50 aa, is tandemly repeated to form superhelical structures (26). Many of these proteins mediate protein–protein interactions and are involved in nucleocytoplasmic transport, vacuolar transport, and cytoskeletal organization. Computer modeling of DOHH predicts a superhelical structure consisting of eight HEAT-repeat domains that is totally different from the β jelly roll structures of Fe(II)- and 2-oxoacid-dependent dioxygenases (including collagen prolyl 4-hydroxylase, HIF1-α prolyl 4-hydroxylase, and lysyl hydroxylase) (27). Our data provide evidence that DOHH is a protein hydroxylase with structure distinct from other known protein hydroxylases.
Materials and Methods
Materials. The yeast GST-ORF library (24, 25) containing 6,144 S. cerevisiae strains, each expressing a unique yeast ORF as a GST fusion protein, was kindly provided by Eric M. Phizicky (University of Rochester, Rochester, NY). The S. cerevisiae strain with a homozygous diploid knockout of YJR070C and the parental strain, BY4743, were purchased from Open Biosystems (Huntsville, AL).
Yeast Culture and Purification of GST-ORF Proteins for Identification of a yDOHH Clone. Expression and isolation of GST fusion proteins from the GST-ORF library strains and pools were carried out as described in refs. 24 and 25 with slight modifications. S. cerevisiae strains in the 64 original pools (each pool containing 96 clones) and in the subpools of plate 18 were cultured by inoculation of 100 μl of the original culture in 10 ml of SD-Ura medium (synthetic dextrose minimal medium lacking uracil). After 36 h, the culture was diluted in 125 ml of SD-Ura-Leu medium. GST fusion protein expression was induced by 1 mM CuSO4 for 3 h. Cells were lysed in 0.6 ml of buffer A (50 mM Tris·HCl, pH 7.5/1 mM DTT/0.1 M NaCl and protease inhibitor mixture) by shearing with glass beads using BioSpec bead beater (Bartlesville, OK). Glutathione Sepharose-4B (0.05-ml bead volume) was added to the clarified cell extract, and the tube was rotated at 4°C for 3 h. The beads were washed three times with 1 ml of buffer B (50 mM Tris·HCl, pH 7.5/1mMDTT/0.1 M NaCl). The GST fusion proteins were eluted by addition of 0.3 ml of buffer C (50 mM Tris·HCl/1mMDTT/0.1 M NaCl/30 mM l-glutathione, final pH 8.0) and rotation at 4°C for 30 min. The GST-ORF proteins were concentrated and freed of glutathione (GSH) by using Microcon YM30 centrifugal filter devices (Millipore), and aliquots were used for SDS/PAGE and DOHH assay.
Construction of Bacterial and Mammalian Expression Vectors for yDOHH and hDOHH. The yeast vector expressing DOHH activity (pYEX 4T-1/yDOHH) was isolated from a single clone from well A7 of plate 18. After confirmation of accuracy of the sequence, this vector was used as a template for PCR amplification. For construction of the bacterial expression vector pGEX-4T-3/yDOHH, the primers, 5′-ctc cag tgt gcg aat tcc ATG TCT ACT AAC TTT GAA AAA CAT TTC-3′ and 5′-ca gct cca gtg tgc tgc ggc cgc CTA ATT AGC AGT TGG AGC ATA TTC TAG-3′ were used. For construction of hDOHH expression vectors, the pOTB7 vector encoding the human gene HLRC1 (MGC4293) (American Type Culture Collection) was used as a template and the primers 5′-ctc cag tgt gct gga tcc ATG GTG ACG GAG CAG GAG GTG GAT GCC-3′ and 5′-ctc cag tgt gct gtc gac CTA GGA GGG GGC CCC GCG CAG CTG CTC-3′ were used for pGEX-4T-3/hDOHH and 5′-ctc cag tgt gct gaa ttc ATG GTG ACG GAG CAG GAG GTG GAT GCC-3′ and 5′-ctc cag tgt gct tct aga CTA GGA GGG GGC CCC GCG CAG CTG CTC-3′ were used for the mammalian expression vector pCEFL/hDOHH.
Preparation of Recombinant GST-DOHH and DOHH from BL21(DE3) Cells. Escherichia coli BL21(DE3) cells transformed with either pGEX-4T-3/hDOHH or pGEX-4T-3/yDOHH were cultured in 1 liter of LB containing 100 μg/ml ampicillin. The protein expression was induced by 1 mM isopropyl β-d-thiogalactoside for 2 h. The cells were harvested and sonicated in 20 ml of buffer A by using an ultrasonic processor (Misonix, Farmingdale, NY). The clarified lysate was incubated with 10 ml of GSH-Sepharose by rotation for 3 h at 4°C; the resin was washed in the column with 50 ml of buffer B. GST-DOHH protein was eluted with buffer C containing 30 mM GSH, yielding ≈30 mg of highly pure GST-DOHH. Free DOHH enzymes were generated from the GSH-resin bound GST-DOHH by cleavage with thrombin by using the Thrombin Cleavage Capture kit (Novagen).
DOHH Assay. A typical DOHH reaction mixture contained 50 mM Tris·HCl (pH 7.5), 6 mM DTT, 25 μg of BSA, 2 pmol of the radiolabeled protein substrate, [3H]eIF5A(Dhp) prepared as described in ref. 28, and enzyme in 20 μl. Human substrate protein was used for human enzyme and the yeast substrate protein for the yeast enzyme. After incubation at 37°C for 1 h, the proteins were precipitated with 10% trichloroacetic acid and the precipitates hydrolyzed in 6 M HCl at 110°C for 18 h. The contents of [3H]hypusine and [3H]deoxyhypusine were determined after ion exchange chromatographic separation as described in refs. 23 and 29.
Computational Analysis of DOHH. The nonredundant database (NRDB) of protein sequences (National Center for Biotechnology Information, National Institutes of Health) was searched by using the blastpgp program (30). Iterative sequence profile searches were done using the psi-blast program (30) either with a single sequence or an alignment used as the query, with a profile inclusion expectation (E) value threshold of 0.01, and were iterated until convergence. For all searches with compositionally biased proteins, the statistical correction for this bias was used. Multiple alignments were constructed using the t_coffee program (31), followed by manual correction based on the psi-blast results. Protein secondary structure was predicted by using a multiple alignment as the input for the jpred program (32). Preliminary clustering of proteins was done by using the blastclust program (33) with empirically determined length and score threshold cut-off values. Structural manipulations and modeling were carried out by using the swiss-pdb viewer (34) and pymol programs (35). Homology modeling of protein structures was performed by using the promod ii program, which uses a gromos energy minimization protocol (36).
Results
Cloning of yDOHH from the S. cerevisiae GST-ORF Library. We initially screened the budding yeast S. cerevisiae GST-ORF Library (24, 25) for the expression of DOHH activity by testing 64 pools, each containing combined strains from a 96-well plate. Only one pool (plate 18) expressed DOHH activity as a GST fusion protein (data not shown). We then tested pools of the rows and columns of plate 18 and identified A7 ORF (YJR070C) to be the gene for S. cerevisiae DOHH. We isolated the pYEX-4T-1/yDOHH plasmid from a single A7 clone expressing DOHH activity in the form of a 62-kDa GST fusion protein (data not shown) and confirmed the nucleotide sequence of the complete ORF. YJR070C encodes a protein of 325 aa (36 kDa) with DOHH activity. Previously it had been reported that YJR070C encoded a protein named Lia1 (ligand of eIF5A) of unknown function identified by yeast two-hybrid screening (37).
Identification of YJR070C as the Single Gene for DOHH in S. cerevisiae. Sequence alignments shown in Fig. 1 illustrate the conservation of DOHH in all eukaryotes from fungi to human. The human homolog sequence HLRC1 is 48% identical (61% similar) to that encoded by YJR070C of S. cerevisiae. Only one homolog is found in each of these eukaryotes.
Fig. 1.
Multiple alignment of DOHH from diverse eukaryotic taxa. The individual HEAT repeats are numbered and shown above the alignment. The two sets of four HEAT repeats, which represent the last internal duplication in this protein, are shown. The secondary structure prediction (H represents α-helix and E an extended configuration) derived by using the jpred program (32) is shown above the alignment. The four characteristic HE motifs are highlighted in reverse shading. The coloring reflects the conservation profile at the 95% consensus. Absolutely conserved residues (capital letter, gray), hydrophobic residues (h: ACFILMVWY), big residues (b: FILMYWKERQ), alcoholic residues (o: T,S), small residues (s: AGSVCDN), tiny subset residues (t: GAS), polar residues (p: STEDKRNQHC), and charged residues (c: DEKHR) are shown in different colors.
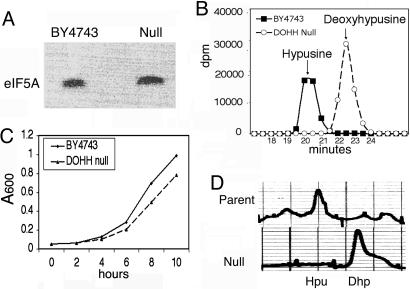
We compared growth and hypusine synthesis in a DOHH homozygous diploid knock-out strain (DOHH null) and its parent (BY4743). As shown in Fig. 2A, the labeling of the eIF5A protein was observed after culture of each strain in minimal SD medium containing [3H]spermidine. As expected, the parental strain contained [3H]hypusine as the labeled component of eIF5A (Fig. 2B). In contrast, only [3H]deoxyhypusine but no [3H]hypusine was found in the trichloroacetic acid-precipitated proteins of the DOHH-null strain (Fig. 2B), indicating that YJR070C indeed is the single gene encoding the DOHH enzyme in S. cerevisiae.
Fig. 2.
Comparison of growth, hypusine synthesis, and hypusine/deoxyhypusine content in DOHH-null and parent strains. The S. cerevisiae DOHH-null strain and the parental strain, BY4743, were cultured at 28°C. (A) SDS/PAGE after labeling with [1,8-3H]spermidine·HCl. (B) Ion exchange chromatographic separation of labeled hypusine and deoxyhypusine from labeled eIF5A proteins. (C) Growth curves. (D) Fluorometric detection of hypusine and deoxyhypusine in total cell protein (29).
The growth rate of the DOHH-null strain was only slightly lower than that of the parent under aerobic culture in rich medium (Fig. 2C) and in minimal medium (data not shown). Because the DHS gene is an essential gene and the first step of hypusine modification is absolutely required for yeast viability (1, 14), it was unexpected that the DOHH-null strain showed nearly normal growth. To rule out the possibility that a new hypusine biosynthetic pathway operates in the DOHH-null strain, we analyzed the amounts of total hypusine and deoxyhypusine in proteins of the two strains by fluorometric detection with O-phthalaldehyde. In accordance with the labeling experiments, only deoxyhypusine was found in the null strain, whereas hypusine and a small amount of deoxyhypusine were detected in the parental strain (Fig. 2D). These findings suggest that the eIF5A intermediate containing deoxyhypusine, eIF5A(Dhp), could fulfill the function of eIF5A in supporting growth of the budding yeast S. cerevisiae in the absence of fully modified eIF5A.
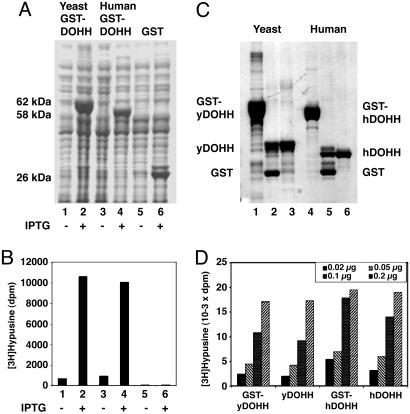
Expression and Purification of the Recombinant yDOHH and hDOHH. We subcloned the ORFs of S. cerevisiae YJR070C and the human homolog, HLRC1 (MGC4293), into the bacterial expression vector, pGEX-4T-3, to overproduce GST fusion proteins. After induction with isopropyl β-d-thiogalactoside, high expression of GST-yDOHH (62 kDa) and GST-hDOHH (58 kDa) was observed in BL21(DE3) cells (Fig. 3A). E. coli lysate expressing the human homolog gene displayed a comparable level of DOHH activity as those expressing the yDOHH, confirming that HLRC1 (MGC4293) is indeed the hDOHH gene. The increase in the amount of GST-DOHH protein in isopropyl β-d-thiogalactoside-treated cells correlated with the increase in DOHH enzyme activity (Fig. 3B). The human enzyme is a smaller protein (32-kDa protein with 302 aa) than the S. cerevisiae enzyme (36-kDa protein with 325 aa) (Fig. 3C). The activities of the GST fusion enzymes were similar to those of the free enzymes, and the purified yeast and human enzymes showed comparable activities (Fig. 3D). These findings confirm that the cloned yeast and human genes indeed encode DOHH enzymes.
Fig. 3.
Expression, purification, and activity determination of human and yeast GST-DOHH and free DOHH enzymes. DOHH proteins were expressed in BL21(DE3) cells as described in Materials and Methods. (A) SDS/PAGE of cell lysate (≈50 μg of protein). (B) DOHH activity assays of the lysate (2 μg of total protein). (C) SDS/PAGE of purified proteins. Lanes: 1–3, yeast enzyme; 4–6, human enzyme; 1 and 4, purified GST fusion proteins before thrombin treatment; 2 and 5, after thrombin treatment of GST-DOHH; 3 and 6, DOHH released after thrombin treatment of GSH-Sepharose-bound GST-DOHH. (D) DOHH activity assays with 0.02, 0.05, 0.1, and 0.2 μg of purified enzymes.
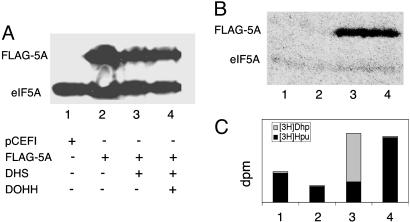
Cotransfection with Vectors Encoding eIF5A, DHS, and DOHH Is Required for Overproduction of Hypusine-Containing, Active eIF5A in Mammalian Cells. We expressed the hDOHH ORF in combination with human eIF5A-1 and DHS ORFs by using mammalian expression vectors. The 293T cells were transfected with p3XFLAG-CMV-7.1/heIF5A-1 alone or in combination with pCEFL/hDHS and pCEFL/hDOHH (Fig. 4). Western blotting with eIF5A-1 Ab showed overexpression of the FLAG-eIF5A-1 in addition to the endogenous eIF5A-1 in all samples transfected with p3XFLAG-CMV-7.1/heIF5A-1 (Fig. 4A, lanes 2–4). To determine how much of the overexpressed FLAG-tagged protein was modified, the transfected cells were cultured in medium containing [3H]spermidine, and the labeled protein and its radioactive components were analyzed (Fig. 4B). In cells transfected with the empty vector, faint labeling of only one protein, endogenous eIF5A-1 (18 kDa) was observed (lane 1). In this case, >90% of the radioactivity was present in [3H]hypusine, suggesting that these cells normally contain sufficient levels of DHS and DOHH to modify most of endogenously translated eIF5A precursor. In cells transfected with FLAG-eIF5A-1 expression vector alone, labeling of FLAG-eIF5A-1 (Fig. 4B, lane 2) was hardly detectable, indicating that the FLAG-eIF5A-1 protein largely exists as the unmodified precursor. This is presumably due to the insufficient level of endogenous DHS activity and/or inefficient modification of the FLAG-eIF5A-1 precursor. Only upon cotransfection with the modification enzymes, DHS (Fig. 4B, lane 3) or DHS plus DOHH (Fig. 4B, lane 4), was enhanced labeling of FLAG-eIF5A-1 manifest. Cells cotransfected with DHS expressing vector alone (Fig. 4C, lane 3) showed increased labeling of deoxyhypusine, indicating the FLAG-h5A-1 exists in the deoxyhypusine form in this case. Increased labeling of hypusine was observed only after cotransfection of FLAG-eIF5A-1 vector with both DHS and DOHH vectors (lane 4). These data demonstrate that the cloned hDOHH gene expresses functional DOHH activity in mammalian cells and that coexpression of all three proteins, eIF5A, DHS, and DOHH, is required for overproduction of fully modified eIF5A in mammalian cells.
Fig. 4.
Overproduction of hypusine-containing eIF5A in 293T cells by cotransfection with three vectors encoding eIF5A, DHS, and DOHH. The 293T cells were transfected with empty pCEFL vector (lane 1), p3XFLAG-CMV-7.1/heIF5A-1 alone (lane 2), together with pCEFL/hDHS (lane 3), or with pCEFL/hDHS plus pCEFL/hDOHH (lane 4). (A) Western blot with eIF5A-1 Ab. This Ab recognizes all three forms of eIF5A-1: the precursor [eIF5A(Lys)], the intermediate [eIF5A-1(Dhp)], and the mature eIF5A-1 (hypusine form) equally well. (B) Fluorogram of SDS gel of proteins of cells cultured with [3H]spermidine. (C) Content of [3H]hypusine and [3H]deoxyhypusine in the labeled proteins.
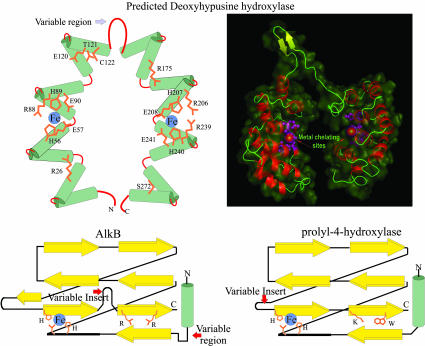
DOHH Is a HEAT-Repeat-Containing Metalloenzyme with a Structure Distinct from Other Known Protein Hydroxylases. Examination of the DOHH sequence alignment reveals that this protein consists of a dyad of four distinctive HEAT repeats (repeats 1–4 in the N-terminal domain and repeats 5–8 in the C-terminal domain, Fig. 1 Upper and Lower, respectively) connected by a variable loop (i.e., between repeat 4 and 5, Fig. 1). Circular dichroism spectral analysis of pure recombinant hDOHH revealed a high content of α-helical structure (80% ± 2.1) (P. McPhie and E.C.W., unpublished data), close to the value calculated from the predicted model (76–78%). The variable region between the dyad repeats does not show strong helical secondary structure prediction. The amino acid composition in this region suggests a loop with small, extended elements. The C-terminal helix of the second and third HEAT repeats in each domain (i.e., HEAT repeats 2, 3, 6, and 7) contains a highly characteristic histidine–glutamate (HE) motif. This arrangement of the conserved residues and the close similarity between the N- and C-terminal domains with four HEAT repeats each suggest that the N- and C-terminal halves form a symmetric shell-like structure with four conserved HE motifs contributing to metal-chelating sites.
To obtain a 3D visualization, we sought to construct a model of DOHH by using crystal structures of preexisting HEAT-repeat proteins as templates. A search of the nonredundant database with the human DOHH (hDOHH) as the query with the psi-blast recovered the E. coli protein YibA with E = 3 × 10–4. The crystal structure of YibA (Protein Data Bank entry 1OYZ) shows that it contains a tandem set of eight HEAT repeats forming a horseshoe like structure. Using YibA as a template, we constructed a homology model of hDOHH (Fig. 5 Upper). This model shows that the C-terminal helices of the HEAT repeats line the “inner” circumference of the curved toroid formed by the repeats. This observation, taken together with the presence of the HE motif on the C-terminal helix, suggests that the predicted metal-chelating sites indeed lie in the interior of the concave structure and that this concavity lined by the C-terminal helices is likely to form the interior cavity that accommodates the substrate protein. The variable region between the two dyads is likely to form a “hinge-like” structure that might allow some mobility for the two symmetric halves to accommodate the substrate. However, it should be stressed that such homology-based structural models only provide an approximate structure for the protein and its possible metal interactions. Although it seems valid that the α-helical HEAT-repeat motifs contribute to a solenoid structure and that the active site is defined by the strictly conserved HE residues, the model depicted in Fig. 5 does not provide a precise distance between the presumed metal-binding HE pairs on the two domains, nor the stoichiometry of bound metal to protein. There may be two active sites, and two metal atoms per DOHH monomer, or the flexible hinge may permit the two HE pairs on each domain to come into close proximity to form one metal-binding, active center per DOHH monomer. Alternatively, two monomers of DOHH could dimerize to form the functional DOHH. In this situation two, four-HEAT repeat dyads from separate adjacent polypeptide could cooperate to bind a metal ion.
Fig. 5.
Comparison of the predicted structure of DOHH with those of two DSBH dioxygenases. (Upper) The predicted structure of DOHH (Left), numbered according to the human protein, and a structural model (Right) based on the known x-ray structure of YibA (Protein Data Bank ID code 1OYZ). These models of DOHH structure are only approximations for its actual structure, and alternative modes of metal chelation and configuration of the HEAT repeats are possible (see text). (Lower) Topological diagrams (27) of two DSBH dioxygenases: AlkB and prolyl-4-hydroxylase (Protein Data Bank ID code 1E5S).
The predicted structure of DOHH is entirely different from those of the majority of previously studied hydroxylases, e.g., Fe(II)- and 2-oxoacid-dependent dioxygenases, which share a common feature in the form of a specific β jelly roll structure [termed the double-stranded β helix (DSBH)] (27). A schematic representation of the structures of two enzymes in this family, alkylated DNA repair protein alkB (ALKB) and prolyl-4 hydroxylase, are shown for comparison (Fig. 5 Lower). It is evident that the all α-helical structure of DOHH is unrelated to any of the diverse DSBH-fold enzymes. Yet both DOHH and DSBH enzymes seem to show similarities in their metal-binding active sites. The majority of the DSBH dioxygenases have a consensus His-1-X-D/E-Xn-His-2 sequence that chelates divalent metal ions by means of appropriately positioned histidine and acidic (E or D) residues in the interior of the cavity formed by the jelly roll structure. DOHH is also predicted to contain analogous metal coordination sites, supported by conserved HE residues (Fig. 5 Upper). Indeed, Fe was consistently found in purified preparations of the yeast and human enzymes upon analysis by high resolution inductively coupled plasma MS. DSBH enzymes contain basic amino acids lysine or arginine that stabilizes α-ketoglutaric acid binding. A potential α-ketoglutaric acid-binding site could not be readily identified in DOHH, consistent with the lack of requirement of this 2-oxoacid for the activity of purified hDOHH or yDOHH (data not shown). Whereas both enzymes involve Fe for catalysis, the reaction mechanism of DOHH may be distinct from that of DSBH enzymes (19, 38).
Discussion
We have identified YJR070C as the single gene for DOHH in S. cerevisiae and demonstrated that this gene and its human homolog, HLRC1 (MGC4293), encode active DOHH enzymes. A single homologous gene is found in eukaryotic organisms but not in eubacteria or archaea, and this DOHH gene is highly conserved from fungi to human. Computational analysis of the DOHH structure reveals that it is a HEAT-repeat protein containing eight helical hairpins (HEAT motif) (Figs. 1 and. 5) and two potential metal coordination sites, which are composed of four strictly conserved HE sequences (Fig. 1). Thus, DOHH appears to be a unique protein hydroxylase distinct from the family of Fe(II)- and 2-oxoacid-dependent dioxygenases.
This study reports on the overexpression of fully modified, hypusine-containing eIF5A in eukaryotes by cotransfection of both DHS and DOHH expression vectors with that encoding eIF5A. Transfection of the eIF5A expression vector alone caused accumulation of unmodified eIF5A precursors in mammalian cells and did not significantly increase the level of hypusine-containing eIF5A, suggesting that DHS and DOHH enzymes are limiting in case of eIF5A precursor overexpression. Thus, coexpression of the DHS and DOHH enzymes is vital for overproduction of mature eIF5A in eukaryotic expression systems for functional and structural studies. Furthermore, overexpression of hypusine-containing eIF5A-1 and -2 isoforms by cotransfection with DHS and DOHH will lead to a better understanding of the role of eIF5A in proliferation, transformation, and apoptosis in mammalian cells.
The emergence and essentiality of the posttranslational modifications of the eIF5A precursor and the hypusine biosynthetic enzymes show an interesting development in the course of evolution. There is currently no experimental evidence suggesting that EF-P, the eubacterial homolog of eIF5A, undergoes modification to the deoxyhypusine- or hypusine-containing form in experimentally studied bacteria. Consistent with this, most commonly studied bacteria lack a DHS gene. However, several cyanobacteria, myxobacteria, Thermus, Chlorobium, Rhodopirellula, Bdellovibrio, Zymomonas, Caulobacter, and Cytophaga contain a DHS cognate that appears to have been laterally transferred from the archaea. It would be of interest, therefore, to determine whether these bacteria modify their EF-Ps through a DHS-mediated reaction. eIF5A and DHS are seen in all archaeal proteomes available to date, suggesting that the first step of modification of lysine to deoxyhypusine is likely to be essential in all archaea (8). However, no orthologs of the eukaryotic DOHH identified in this study are seen in the available archaeal proteomes. Yet, certain branches of archaea were reported to contain hypusine, whereas others contain deoxyhypusine only (7). It is not clear how hypusine was produced in these species, given that no DOHH homolog is found in archaea. DOHH homologs do exist in all species of yeast but their functional significance seems to vary. A S. cerevisiae DOHH-null strain grew only slightly slower than its parent strain. In contrast, DOHH seems to be functionally more significant in the fission yeast, S. pombe. The S. pombe homolog of the DOHH gene, Mmd1, was recently reported to be important for normal mitochondrial morphology and distribution in S. pombe (39). An mmd1 mutation (E66K) caused temperature-sensitive growth and defects in mitochondrial morphology and distribution at nonpermissive temperature. These phenotypes were attributed to the defects of mmd1 mutant protein in promoting microtubule assembly at the nonpermissive temperature. It is not known whether DOHH itself or its product of modification, namely eIF5A (the hypusine-containing form), is directly or indirectly involved in microtubule stability and function. The site of the temperature-sensitive mutation in the mmd1 protein (E66) corresponds to the E57 of the hDOHH, at one of the strictly conserved HE motifs proposed for metal chelation (Figs. 1 and 5). In higher organisms, deoxyhypusine hydroxylation seems to be a mandatory requirement for survival and growth because inactivation of the DOHH gene is recessively lethal in higher organisms, i.e., Caenorhabditis elegans (40) or Drosophila melanogaster (41). Thus, the essentiality of the final step of hypusine modification has probably evolved only in the multicellular eukaryotes.
Whereas the DOHH homologs belong to a family of HEAT-repeat proteins, their sequence conservation is much greater than that observed for other conserved HEAT-repeat proteins, namely karyopherins. The majority of HEAT-repeat domains known to date, such as the archetypal karyopherins, are noncatalytic proteins with roles in protein–protein interactions. The closest relatives of the HEAT repeats found in DOHH are the HEAT repeats of the phycobilisome lyase family, which includes the phycocyanobilin and phycoerythrobilin lyases encoded by the CpcE/F, CpcY/Z, and MpeU/V genes of photosynthetic bacteria (26). In addition to DOHH, these are the only other HEAT-repeat catalytic domains in enzymes known to date. It is likely that this family had a preadaptation to bind organic substrates, which favored the evolution of unique catalytic activities amongst them.
The structural fold of DOHH is unrelated to the β jelly roll structure termed DSBH of other protein hydroxylases (Fig. 5). DSBH is observed or predicted through sequence profile analysis in a number of distinct forms of dioxygenases modifying proteins, nucleic acids, and other substrates (27). Despite the structural differences, the chemical mechanism of DOHH may resemble the metal-dependent DSBH dioxygenases (19, 38); thus, it represents a remarkable example of convergent evolution, where similar mechanisms and active sites have emerged independently in totally different structural scaffolds. DOHH contains highly conserved HE motifs, the potential metal coordination sites consisting of four strictly conserved HE motifs. Unlike the DSBH dioxygenases, DOHH is the first and only known representative of a HEAT-repeat Fe-dependent dioxygenase. Hence, it is likely that it is a relatively late evolutionary innovation, where a catalytic activity arose in a very specific context in the translation apparatus. In light of this, it is tempting to speculate that DOHH probably arose from a HEAT protein that originally merely bound the deoxyhypusine-containing eIF5A and possibly served as an adaptor mediating certain protein–protein interactions. Subsequently, key active site residues appear to have evolved that allowed DOHH catalysis. Therefore, in addition to its enzymatic activity, DOHH may have an evolutionarily conserved function in protein–protein interaction, perhaps critical for translation or cytoskeletal organization.
DOHH and mature eIF5A (hypusine form) are essential for proliferation of mammalian cells. Various metal-chelating inhibitors of DOHH, including mimosine and ciclopirox olamine, cause inhibition of cell proliferation by causing cell cycle arrest at the G1/S boundary (21). Ciclopirox olamine and other inhibitors exhibited strong inhibition of human umbilical vein endothelial cells proliferation and angiogenesis in model assays, suggesting the potential utility of DOHH inhibitors as antitumor agents (23). Although eIF5A and DOHH have been proposed as potential targets of antitumor therapy and anti-HIV-1 therapy, no specific inhibitors of DOHH are currently available. Molecular cloning of DOHH is the first critical step toward characterization of structure and mechanism and toward development of specific inhibitors of this important eukaryotic enzyme.
Acknowledgments
We thank Dr. Kyung Sang Lee (National Cancer Institute, National Institutes of Health, Bethesda) for making the Biospec Bead Beater available and for advice on the yeast techniques and Drs. Myong-Hee Sung (National Cancer Institute), Hans E. Johansson (Biosearch Technologies, Novato, CA), and Jessica Bell (National Institute of Diabetes and Digestive and Kidney Diseases) for helpful discussions. This research was supported by the Intramural Research Program of the National Institutes of Health (National Institute of Dental and Craniofacial Research, National Center for Biotechnology Information/National Library of Medicine).
Author contributions: M.H.P. designed research; J.-H.P., E.C.W., J.K., Y.S.K., and M.H.P. performed research; L.A. contributed new reagents/analytical tools; E.C.W. and M.H.P. analyzed data; L.A. and M.H.P. wrote the paper; and L.A. performed the sequence/structure analysis of the DOHH protein and the computational prediction of active site residues.
Conflict of interest statement: No conflicts declared.
Abbreviations: eIF5A, eukaryotic initiation factor 5A; DHS, deoxyhypusine synthase; DOHH, deoxyhypusine hydroxylase; DSBH, double-stranded β-helix; GSH, glutathione; HE, histidine–glutamate.
References
- 1.Park, M. H., Lee, Y. B. & Joe, Y. A. (1997) Biol. Signals 6, 115–123. [DOI] [PubMed] [Google Scholar]
- 2.Chen, K. Y. & Liu, A. Y. (1997) Biol. Signals 6, 105–109. [DOI] [PubMed] [Google Scholar]
- 3.Joe, Y. A., Wolff, E. C. & Park, M. H. (1995) J. Biol. Chem. 270, 22386–22392. [DOI] [PubMed] [Google Scholar]
- 4.Abbruzzese, A., Park, M. H. & Folk, J. E. (1986) J. Biol. Chem. 261, 3085–3089. [PubMed] [Google Scholar]
- 5.Byers, T. L., Lakanen, J. R., Coward, J. K. & Pegg, A. E. (1994) Biochem. J. 303, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay, M. K., Tabor, C. W. & Tabor, H. (2003) Proc. Natl. Acad. Sci. USA 100, 13869–13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartig, D., Schümann, H. & Klink, F. (1990) Syst. Appl. Microbiol. 13, 112–116. [Google Scholar]
- 8.Jansson, B. P. M., Malandrin, L. & Johansson, H. E. (2000) J. Bacteriol. 182, 1158–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, K. K., Hung, L. W., Yokota, H., Kim, R. & Kim, S. H. (1998) Proc. Natl. Acad. Sci. USA 95, 10419–10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peat, T. S., Newman, J., Waldo, G. S., Berendzen, J. & Terwilliger, T. C. (1998) Structure 6, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 11.Hanawa-Suetsugu, K., Sekine, S., Sakai, H., Hori-Takemoto, C., Terada, T., Unzai, S., Tame, J. R., Kuramitsu, S., Shirouzu, M. & Yokoyama, S. (2004) Proc. Natl. Acad. Sci. USA 101, 9595–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnier, J., Schwelberger, H. G., Smit-McBride, Z., Kang, H. A. & Hershey, J. W. (1991) Mol. Cell. Biol. 11, 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wöhl, T., Klier, H. & Ammer, H. (1993) Mol. Gen. Genet. 241, 305–311. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki, K., Abid, M. R. & Miyazaki, M. (1996) FEBS Lett. 384, 151–154. [DOI] [PubMed] [Google Scholar]
- 15.Park, M. H., Joe, Y. A. & Kang, K. R. (1998) J. Biol. Chem. 273, 1677–1683. [DOI] [PubMed] [Google Scholar]
- 16.Wolff, E. C., Folk, J. E. & Park, M. H. (1997) J. Biol. Chem. 272, 15865–15871. [DOI] [PubMed] [Google Scholar]
- 17.Umland, T. C., Wolff, E. C., Park, M. H. & Davies, D. R. (2004) J. Biol. Chem. 279, 28697–28705. [DOI] [PubMed] [Google Scholar]
- 18.Wolff, E. C., Wolff, J. & Park, M. H. (2000) J. Biol. Chem. 275, 9170–9177. [DOI] [PubMed] [Google Scholar]
- 19.Hausinger, R. P. (2004) Crit. Rev. Biochem. Mol. Biol. 39, 21–68. [DOI] [PubMed] [Google Scholar]
- 20.Kivirikko, K. I. & Pihlajaniemi, T. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 325–398. [DOI] [PubMed] [Google Scholar]
- 21.Hanauske-Abel, H. M., Park, M. H., Hanauske, A. R., Popowicz, A. M., Lalande, M. & Folk, J. E. (1994) Biochim. Biophys. Acta 1221, 115–124. [DOI] [PubMed] [Google Scholar]
- 22.Csonga, R., Ettmayer, P., Auer, M., Eckerskorn, C., Eder, J. & Klier, H. (1996) FEBS Lett. 380, 209–214. [DOI] [PubMed] [Google Scholar]
- 23.Clement, P. M., Hanauske-Abel, H. M., Wolff, E. C., Kleinman, H. K. & Park, M. H. (2002) Int. J. Cancer 100, 491–498. [DOI] [PubMed] [Google Scholar]
- 24.Martzen, M. R., McCraith, S. M., Spinelli, S. L., Torres, F. M., Fields, S., Grayhack, E. J. & Phizicky, E. M. (1999) Science 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 25.Phizicky, E. M., Martzen, M. R., McCraith, S. M., Spinelli, S. L., Xing, F., Shull, N. P., Van Slyke, C., Montagne, R. K., Torres, F. M., Fields, S., et al. (2002) Methods Enzymol. 350, 546–559. [DOI] [PubMed] [Google Scholar]
- 26.Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W. & Bork, P. (2001) J. Mol. Biol. 309, 1–18. [DOI] [PubMed] [Google Scholar]
- 27.Aravind, L. & Koonin, E. V. (2001) Genome Biol. 2, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, J. H., Wolff, E. C., Folk, J. E. & Park, M. H. (2003) J. Biol. Chem. 278, 32683–32691. [DOI] [PubMed] [Google Scholar]
- 29.Park, M. H., Liberato, D. J., Yergey, A. L. & Folk, J. E. (1984) J. Biol. Chem. 259, 12123–12127. [PubMed] [Google Scholar]
- 30.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 32.Cuff, J. A., Clamp, M. E., Siddiqui, A. S., Finlay, M. & Barton, G. J. (1998) Bioinformatics 14, 892–893. [DOI] [PubMed] [Google Scholar]
- 33.Lespinet, O., Wolf, Y. I., Koonin, E. V. & Aravind, L. (2002) Genome Res. 12, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex, N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 35.DeLano, W.L. (2002) pymol: A Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 36.Peitsch, M. C. (1996) Biochem. Soc. Trans. 24, 274–279. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, G. M., Cano, V. S. & Valentini, S. R. (2003) FEBS Lett. 555, 464–468. [DOI] [PubMed] [Google Scholar]
- 38.Hanauske-Abel, H. M. & Gunzler, V. (1982) J. Theor. Biol. 94, 421–455. [DOI] [PubMed] [Google Scholar]
- 39.Weir, B. A. & Yaffe, M. P. (2004) Mol. Biol. Cell 15, 1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugimoto, A. (2004) Differentiation 72, 81–91. [DOI] [PubMed] [Google Scholar]
- 41.Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]