Abstract
Many human diseases are characterized by the development of tissue hypoxia. Inadequate oxygenation can cause cellular dysfunction and death. Tissues use many strategies, including induction of angiogenesis and alterations in metabolism, to survive under hypoxic conditions. The heterodimeric transcription factor hypoxia-inducible factor (HIF) is a master regulator of genes that promote adaptation to hypoxia. HIF activity is linked to oxygen availability because members of the EGLN family hydroxylate HIFα subunits on specific prolyl residues when oxygen is present, which marks them for ubiquitination and proteasomal degradation. We created a mouse that ubiquitously expresses a bioluminescent reporter consisting of firefly luciferase fused to a region of HIF that is sufficient for oxygen-dependent degradation. Our validation studies suggest that this mouse will be useful for monitoring hypoxic tissues and evaluating therapeutic agents that stabilize HIF. One such agent, the HIF prolyl hydroxylase inhibitor FG-4383, was active in the liver and kidney after systemic administration as determined by bioluminescence imaging, transcription profiling, and production of erythropoietin, indicating that the HIF transcriptional program can be manipulated in vivo with orally active organic small molecules.
Keywords: bioluminescence, imaging, von Hippel-Lindau
A number of imaging modalities, including positron emission tomography, magnetic resonance imaging (MRI), and optical imaging, can provide anatomical and functional information about normal and diseased tissue (1). Optical imaging is the least expensive of these three modalities and, hence, the most widely used by academic and pharmaceutical research laboratories. Regardless of the modality used, noninvasive imaging is likely to become an increasingly important part of basic and translational research as well as drug development and medical care by enabling the detection of molecular signatures indicative of specific disease states or the actions of particular therapeutics.
A number of bioluminescent and fluorescent reporters have been used for optical imaging including firefly luciferase, green fluorescent protein, and caged near infrared probes (1, 2). Bioluminescent and fluorescent proteins can be genetically reengineered so that particular enzymes affect their abundance or specific activity. For example, such proteins can be rendered unstable by fusion to polypeptides that are recognized by specific ubiquitin ligases complexes, which act to target proteins for proteasomal degradation (reviewed in ref. 2). The resulting fusion proteins can then be used to interrogate molecular events that affect their respective ubiquitin ligases. Bioluminescent reporters, such as luciferases, have the advantage that they do not require excitation by an external source, and, hence, problems related to tissue autofluorescence are minimized.
Oxygen plays a critical role in cellular homeostasis and many human diseases, including atherosclerotic diseases and cancer, are characterized by inadequate tissue oxygenation. In the former, lack of oxygen contributes to cell death and, in the latter, lack of oxygen is an early signature of disease that might also affect malignant cell behavior. The transcription factor hypoxia-inducible factor (HIF), which consists of a labile α subunit and a stable β subunit, plays a pivotal role in adaptation to hypoxia. In the presence of oxygen, members of the EGLN family hydroxylate HIFα subunits on conserved prolyl residues (3, 4). Hydroxylated HIFα species are polyubiquitinated and, hence, marked for destruction, by an ubiquitin ligase that contains the pVHL tumor suppressor protein (3, 4). Under low-oxygen conditions, or in the absence of pVHL, HIF accumulates in its active form and transcriptionally activates genes involved in acute or chronic adaptation to hypoxia. EGLN belongs to a superfamily of enzymes that require oxygen, Fe2+, and 2-oxoglutarate for activity (5, 6). EGLN activity is sensitive to changes in oxygen over a physiologically relevant concentration range, suggesting that it is capable of acting as an oxygen sensor (3, 4). Small-molecule EGLN inhibitors that interfere with iron or 2-oxoglutarate utilization are being developed for the treatment of ischemic diseases (7). We reasoned that fusing luciferase to the region of HIF1α that binds to pVHL in an oxygen-dependent manner would generate a reporter for EGLN activity that might aid the preclinical development of EGLN antagonists. In addition, such a reporter might be used to monitor oxygen availability in intact cells.
Materials and Methods
Plasmids. A HIF1α cDNA was PCR amplified by using PFU DNA polymerase and primers (HIF530F: 5′CCCAAGCTTGGATCCGAATTCGCCACCATGGAATTCAAGTTGGAATTGGTAG 3′ and HIF653B 5′TAGAATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCAGTAGTTTCTTTATGTATGTGGG 3′) that introduced a 5′ HindIII site and 3′ NarI site. The PCR product was subcloned into pGL3-control (Promega) cut with these two enzymes to make pGL3-ODD-Luc. The ODD-Luc cDNA was excised with HindIII and XbaI and inserted into pcDNA3 (Invitrogen) cut with these two enzymes to make pcDNA3-ODD-Luc.
The Rosa-26 PA and the pBig-T plasmids were a gift of F. Constantini (Columbia University, College of Physicians and Surgeons, New York). pBigT contains two LoxP sites that flank a Neo expression cassette and a strong transcriptional stop sequence. After the second LoxP site, there is a multiple cloning site. All these elements are located between unique PacI and AscI restriction sites. The Rosa-26 PA plasmid contains the Rosa-26 genomic sequence within which is inserted a linker that contains PacI and AscI sites. In addition, this plasmid contains a diphtheria toxin cassette (PGK-DTA) 3′ of the ROSA26 genomic DNA to facilitate selection against nonhomologous recombinants (8).
To make pBigT-ODD-Luc, pcDNA3-ODD-Luc was cut with HindIII (followed by fill-in with Klenow) and ApaI. The liberated luciferase cDNA was then ligated into pBig T that had been cut with XhoI (followed by fill-in with Klenow) and ApaI. The PacI-AscI fragment from pBigT-luciferase containing the floxed neomycin cassette and ODD-Luc cDNA were ligated into pRosa-26PA cut with the same enzymes to make the L-S-L ODD-Luciferase ROSA26 targeting construct.
pVHL Binding Studies. pVHL binding studies, including production of GST-pVHL/elongin B/elongin C complexes, were performed as described in ref. 9.
Cell Culture Luciferase Assays. 786-O cells and their derivatives were transfected with 1 μg of either pcDNA3-luciferase or pcDNA3-ODD-Luc, 4 μg of pcDNA3, and 50 ng of the Renilla luciferase expression plasmid pRL-CMV (Promega) by using Fugene transfection reagent according to the manufacturer's instructions. HeLa cells were transfected with 0.5 μg of either pcDNA3-luciferase or pcDNA3-ODD-Luc, 4.5 μg of pcDNA3, and 50 ng of pRL-CMV by using the calcium phosphate method. The next day, cells were split to 60-mm plates and allowed to recover for 6 h before treatment with hypoxia or hypoxia mimetics. Twenty-four hours later, luciferase activity of cells extracts was measured with the Dual Luciferase Assay System (Promega) according to the manufacturer's instructions and a Berthold Technologies luminometer.
Targeting of the Rosa-26 ODD-Luciferase in ES Cells. The ROSA26 ODD-Luc targeting construct was linearized with KpnI and electroporated into TC1 embryonic stem (ES) cells (derived from 129SvEv strain) by using standard techniques. Two of 100 G418-resistant ES clones underwent successful homologous recombination, as determined by Southern blot with a 5′ ROSA26 probe (External Probe in Fig. 6A, which is published as supporting information on the PNAS web site), and were microinjected into C57/BL6 blastocysts. High-percentage chimeric mice were obtained and bred to FVB-EIIA-Cre mice (10).
Mice Genotyping. PCR of genomic DNA was performed with AmpliTaq Gold DNA (Applied Biosystems) according to the manufacturer's instruction with forward primer 5′-CGGTATCGTAGAGTCGAGGCC-3′ and reverse primer 5′-GGTAGTGGTGGCATTAGCAGTAG-3′ to detect the ODD-Luc cDNA or reverse primer 5′-CAGGGCGTATCTCTTCATAGCC-3′ to detect either ODD-Luc or luciferase cDNA.
Detecting Luciferase Expression in Vivo. Mice were given a single i.p. injection of a mixture of luciferin (50 mg/kg) ketamine (150 mg/kg) and xylazine (12 mg/kg) in sterile water. Fifteen minutes later, mice were placed in a light-tight chamber equipped with a charge-coupled device IVIS imaging camera (Xenogen, Alameda, CA). Photons were collected for a period of 5–20 s, and images were obtained by using living image software (Xenogen) and igor image analysis software (WaveMatrics, Lake Oswego, OR).
Exposure of Mice to Hypoxic Environment. Mice were placed in hypoxia chamber (INVIVO2 from Biotrace, Bridgend, U.K.) containing 21% O2. The oxygen levels were decreased 2% every 10 min until they reached 8%. The mice were left at 8% O2 for 4 h before imaging.
Nephrectomies. Nephrectomies were performed essentially as described in ref. 11. Briefly, mice were anesthetized with isoflurane, and a midline abdominal incision was made under sterile conditions. The kidney capsules were removed, and both kidneys were removed after pedicle ligation. The abdomen was then closed with surgical staples, and the animals were allowed to recover for 2 h before treatment with FG-4383.
Erythropoietin (EPO) ELISA. Blood was collected via the abdominal vein into a heparinized tube 6 h after a single oral dose. The tubes were spun at 10,000 rpm in a Microfuge 22R centrifuge (Beckman Coulter) for 10 min at 4°C, and the plasma was transferred to a 1.5-ml tube for EPO analysis by using a commercial ELISA kit specific for mouse EPO (R & D Systems) according to the manufacturer's instructions.
Results
HIF1α contains two potential prolyl hydroxylation sites at residues 402 and 564 (3, 4). Previous studies showed that a peptide corresponding to HIF1α residues 556–575 could bind to pVHL in a hydroxylation-dependent manner and that a polypeptide corresponding to HIF1α residues 530–652 could target a heterologous protein for oxygen-dependent degradation in cis (12–16). Therefore, we made expression plasmids that encode fusion proteins consisting of either HIF1α (556–575) or HIF1α (530–652) fused to the N terminus of firefly luciferase. Pilot experiments suggested that the HIF1α (530–652) containing fusion protein, hereafter called ODD-Luc, bound to pVHL with higher affinity than the fusion protein with the HIF1α (556–575) element (data not shown). Therefore we focused our attention on ODD-Luc.
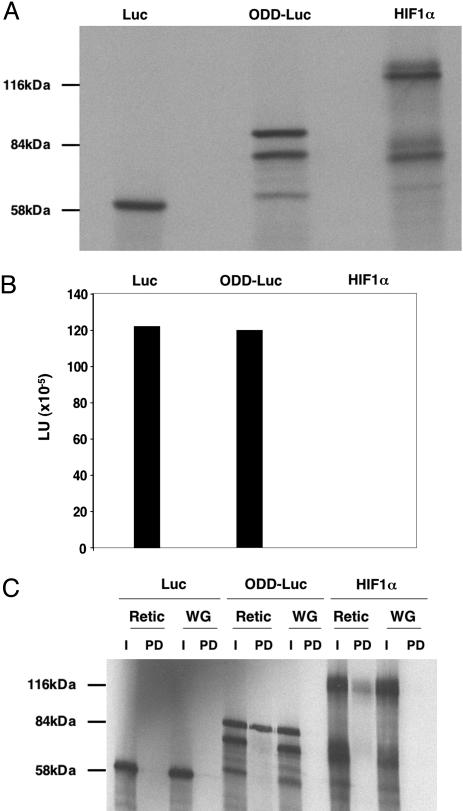
Luciferase activity of ODD-Luc made by in vitro translation was comparable to that of the wild-type firefly luciferase (Fig. 1 A and B). In addition, ODD-Luc, like full-length HIF1α, bound to recombinant pVHL (Fig. 1C). Elongin B and elongin C were included in these binding assays to promote the proper folding of pVHL (17, 18). Earlier studies showed that rabbit reticulocyte lysate, in contrast to wheat germ extract, contains HIF prolyl hydroxylase activity (7, 12). pVHL bound to ODD-Luc and HIF produced in rabbit reticulocyte lysates but not in wheat germ extracts (Fig. 1C). Moreover, binding of ODD-Luc to pVHL was blocked by a HIF1α-derived pVHL-binding peptide, provided that Pro-564 was hydroxylated (data not shown). Collectively, these experiments indicated that ODD-Luc displays the key functional attributes of both fusion partners in vitro.
Fig. 1.
ODD-Luc fusion protein retains luciferase activity and binds to pVHL. (A) Autoradiogram of the indicated 35S-labeled proteins after in vitro translation and SDS/polyacrylamide gel electrophoresis. (B) Luciferase activity (LU, light units) corresponding to 50% (by volume) of the in vitro translation products studied in A. Data shown are representative of two independent experiments. (C) Autoradiogram of the indicated 35S-labeled proteins made by in vitro translation in rabbit reticulocyte lysate (Retic) or wheat germ extract (WG). Proteins were incubated with glutathione Sepharose preloaded with GST-pVHL, elongin B, and elongin C. Specifically bound proteins were released by boiling in SDS-containing sample buffer and resolved by SDS-polyacrylamide gel electrophoresis (PD, pulldown). For comparison, 10% of the protein used for the binding studies was load directly and resolved in the same gel (I, input).
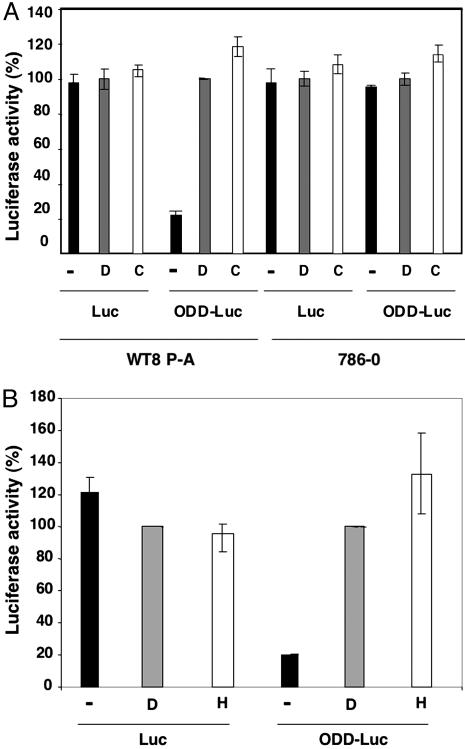
Next, 786-O VHL–/– renal carcinoma cells and WT8 HIF2α (P → A) renal carcinoma cells were transiently transfected with a plasmid-encoding renilla luciferase and a plasmid-encoding wild-type firefly luciferase or ODD-Luc. WT8 (P → A) cells are 786-O derivatives that were stably transfected to produce wild-type pVHL and retrovirally infected to produce a HIF2α variant lacking Pro-564, which escapes recognition by pVHL (19). After a 24-h recovery period, the cells were treated with the hypoxia mimetics deferoxamine (DFO) or cobalt chloride, which inhibit HIF prolyl hydroxylase activity. Both DFO and cobalt chloride significantly induced ODD-Luc activity in WT8 (P → A) cells, which produce wild-type pVHL, but not in 786-O cells, which lack wild-type pVHL, (Fig. 2A). In contrast, neither DFO nor cobalt chloride significantly affected wild-type firefly luciferase activity or that of the renilla luciferase included for normalization purposes. Similarly, treatment with DFO or low (0.2%) oxygen increased firefly luciferase activity in HeLa VHL+/+ cervical carcinoma cells that were transiently transfected to produce ODD-Luc but not in cells transfected to produce wild-type firefly luciferase (Fig. 2B). Together, these results indicated that ODD-Luc is sensitive to changes in HIF prolyl hydroxylase activity in a well defined cell culture-based system. The observed induction of ODD-Luc might underestimate the potential dynamic range of this reporter in vivo because certain proteins required for the polyubiquitylation and destruction of ODD-Luc might become limiting at the ODD-Luc concentrations achieved after transient transfection.
Fig. 2.
ODD-Luc is induced by hypoxia and hypoxia mimetics. (A) Firefly luciferase activity, corrected for renilla luciferase, of WT8 P → A cells [pVHL(+)] and 786-O [pVHL(–)] renal carcinoma cells transfected to produce wild-type firefly luciferase or ODD-Luc. Where indicated, 500 μM deferoxamine (D) or 100 μM cobalt chloride (C) was added to media 24 h before analysis. Luciferase data were normalized to D for each cell line-reporter pair. (B) Normalized luciferase values for HeLa VHL+/+ cervical carcinoma cells transfected to produce wild-type firefly luciferase or ODD-Luc. Where indicated, cells were exposed to 500 μM deferoxamine (D) or 0.2% oxygen (H) for 24 h before analysis. Error bars indicate one standard error.
Encouraged by these cell culture data, the ODD-Luc cDNA was introduced into the ubiquitously expressed mouse ROSA26 locus by homologous recombination (20, 21). Mouse embryonic stem cells that had undergone successful recombination, as determined by Southern blot analysis and PCR, were microinjected into C57/BL6 blastocysts to generate germline-transmitting chimeric mice, which were then bred to FVB-EIIA Cre mice to remove the neomycin-resistance cassette present in the targeting vector (Fig. 6). In parallel, we generated similar mice, which have been described in ref. 10, in which a wild-type luciferase cDNA was inserted into the ROSA26 locus.
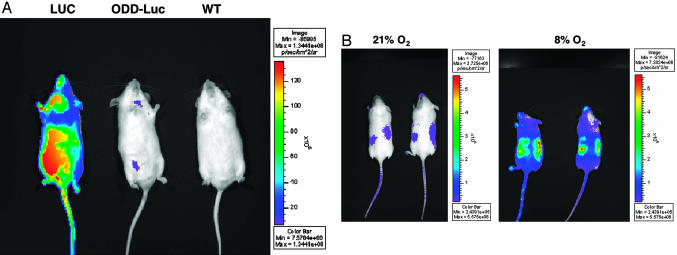
As expected, ROSA26 ODD-Luc/+ mice emitted much less light than ROSA26 Luc/+ mice after i.p. administration of luciferin (Fig. 3A). In these and subsequent experiments, emitted light was measured by using a photon-counting charge-coupled device camera and software was used to generate a pseudocolorized image that reflected signal strength. Although a number of variables can influence regional light emission in such assays (see Discussion), we noted that a light signal was detectable in the region of the kidneys in ROSA26 ODD-Luc/+ mice that were breathing room air (Fig. 3B). This signal was eliminated by ligature of the renal artery, and 2-nitroimidazole (HypoxyProbe) staining confirmed earlier reports that the kidneys of mammals breathing room air contain hypoxic zones (Figs. 7 and 8, which are published as supporting information on the PNAS web site). A 5- to 10-fold increase in light emission was observed when ROSA26 ODD-Luc/+ mice were placed in a low oxygen (8%) environment, unlike ROSA26 Luc/+ mice (Fig. 3B; see also Fig. 9, which is published as supporting information on the PNAS web site) (see Discussion regarding oxygen concentration required for luciferase to produce light). In our hands, the available luciferase and HIF1α antibodies are not sensitive enough to detect the induction of these proteins in immuohistochemical and immunoblot assays of different tissues obtained from mice breathing 8% oxygen, and luciferase assays of crude tissue extracts might be confounded because of changes occurring ex vivo (such as due to exposure to oxygen). For these reasons, additional studies will be needed to more formally characterize the ability to image hypoxia in different tissues by using ROSA26 ODD-Luc/+ mice.
Fig. 3.
Imaging hypoxia by using ODD-Luc mice. (A) Bioluminescent images (anterior view) of ROSA26 Luc/+, ODD-Luc/+, and +/+ FVB mice. (B) Bioluminescent images (posterior view) of the same two ROSA26 ODD-Luc/+ mice breathing 21% or 8% oxygen. For the latter, the ambient oxygen was reduced from 21% to 8% over 1 h and maintained at 8% for an additional 4 h. Color bar indicates photons/(cm2·sec·steradian) with minimum and maximum threshold values.
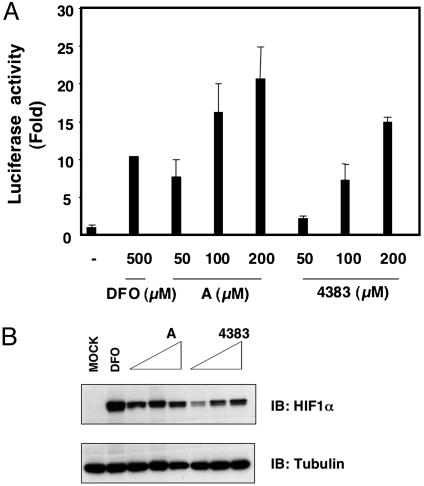
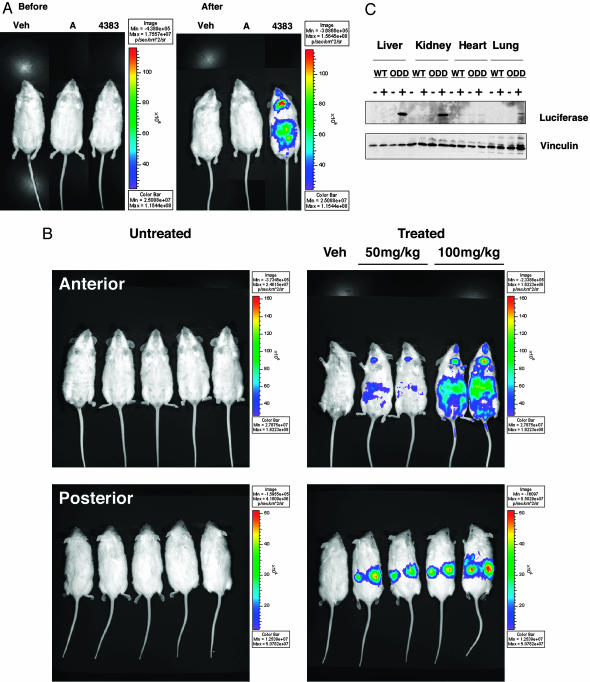
Preclinical data suggest that HIF agonists might be useful adjuncts for the treatment of ischemic diseases, and HIF prolyl hydroxylation can be inhibited with small organic molecules with drug-like properties, leading to increased HIF activity (7). Two such molecules, Compound A and FG-4383, were further characterized by using biochemical and cell-based screens similar to those depicted in Fig. 2. The EGLN1 IC50 values of these two compounds were 3.8 μM and 2.6 μM, respectively, in biochemical assays, although Compound A was clearly superior to FG-4383 in cell-based assays (Fig. 4). In the ROSA26 ODD-Luc/+ mouse assay, however, FG-4383 was far more active than Compound A (Fig. 5A and data not shown) and, therefore, was chosen for further study. Administration of FG-4383 to ROSA26 ODD-Luc/+ mice by oral gavage caused a dose-dependent increase in light emission, especially in the regions of the kidneys and liver (Fig. 5B). Induction of ODD-Luc in these two organs, and to a much lesser extent in the lungs, was confirmed by immunoblot analysis (Fig. 5C) and was associated with activation of a subset of HIF target genes as determined by real-time PCR (data not shown). To date, we have not observed ODD-Luc stabilization in mice treated with DFO or the hydroxylase inhibitor dimethyloxalylglycine, probably because these agents are far less potent than FG-4383 (data not shown).
Fig. 4.
Inhibition of HIF prolyl hydroxylase by small molecules in cell-based assays. (A) Fold increase in normalized luciferase values of HeLa cells transfected to produce ODD-Luc and exposed to compound A, FG-4383, or deferoxamine (DFO) at the indicated concentrations relative to untreated cells. Error bars indicate one standard error. (B) Immunoblot analysis of HeLa cells treated as in A.
Fig. 5.
Pharmacodynamic monitoring of HIF prolyl hydroxylase inhibitor. (A) Bioluminescent images of ROSA26 ODD-Luc/+ mice 6 h after administration of vehicle, Compound A (100 mg/kg), or FG-4383 (100 mg/kg) by oral gavage. (B) Bioluminescent images of ROSA26 ODD-Luc/+ mice before and 6 h after administration of FG-4383 by oral gavage at indicated doses. Color bar indicates photons/(cm2·sec·steradian) with minimum and maximum threshold values. (C) Immunoblot analysis of tissue extracts prepared from wild-type (WT) and ROSA26 ODD-Luc/+ (ODD) mice 6 h after treatment with 100 mg/kg FG-4383.
The kidney is the major source of EPO, which is HIF-regulated and stimulates red blood cell production in response to renal hypoxia. EPO mRNA levels were also increased in the kidney (data not shown) in response to FG-4383, leading to increased circulating EPO levels (Table 1). The kidneys were primarily responsible for this increase because EPO production was significantly diminished in anephric mice treated with this compound. Nonetheless, FG-4383 did increase EPO levels in anephric animals, presumably due to hepatic production of this hormone. Therefore, ROSA26 ODD-Luc/+ mice can be used to rapidly determine whether a potential HIF prolyl hydroxylase inhibitor is biologically active in animals and can be used to gather spatial and temporal information about drug action in vivo.
Table 1. EPO induction by FG-4383.
| Treatment group | No. of animals | Individual serum EPO, pg/ml |
|---|---|---|
| Sham/vehicle | 3 | 169; 317; 290 |
| Sham/FG-4383 | 3 | 54,275; 41,273; 48,434 |
| Nephrectomy/vehicle | 3 | 117; 110; 84 |
| Nephrectomy/FG-4383 | 4 | 1,344; 151; 515; 459 |
Discussion
We fused a fragment of HIF that is subject to oxygen-dependent degradation to firefly luciferase and showed that the resulting fusion protein (ODD-Luc) is responsive to hypoxia and hypoxia mimetics in live cells grown in culture. Moreover, we engineered a mouse that ubiquitously expresses the ODD-Luc reporter and showed that this mouse strain, ROSA26 ODD-Luc/+, can be used to image the development of tissue hypoxia and the action of small molecule inhibitors of HIF prolyl hydroxylase activity.
Firefly luciferase requires oxygen, ATP, and luciferin to emit light. Therefore, changes in these three factors could potentially complicate the interpretation of bioluminescent images obtained with the ROSA26 ODD-Luc/+ mice. Fortunately, the firefly luciferase Km for oxygen and ATP are ≈1–10 μM and ≈50 μM respectively (22, 23, and A. Campbell, personal communication). In contrast, the oxygen Km for the HIF prolyl hydroxylases are ≈200 μM, and cellular ATP levels are estimated to be ≈1 mM (24). Although firefly luciferase in mammalian cells may behave differently than the purified enzyme used to determine the Km values listed above (25), these considerations suggest that even relatively severe degrees of hypoxia should induce the accumulation of ODD-Luc without compromising light production. Consistent with this hypothesis, we did not observe a decrease in light produced by animals producing wild-type luciferase when breathing 8% oxygen. Luciferin is widely distributed after i.p. or i.v. injection with estimated tissue levels of 60–180 μM, with somewhat lower levels in the brain, at the doses used here (26, 27). These levels exceed the firefly luciferase luciferin Km (22), and luciferase activity has been successfully imaged in a wide variety of tissues (including brain) by using protocols similar to the one used by us (10, 28, 29). Nonetheless, the intracellular concentrations of luciferin achieved in different tissues after systemic administration are not known, and differences in luciferin availability might confound luciferase-based imaging in certain settings.
HIF transcriptionally activates a battery of genes that play roles in acute and chronic adaptation to hypoxia, including EPO. Preclinical studies suggest that HIF agonists might be beneficial in settings characterized by tissue hypoxia, such as myocardial infarction and stroke (7). The mouse strain described here should facilitate the preclinical development of HIF agonists that inhibit HIF prolyl hydroxylase activity or otherwise prevent the polyubiquitination of HIF by pVHL. As shown here, this strain can be used to rapidly compare the pharmacodynamics of compounds that otherwise behave similarly in biochemical and cell-based assays. A particularly desirable feature of our model is the ability to do repeated, noninvasive, measurements over time in the same animal. Such assays, in conjunction with classical pharmacokinetic studies, should provide useful information with respect to the choice and optimization of lead compounds.
Bioluminescent imaging of ROSA26 ODD-Luc/+ mice breathing in a low-oxygen environment supports the idea that the kidneys are important oxygen sensors in mammals. Earlier work indicated that kidneys are borderline hypoxic at baseline and, therefore, are likely poised to respond to further decrements in oxygen availability (30–33). Consistent with this view, we detect renal signals above the whole body background in ROSA26 ODD-Luc/+ mice breathing room air, although it remains possible that these signals are due to hypoventilation caused by the anesthetics needed for imaging. It is possible that other factors contribute to renal sensitivity to hypoxia, including changes in cellular pH and levels of the different EGLN isoforms. In most cells examined, EGLN1 appears to be the primary HIF prolyl hydroxylase, with recruitment of EGLN2 and EGLN3 after prolonged hypoxia (3, 4). Some of these same considerations, in conjunction with differences in tissue distribution (bioavailability), are likely to influence the sensitivity of various tissues to small-molecule HIF agonists.
Most solid tumors are thought to contain regions that are hypoxic. It will be of interest to see whether tumors arising in different cancer-prone mouse strains can be imaged based on accumulation of the ODD-Luc reporter. Similarly, it will be important to determine whether the signals observed in such tumors are altered after treatment with agents that affect tumor angiogenesis. The ROSA26 ODD-Luc/+ strain should be particularly useful for imaging tumors that arise as a consequence of pVHL inactivation, such as those that occur stochastically in VHL +/– mice (34).
The interaction of pVHL and HIF illustrates the principle that many ubiquitin ligases recognize modular degrons and that signals impinging on the ligase, the degron, or both influence whether successful engagement of the substrate by the ligase will take place. Hundreds of potential ubiquitin ligases have been identified and linked to many aspects of biology. For example, SCF (SKP1/Cullin/F-Box) ubiquitin ligases and A PC (Anaphase Promoting Complex) ubiquitin ligases have been exploited to make cell-cycle reporters suitable for imaging (for review, see ref. 2). Fusion of degrons to bioluminescent or fluorescent proteins should be a generally useful method for making reporters for different biological processes. Mice engineered to express such reporters should be valuable tools for physiological and pharmacological studies.
Supplementary Material
Acknowledgments
We thank Andrew Kung for help with bioluminescent imaging; Massimo Loda for help with immunohistochemical analysis; Shyam Sunder Sirasanagandla for technical help; members of the Kaelin Laboratory and Chris Contag for useful discussions; Guangjie Guo and QingJian Wang from FibroGen, Inc., for the animal EPO studies; and Mitch Brenner, Todd Seeley, Robert Stevenson, and Jenny Tsao from Fibrogen, Inc., for the enzyme assays and gene expression studies. This work was supported by grants from the National Institutes of Health (M.S. and W.G.K.) and the Murray Foundation. W.G.K. is a Howard Hughes Medical Institute Investigator.
Author contributions: M.S., V.G., R.A.D., and W.G.K. designed research; M.S., W.Y.K., F.O., L.F., and J.W.H. performed research; M.S., L.F., and V.G. contributed new reagents/analytic tools; M.S., V.G., and W.G.K. analyzed data; M.S. and W.G.K. wrote the paper; J.W.H. assisted in the construction of knock-in mice; and R.A.D. helped to design knock-in mice strategy.
Conflict of interest statement: W.G.K. is a founder of Imigen Systems, which owns patents surrounding the development of HIF prolyl hydroxylase inhibitors and the use of luciferase fusion proteins, including the HIF-luciferase fusion protein described here, for imaging. L.F. and V.G. are Fibrogen employees. Fibrogen owns the two compounds used in this work and is developing prolyl hydroxylase inhibitors as potential therapeutics.
Abbreviations: DFO, deferoxamine; EPO, erythropoietin; HIF, hypoxia-inducible factor.
References
- 1.Weissleder, R. & Ntziachristos, V. (2003) Nat. Med. 9, 123–128. [DOI] [PubMed] [Google Scholar]
- 2.Gross, S. & Piwnica-Worms, D. (2005) Cancer Cell 7, 5–15. [DOI] [PubMed] [Google Scholar]
- 3.Schofield, C. J. & Ratcliffe, P. J. (2004) Nat. Rev. Mol. Cell Biol. 5, 343–354. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin Jr., W. G. (2005) Annu. Rev. Biochem. 74, 115–128. [DOI] [PubMed] [Google Scholar]
- 5.Taylor, M. S. (2001) Gene 275, 125–132. [DOI] [PubMed] [Google Scholar]
- 6.Aravind, L. & Koonin, E. V. (2001) Genome Biol. 2, research0007.1–0007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivan, M., Haberberger, T., Gervasi, D. C., Michelson, K. S., Gunzler, V., Kondo, K., Yang, H., Sorokina, I., Conaway, R. C., Conaway, J. W. & Kaelin, W. G., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivas, S., Watanabe, T., Lin, C. S., William, C. M., Tanabe, Y., Jessell, T. M. & Costantini, F. (2001) BMC Dev. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, H., Ivan, M., Min, J., Kim, W. & Kaelin, W. (2004) Methods Enzymol. 381, 320–335. [DOI] [PubMed] [Google Scholar]
- 10.Safran, M., Kim, W. Y., Kung, A. L., Horner, J. W., DePinho, R. A. & Kaelin, W. G., Jr. (2003) Mol. Imaging 2, 297–302. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, L. O., Goldwasser, E., Fried, W. & Plzak, L. (1957) Nature 179, 633–634. [DOI] [PubMed] [Google Scholar]
- 12.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J., Lane, W. & Kaelin, W. J. (2001) Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola, P., Mole, D., Tian, Y., Wilson, M., Gielbert, J., Gaskell, S., Kriegsheim, A., Hebestreit, H., Mukherji, M., Schofield, C., et al. (2001) Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- 14.Pugh, C., O'Rourke, J., Nagao, M., Gleadle, J. & Ratcliffe, P. (1997) J. Biol. Chem. 272, 11205–11214. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L. E., Gu, J., Schau, M. & Bunn, H. F. (1998) Proc. Natl. Acad. Sci. USA 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallio, P., Wilson, W., O'Brien, S., Makino, Y. & Poellinger, L. (1999) J. Biol. Chem. 274, 6519–6525. [DOI] [PubMed] [Google Scholar]
- 17.Stebbins, C. E., Kaelin, W. G. & Pavletich, N. P. (1999) Science 284, 455–461. [DOI] [PubMed] [Google Scholar]
- 18.Schoenfeld, A. R., Davidowitz, E. J. & Burk, R. D. (2000) Proc. Natl. Acad. Sci. USA 97, 8507–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, K., Klco, J., Nakamura, E., Lechpammer, M. & Kaelin, W. G. (2002) Cancer Cell 1, 237–246. [DOI] [PubMed] [Google Scholar]
- 20.Zambrowicz, B. P., Imamoto, A., Fiering, S., Herzenberg, L. A., Kerr, W. G. & Soriano, P. (1997) Proc. Natl. Acad. Sci. USA 94, 3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soriano, P. (1999) Nat. Genet. 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 22.Lembert, N. & Idahl, L. A. (1995) Biochem. J. 305, 929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell, A. K., Hallett, M. B. & Weeks, I. (1985) Methods Biochem. Anal. 31, 317–416. [DOI] [PubMed] [Google Scholar]
- 24.Hirsila, M., Koivunen, P., Gunzler, V., Kivirikko, K. I. & Myllyharju, J. (2003) J. Biol. Chem. 278, 30772–30780. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, H. J., Pouli, A. E., Ainscow, E. K., Jouaville, L. S., Rizzuto, R. & Rutter, G. A. (1999) J. Biol. Chem. 274, 13281–13291. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. H., Byun, S. S., Paik, J. Y., Lee, S. Y., Song, S. H., Choe, Y. S. & Kim, B. T. (2003) Nucl. Med. Commun. 24, 1003–1009. [DOI] [PubMed] [Google Scholar]
- 27.Contag, C. H., Spilman, S. D., Contag, P. R., Oshiro, M., Eames, B., Dennery, P., Stevenson, D. K. & Benaron, D. A. (1997) Photochem. Photobiol. 66, 523–531. [DOI] [PubMed] [Google Scholar]
- 28.Rehemtulla, A., Stegman, L., Cardozo, S., Gupta, S., Hall, D., Contag, C. & Ross, B. (2000) Neoplasia 2, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook, S. H. & Griffin, D. E. (2003) J. Virol. 77, 5333–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donnelly, S. (2003) Adv. Exp. Med. Biol. 543, 73–87. [DOI] [PubMed] [Google Scholar]
- 31.Zhong, Z., Arteel, G. E., Connor, H. D., Yin, M., Frankenberg, M. V., Stachlewitz, R. F., Raleigh, J. A., Mason, R. P. & Thurman, R. G. (1998) Am. J. Physiol. 275, F595–F604. [DOI] [PubMed] [Google Scholar]
- 32.Heyman, S. N., Rosen, S. & Brezis, M. (1997) Blood Purif. 15, 232–242. [DOI] [PubMed] [Google Scholar]
- 33.Brezis, M. & Rosen, S. (1995) N. Engl. J. Med. 332, 647–655. [DOI] [PubMed] [Google Scholar]
- 34.Haase, V. H., Glickman, J. N., Socolovsky, M. & Jaenisch, R. (2001) Proc. Natl. Acad. Sci. USA 98, 1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.