Abstract
Atherosclerosis is a complex disease that is affected by environmental as well as genetic factors. The aim of the present study was to identify loci of atherosclerosis susceptibility in a cross of atherosclerosis-susceptible C57BL/6 and atherosclerosis-resistant FVB/N mice on the low-density lipoprotein (LDL) receptor (LDLR)-deficient background (LDLR–/–) and to test whether these loci are affected by lineage. A total of 459 F2s were generated in two ways: In cross “mB6xfFVB,” male B6.LDLR–/– mice were crossed to female FVB.LDLR–/– mice to generate 107 female and 112 male F2s. In cross “mFVBxfB6,” male FVB.LDLR–/– mice were crossed to female B6.LDLR–/– mice to generate 120 female and 120 male F2s. Animals were phenotyped for cross-sectional atherosclerotic lesion area at the aortic root, and a genome scan was carried out with 192 microsatellite markers. Quantitative trait locus mapping revealed significant loci of atherosclerosis susceptibility on chromosomes 3, 10, and 12. On chromosome 10 maximal logarithm of odds (LOD) scores of 13.1 (D10Mit16, 16 cM) and 5.7 (D10Mit168, 9 cM) were found in female and male mice, respectively. On chromosome 3, a maximal LOD score of 5.1 (D3Mit45, 79 cM) was detected only in females. On proximal chromosome 12 significant LOD scores were lineage-dependent, with maximal LOD scores of 3.9 (D12Mit82, 3 cM) and 4.8 (D12Mit189, 24 cM) present only in female mice of cross mB6xfFVB and male mice of cross mFVBxfB6, respectively. We conclude that, in this intercross, loci of atherosclerosis susceptibility are in part sex- and lineage-dependent. Awareness of these complexities may have major consequences for the identification of atherosclerosis susceptibility genes by quantitative trait locus mapping.
Keywords: genetics, quantitative trait locus mapping, mouse
Atherosclerosis is a complex disease that is affected by environmental as well as genetic factors (1–3). Heritability estimates in humans demonstrate that genetic factors explain ≈50% of the variability to develop atherosclerosis (4, 5). Genetic differences of atherosclerosis susceptibility have also been noted in animal models of pigeons (6), rabbits (7), and inbred mice (8). Because of their genetic homogeneity, inbred mice are particularly useful for the identification of genetic factors of disease. In addition, mice offer a viable alternative to human studies in the dissection of the complex genetic etiology of atherosclerosis, because experimental parameters such as environment, breeding scheme, and detailed phenotyping can be controlled (9). However, attempts to identify genetic factors of atherosclerosis susceptibility in the mouse have been hampered by its relative resistance to atherosclerosis as a species, which required feeding high-fat, high-cholesterol, cholate-containing diets to induce relatively small lesions (8, 10). Despite these difficulties, a number of loci of atherosclerosis susceptibility have been identified in the diet-induced mouse model using recombinant inbred strains and quantitative trait locus (QTL) analysis (11–17).
Since 1992, induced mutant mouse models of atherosclerosis have been created and used to study the mechanisms of lesion formation, diet and drug effects on the extent of lesions, and the role of candidate genes in atherosclerosis susceptibility. These studies have generally used apolipoprotein E-deficient (ApoE–/–) (18, 19) and low-density lipoprotein (LDL) receptor-deficient (LDLR–/–) mice (20). ApoE–/– and LDLR–/– mice have significantly greater lesion sizes than the diet-induced model, and lesions have been shown to represent most characteristics of human atherosclerosis (21, 22). In addition, the extent of atherosclerosis in ApoE–/– and LDLR–/– mice depends largely on the genetic background: Animals were atherosclerosis-susceptible when crossed to the C57BL/6 (B6) background and atherosclerosis-resistant when crossed to the FVB/N (FVB) background (23, 24).
Few studies have been done to identify loci of atherosclerosis susceptibility on the ApoE–/– and LDLR–/– backgrounds. In an F2 intercross of B6 and FVB mice on the ApoE–/– background, we identified one significant locus on proximal chromosome 10 and two additional loci on chromosomes 14 and 19 (25). Welch et al. (26) and Bretschger-Seidelmann et al. (27) used a backcross strategy of MOLF/Ei to B6.LDLR–/– and PERA to B6.LDLR–/–, respectively, and identified loci of atherosclerosis susceptibility on chromosomes 2, 4, and 6.
Our current study had two aims: The first was to test whether loci of atherosclerosis susceptibility would differ depending on the mode of lesion induction by the ApoE–/– and LDLR–/– backgrounds. To this end, we set up an F2 intercross of atherosclerosis-susceptible B6 and atherosclerosis-resistant FVB mice on the LDLR–/– background, the same strains as were previously used in an F2 intercross on the ApoE–/– background (25). The second aim was to test the potential effects of lineage and gender on loci of atherosclerosis susceptibility by using reciprocal crosses to generate male and female F2 animals.
Methods
Mice. LDLR–/– mice on the C57BL/6 background were obtained from The Jackson Laboratory (B6.129S7-Ldlrtm1Her/J, stock no. 002207, henceforth called B6.LDLR–/–). LDLR–/– mice on the FVB background (henceforth called FVB.LDLR–/–) were obtained from our own colony; the generation of these animals was described in ref. 24. F1 and F2 animals were generated in two ways: In cross “mB6xfFVB,” male B6.LDLR–/– mice were crossed to female FVB.LDLR–/– mice and the resulting F1s were intercrossed to generate 107 female and 112 male F2s. In cross “mFVBxfB6,” male FVB.LDLR–/– mice were crossed to female B6.LDLR–/– mice and the F1s were intercrossed to generate 120 female and 120 male F2s. Mice used for phenotyping were weaned at 28 days of age and fed a semisynthetic modified AIN76 diet containing 0.02% cholesterol until they were killed at 20 weeks of age (24). On the day of euthanasia, food was removed from the cage at 9 a.m. and mice were allowed access to water. Approximately 8 h later, glucose was measured from tail blood by using a glucometer (Bayer), and the mice were killed as described in ref. 28. Animals were housed in The Rockefeller University's Laboratory Animal Research Center in a specific pathogen-free environment in rooms with a 7 a.m. to 7 p.m. light/dark cycle. The Rockefeller University's Institutional Animal Care and Use Committee approved all procedures involving mice.
Blood Analyses. Lipoproteins were isolated by sequential ultracentrifugation from 60 μl of plasma at d <1.006 g/ml [very low-density lipoprotein (VLDL)], 1.006 ≤ d ≤ 1.063 g/ml (intermediate-density lipoprotein and LDL), and d >1.063 g/ml (high-density lipoprotein) in a TL-100 ultracentrifuge (Beckman). Cholesterol was determined enzymatically by using a colorimetric method (Roche) (28).
Quantification of Atherosclerosis. To quantify atherosclerosis at the aortic root, formalin-fixed hearts were processed as described in refs. 24 and 28.
Genome Scan. Genomic DNA was isolated from tail tip or liver tissue by digestion with proteinase K and precipitation with ethanol. Genotyping was performed using a set of microsatellite markers with known polymorphisms between B6 and FVB (25), supplemented with additional markers tested for polymorphisms on polyacrylamide gels. Forward primers were fluorescently labeled with the dyes 6-carboxyfluorescein (FAM), 4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein (HEX), and 2,7′,8′-benzo-5′-fluoro-2′,4,7-trichloro-5-carboxyfluorescein (NED). The reverse primers were unlabeled, but a GCTTCT sequence was attached to the 5′ end to reduce stutter bands (29). The markers were grouped into 14 panels of 8–12 markers each. The initial genome scan was done with 154 markers spaced at ≈10-cM intervals. A second set of 46 markers grouped into five panels was used for fine mapping and to repeat markers that failed in the first genome scan. All marker names, positions, fluorescent labels, and paneling information can be found in Tables 4 and 5, which are published as supporting information on the PNAS web site. Genotyping was performed at the Genomics Resource Center of The Rockefeller University. The PCRs were done in 384-well plates with 10–50 ng of dried-down genomic DNA. The PCR mastermix contained 200 μM each dNTP, 0.02 units/μl Taq polymerase (Fisher), and 0.3 μl of an 8 μM primer mix of each forward and reverse primers in a final volume of 5 μl. PCR products were pooled according to the panels, resolved by capillary electrophoresis on a 3700 DNA sequencer, and analyzed with genotyper software (Applied Biosystems).
Statistical Analysis. All data were expressed as mean ± SD unless otherwise indicated. Normality of distribution was assessed by using the Kolmogorov–Smirnov test implemented in prism statistical software (GraphPad, San Diego). Comparison of multiple groups was done using the Kruskal–Wallis test and Dunn's multiple-comparison test as a posttest, and comparison of two groups was done by using the Mann-Whitney test. For linkage analysis, atherosclerotic lesion measurements were log-transformed to achieve normality. Linkage analysis for single QTLs and interacting QTLs were done using mapmanager qtx b20, freely available at www.mapmanager.org (30). Levels of significance were determined empirically by permutation testing in 1-cM steps.
Results
To identify atherosclerosis susceptibility loci 459 F2 mice were generated in a reciprocal intercross of atherosclerosis-susceptible B6.LDLR–/– and atherosclerosis-resistant FVB.LDLR–/– mice. Atherosclerosis was determined at 20 weeks of age after mice had been fed a semisynthetic diet for 16 weeks. Mean lesion areas at the aortic root are shown in Table 1, and individual data points are shown in Fig. 5, which is published as supporting information on the PNAS web site. Because lesion areas were significantly greater in female F2s than in male F2s (89,137 ± 74,674 μm2 vs. 59,739 ± 46,141 μm2; P < 0.0001), genders were analyzed separately. To account for the inheritance of the sex chromosomes, 219 F2s were generated by crossing male B6.LDLR–/– mice to female FVB.LDLR–/– mice and crossing the resulting F1s (cross mB6xfFVB). The remaining 240 F2s were generated by crossing male FVB.LDLR–/– mice to female B6.LDLR–/– mice and crossing the resulting F1s (cross mFVBxfB6). In females, no significant differences were observed in the mean atherosclerotic lesion area between F1 and F2 mice from cross mB6xfFVB or cross mFVBxfB6 (P = not significant). In males, lesion areas in F1 animals from cross mB6xfFVB and cross mFVBxfB6 were comparable (P = not significant); however, F2s from cross mB6xfFVB had 43% larger lesions than did F2s from cross mFVBxfB6 (70,663 ± 47,958 μm2 vs. 49,555 ± 42,084 μm2; P < 0.0001).
Table 1. Atherosclerotic lesion size at the aortic root of parental B6.LDLR–/– and FVB.LDLR–/– mice and F1 and F2 offspring.
| Genetic background
|
Lesion size, μm2
|
|
|---|---|---|
| Females | Males | |
| B6.LDLR-/- | 289,708 ± 49,942 (9) | 122,457 ± 36,137 (10) |
| FVB.LDLR-/- | 10,344 ± 2,807 (9) | 18,268 ± 5,782 (12) |
| F1 (mB6xfFVB) | 75,086 ± 24,173 (12) | 49,704 ± 27,765 (13) |
| F1 (mFVBxfB6) | 80,412 ± 19,460 (20) | 50,463 ± 22,682 (16) |
| F2 (mB6xfFVB) | 91,698 ± 78,986 (106) | 70,663 ± 47,958* (110) |
| F2 (mFVBxfB6) | 86,668 ± 70,544 (110) | 49,555 ± 42,084* (118) |
F1 and F2 animals were generated by crossing male B6.LDLR-/- to female FVB.LDLR-/- mice (mB6xfFVB) and by intercrossing the resulting F1 or by crossing male FVB.LDLR-/- to female B6.LDLR-/- mice (mFVBxfB6) and intercrossing the resulting F1. Values in parentheses indicate the number of mice with measurements of atherosclerosis per group. *, P < 0.001 between groups.
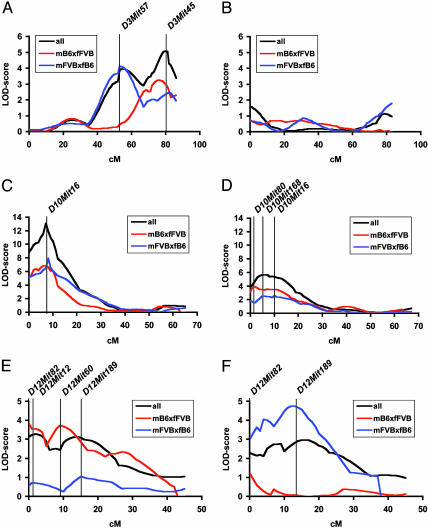
QTL mapping based on the initial ≈10-cM genome scan using mapmanager qtx b20 revealed significant loci of atherosclerosis susceptibility on chromosomes 3, 10, and 12. These loci were subsequently fine-mapped with additional markers. The interval maps of these chromosomes are shown in Fig. 1. The highest maximal logarithm of odds (LOD) scores were observed on proximal chromosome 10 with maximal LODs of 13.1 at D10Mit16 (16 cM) and 5.7 at D10Mit168 (9 cM) in female and male mice, respectively (cM positions correspond to the Mouse Genome Informatics database) (Fig. 1 C and D and Tables 2 and 3). On chromosome 3, significant linkage was detected in females but not in males (Fig. 1 A and B and Tables 2 and 3). When all female mice were analyzed as one group, maximal linkage (LOD 5.1) was observed at D3Mit45 (79 cM) with an additional linkage peak at D3Mit57 (55 cM) (LOD 3.9). Analyzing females from crosses mB6xfFVB and mFVBxfB6 separately revealed that the locus at D3Mit45 originated from both crosses, whereas the locus at D3Mit57 originated predominantly from cross mFVBxfB6. These data suggest that the locus at chromosome 3 results from at least two genes with different lineage dependence in female mice. A strong lineage effect was also seen for the locus on proximal chromosome 12. This locus was present in male and female mice. However, a significant LOD score was observed only in female mice of cross mB6xfFVB (LOD 3.9; D12Mit82, 3 cM) and in male mice of cross mFVBxfB6 (LOD 4.8; D12Mit189, 24 cM) (Fig. 1 E and F and Tables 2 and 3). A summary of all significant and suggestive QTLs for atherosclerosis revealed in this cross is given in Tables 5 and 6, which are published as supporting information on the PNAS web site.
Fig. 1.
Chromosome interval maps for atherosclerotic lesion area at the aortic root on chromosome 3 (A and B, females and males, respectively), chromosome 10 (C and D, females and males, respectively), and chromosome 12 (E and F, females and males, respectively). Data are shown for the F2 cohorts of all mice, cross mB6xfFVB and cross mFVBxfB6.
Table 2. Significant single QTL identified in female F2s.
| All female F2s (n = 216)
|
Cross mB6xfFVB, female F2s (n = 106)
|
Cross mFVBxfB6, female F2s (n = 110)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Peak | cM | LOD | Variance, % | Peak | cM | LOD | Variance, % | Peak | cM | LOD | Variance, % |
| Chr. 3 | D3Mit45 | 78.5 | 5.1* | 10 | D3Mit45 | 78.5 | 3.3† | 13 | D3Mit57 | 55.0 | 4.1* | 16 |
| Chr. 10 | D10Mit16 | 16.0 | 13.1* | 24 | D10Mit16 | 16.0 | 6.8* | 26 | D10Mit16 | 16.0 | 7.9* | 28 |
| Chr. 12 | D12Mit12 | 6.0 | 3.3† | 7 | D12Mit82 | 3.0 | 3.9* | 15 | D12Mit189 | 24.0 | 1.0 | 4 |
Peak, marker nearest to peak LOD score; cM, location of marker from Mouse Genome Informatics database (www.informatics.jax.org); LOD, peak LOD score; variance, percentage of the total phenotypic variance detected in the F2s and associated with the peak LOD; Chr., chromosome.
Significant LOD scores determined by permutation testing (2,000 permutations).
Suggestive LOD scores determined by permutation testing (2,000 permutations).
Table 3. Significant single QTL identified in male F2s.
| All male F2s (n = 228)
|
Cross mB6xfFVB, male F2s (n = 110)
|
Cross mFVBxfB6, male F2s (n = 118)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Peak | cM | LOD | Variance, % | Peak | cM | LOD | Variance, % | Peak | cM | LOD | Variance, % |
| Chr. 10 | D10Mit168 | 9.0 | 5.7* | 11 | D10Mit80 | 4.0 | 3.9* | 15 | D10Mit16 | 16.0 | 2.6† | 10 |
| Chr. 12 | D12Mit189 | 24.0 | 3.0† | 6 | D12Mit82 | 3.0 | 1.2 | 5 | D12Mit189 | 24.0 | 4.8* | 17 |
Peak, marker nearest to peak LOD score; cM, location of marker from Mouse Genome Informatics database (www.informatics.jax.org); LOD, peak LOD score; variance, percentage of the total phenotypic variance detected in the F2s and associated with the peak LOD; Chr., chromosome.
Significant LOD scores determined by permutation testing (2,000 permutations).
Suggestive LOD scores determined by permutation testing (2,000 permutations).
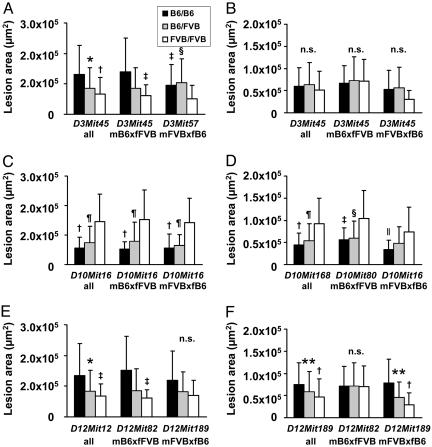
The allelic contributions of genotypes in the F2 mice at the chromosome 3, 10, and 12 loci to atherosclerotic lesion areas at the aortic root are shown in Fig. 2. On chromosomes 3 and 12, inheritance of the B6 allele conferred atherosclerosis susceptibility, whereas inheritance of the FVB allele conferred atherosclerosis resistance. The strongest phenotypic effect of the locus on chromosome 3 was seen at D3Mit45 in female mice of cross mB6xfFVB. At this locus, mean lesion size at the aortic root was 2.3-fold larger in homozygotes for the B6 allele than in homozygotes for the FVB allele, whereas lesion size in heterozygotes was intermediate (Fig. 2A). In contrast, the mode of inheritance at the second locus at D3Mit57 in cross mFVBxfB6 was strikingly different. Whereas homozygous and heterozygous carriers of the B6 allele had comparable atherosclerosis, lesion size was significantly higher than in FVB homozygotes. This finding suggested a dominant effect of the B6 allele at D3Mit57, whereas the effect was codominant at D3Mit45. In males, no significant allelic effects were observed on chromosome 3, as expected from the nonsignificant LOD scores in these animals (Fig. 2B). The strongest phenotypic effect of the locus on chromosome 12 in female mice was observed in cross mB6xfFVB at D12Mit82. Mean lesion size at the aortic root was 2.5-fold larger in homozygotes for the B6 allele than in homozygotes for the FVB allele, whereas lesion size in heterozygotes was intermediate (Fig. 2E). In male mice, a strong effect on atherosclerosis was seen in cross mFVBxfB6 at D12Mit189 with 2.7-fold larger lesion areas in homozygotes for the B6 allele than in homozygotes for the FVB allele, whereas no significant allelic effects were observed in males from cross mB6xfFVB (Fig. 2F). In contrast, allelic contributions of the genotypes of the locus on chromosome 10 showed that the FVB allele conferred atherosclerosis susceptibility and that the B6 allele conferred atherosclerosis resistance at the aortic root. This effect was seen in male and female mice of both crosses. In females, FVB homozygotes had 2.5-fold larger lesions than did B6 homozygotes; in males, FVB homozygotes had 2.1-fold larger lesions than did B6 homozygotes (Fig. 2 C and D).
Fig. 2.
Allelic contributions of genotypes to atherosclerosis at the aortic root. (A) Females at D3Mit45 (all and cross mB6xfFVB) and D3Mit57 (cross mFVBxfB6). (B) Males at D3Mit45. (C) Females at D10Mit16. (D) Males at D10Mit168 (all), D10mit80 (cross mB6xfFVB), and D10Mit16 (cross mFVBxfB6). (E) Females at D12mit12 (all), D12Mit82 (cross mB6xfFVB), and D12Mit189 (cross mFVBxfB6). (F) Males at D12Mit189 and D12Mit82. *, P < 0.01 (B6/B6 vs. B6/FVB); †, P < 0.001 (B6/B6 vs. FVB/FVB); ‡, P < 0.01 (B6/B6 vs. FVB/FVB); §, P < 0.01 (B6/FVB vs. FVB/FVB); ¶, P < 0.001 (B6/FVB vs. FVB/FVB); ∥, P < 0.05 (B6/B6 vs. FVB/FVB); **, P < 0.05 (B6/B6 vs. B6/FVB).
We next tested the allelic contributions of plasma total cholesterol, VLDL cholesterol, LDL cholesterol, high-density lipoprotein cholesterol, and glucose at the atherosclerosis loci. These data are summarized in Table 7, which is published as supporting information on the PNAS web site. No significant effects were seen at the loci on chromosomes 3 and 10. However, at the locus on chromosome 12, the FVB allele conferred significantly lower plasma cholesterol concentrations in male and female mice (Kruskal–Wallis test) because of an ≈2-fold reduction of VLDL cholesterol in these animals.
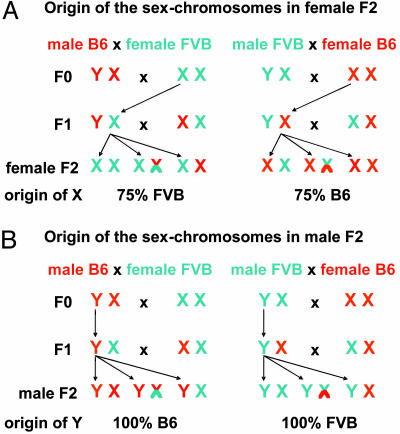
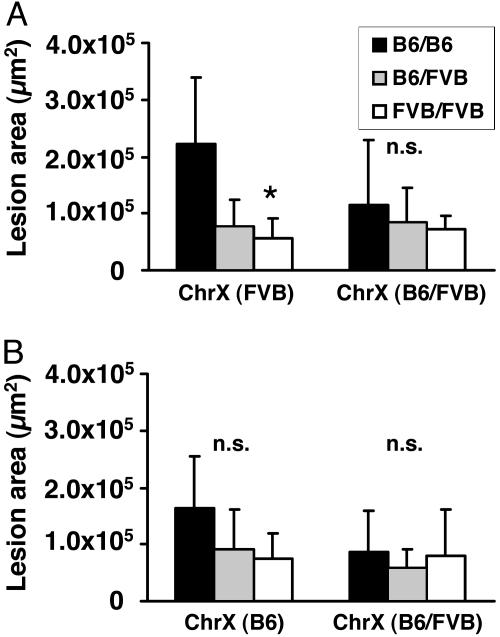
As shown in Fig. 3, the direction of the cross (lineage) has important effects on the distribution of the sex chromosomes. In male F2s, the Y chromosome is inherited exclusively from B6 in cross mB6xfFVB and exclusively from FVB in cross mFVBxfB6. The situation is more complex in female F2s, where the content of FVB on the X chromosome is 50–100% (average 75%) in cross mB6xfFVB and 0–50% (average 25%) in cross mFVBxfB6. In view of the strong lineage dependence of the chromosome 12 locus, a subgroup analysis was performed by comparing lesion area in F2 female mice in cross mB6xFVB with full FVB homozygosity or heterozygosity throughout the X chromosome (Fig. 4A) and in cross mFVBxB6 with full B6 homozygosity or heterozygosity throughout the X chromosome (Fig. 4B). This analysis revealed that homozygosity for FVB at the X chromosome potentiated the effect of D12Mit82 on atherosclerosis susceptibility, leading to 4-fold more atherosclerosis in B6/B6 vs. FVB/FVB homozygotes at D12Mit82 (P < 0.05).
Fig. 3.
Distribution of sex chromosomes in female (A) and male (B) F2s depending on male or female F0 lineage. F1 and F2 animals were generated in two ways: (i) by crossing male B6.LDLR–/– mice to female FVB.LDLR–/– mice and (ii) by crossing male FVB.LDLR–/– mice to female B6.LDLR–/– mice. Arrows indicate X and Y chromosomes that are passed through from the F0 via male F1s to the F2 generation without recombination, being of F0 lineage and thus 100% FVB or 100% B6. The X chromosome, inherited from the female F1, will carry on average 50% FVB and 50% B6 alleles. Thus, in females (A) 75% of the alleles on X will originate from the female F0, and in males (B) 100% of the alleles on Y will originate from the male F0.
Fig. 4.
Effect of X-chromosomal genotype on allelic contributions to atherosclerosis at the aortic root at D12mit82 in female mice. (A) Cross mB6xfFVB. (B) Cross mFVBxfB6. Animals were selected to be homozygous FVB, heterozygous (B6/FVB), or homozygous B6 throughout chromosome X. *, P < 0.05 (B6/B6 vs. FVB/FVB).
To evaluate the possibility of gene–gene interactions mapmanager qtx b20 was used to do a systematic pair wise genome scan (Table 8, which is published as supporting information on the PNAS web site). Most significant interactions involved loci at chromosomes 3, 10, and 12 because of the additive effects of these loci. The strongest LOD of interaction (5.4) (i.e., nonadditive) was observed between loci on chromosomes 8 and 18 (D8Mit166 and D18Mit121), resulting in a total LOD of 9.2. A summary of the allelic effects of these gene interactions on atherosclerosis at the aortic root is shown in Fig. 6, which is published as supporting information on the PNAS web site.
Discussion
In the present study we have identified significant atherosclerosis susceptibility loci on chromosomes 3, 10, and 12 in an intercross of atherosclerosis-susceptible B6 and atherosclerosis-resistant FVB mice on the LDLR–/– background. The loci on chromosomes 12 and 3 showed prominent sex- and/or lineage-dependent modes of inheritance. Lineage dependence was particularly strong at the chromosome 12 locus, which was found in only one of each of the reciprocal crosses in males and females (Figs. 1 E and F and 2 E and F and Table 1). The locus on chromosome 3 had not been mapped in other mouse crosses.
Lineage-dependent traits have been described for a number of phenotypes, including behavioral traits in the rat (31, 32). To the best of our knowledge, lineage dependence of atherosclerosis susceptibility has not yet been described. In the published literature, crosses have been set up only with male F0s from one strain and female F0s from another strain, or the gender of the F0s has not been specified. As shown in Fig. 3, the design of a cross has important effects on the distribution of the sex chromosomes in the F2s. In male F2s, the Y chromosome originates exclusively from B6 if the male F0 grandparent was B6 and exclusively from FVB if the male F0 grandparent was FVB. The genotypic distribution of the autosomes is not affected in male F2s. In female F2s, one X chromosome originating from the F0 female grandparent is transmitted unchanged through the F1 males onto all offspring. The other X chromosome originating from the F1 females is B6, FVB, or recombinant, with a 50% chance of inheriting B6 or FVB alleles at a given marker. Thus, in female F2s, the X chromosome is on average 75% B6 if the female F0 grandparent was B6 and on average 75% FVB if the female F0 grandparent was FVB. This prediction was confirmed experimentally in the F2 mice in the current study by genotyping markers on the X chromosome (data not shown). As in males, the genotypic distribution of the autosomes is unaffected in female F2s. At the chromosome 12 locus we observed that the Y chromosome originating from FVB was permissive to bring out differences in atherosclerosis susceptibility at the chromosome 12 locus in male mice, whereas in female mice homozygosity for FVB at the X chromosomes was particularly effective (Fig. 4). A lineage-dependent effect was also present at D3Mit57, which was the strongest locus in females from cross mFVBxfB6 on chromosome 3 but did not reach statistical significance in females from cross mB6xfFVB (LOD < 1). In addition, the mode of inheritance was quite different at D3Mit57 and neighboring D3Mit45. Whereas B6 was dominant at D3Mit57, it appeared to be recessive at D3Mit45. Some lineage dependence was also observed at the locus on chromosome 10. Even though this locus was most consistently seen in all crosses, there were marked differences in the height of the LOD score, which was significant in all female crosses and males from cross mB6xfFVB but only suggestive in males from cross mFVBxfB6.
In addition to the differences in the distribution of the sex chromosomes, a number of other factors may underlie the basis of these lineage effects. Mitochondria are maternally inherited, and potential differences between the B6 and FVB mitochondrial DNA may be relevant. In addition, epigenetic effects may play an important role (33). In any case, we believe that lineage effects have important consequences for the experimental design in gene discovery using congenic mice and induced mutant mouse models.
Another aim of this study was to test whether loci of atherosclerosis susceptibility would differ depending on the mode of lesion induction on the ApoE–/– and LDLR–/– backgrounds. Here, we confirmed the locus at chromosome 10, previously identified on the ApoE–/– background (25). The mapping position of the locus on chromosome 10 in the present cross was comparable with the position in our previous cross. Moreover, the mode of inheritance was comparable with a dominant effect of the B6 allele conferring atherosclerosis resistance at this locus. The identification of genes underlying this locus is presently underway, and congenic animals have been made. Two other loci, one suggestive locus at chromosome 14 (D14Mit60, LOD 3.2) and a significant locus at chromosome 19 (D19Mit120, LOD 3.8), were not present in the present cross. Instead, loci on chromosome 3 and chromosome 12 were identified that were not seen in our previous cross on the ApoE–/– background. One could speculate that these loci indeed depended on the induction of atherosclerosis by the ApoE–/– and LDLR–/– backgrounds and related differences in the lipoprotein profile (23, 24). Specifically, total cholesterol was lower, with a dominance of LDL cholesterol in the present cross, compared with higher total cholesterol levels and a dominance of VLDL in the previous cross (25). It is of interest that the loci on chromosomes 3, 10, and 12 were not specific for the LDLR–/– background, because, in studies using backcrosses of MOLF/Ei to B6.LDLR–/– and PERA to B6.LDLR–/–, Welch et al. (26) and Bretschger-Seidelmann et al. (27) identified loci of atherosclerosis susceptibility on chromosomes 2, 4, and 6.
Because the chromosome 3 locus has not been mapped in previous mouse crosses, we next determined the syntenic region in the human genome. The mouse chromosome 3 QTL interval maps to human chromosomes 1p and 4q (www.ensembl.org/Mus_musculus/syntenyview?otherspecies=Homo_sapiens;chr=3); however, no loci of atherosclerosis susceptibility have been mapped to this region in human studies. The Ensembl mouse genome database (version 33, September 2005) was searched for known genes between 100 Mb and 159 Mb on chromosome 3 (D3Mit57 and D3Mit45 correspond to 114 Mb and 147 Mb, respectively). Candidate genes of atherosclerosis in that region include vascular cell adhesion molecule 1 (115 Mb), NF-κB1 (135 Mb), and microsomal triglyceride transfer protein (137 Mb).
The QTL mapped to proximal chromosome 12 corresponded to the position of a QTL (Ath6) previously mapped by Paigen and colleagues (14) in a cross between B6-db/db and C57BLKS/J mice. In that cross, lesion size was affected by a locus at D12Mit49 with a LOD score of 2.5. Candidate genes in that region, syntenic to human chromosomes 2p, 7, and 14q, include apolipoprotein B (7 Mb), syndecan 1 (8 Mb), and a disintegrin and metalloproteinase domain 17 (ADAM17, 19 Mb). In subsequent work, Paigen and colleagues have narrowed the QTL interval to 1.07 ± 0.26 cM. Three ESTs were mapped to the currently defined locus, ruling out the aforementioned candidate genes (34). However, we believe that these results do not preclude the importance of those genes in our cross, which was done with different strains under different conditions. In particular, increased VLDL cholesterol in mice carrying the B6 allele at this locus (Table 7) may indicate that strain differences in apolipoprotein B may play a role in atherosclerosis susceptibility.
In summary, we have identified significant loci of atherosclerosis susceptibility on chromosomes 3, 10, and 12 in a reciprocal F2 intercross of FVB.LDLR–/– and B6.LDLR–/– mice. Loci on chromosomes 12, 3, and (to a lesser extent) 10 showed a sex- and lineage-dependent mode of inheritance. This finding may have major consequences for studies of gene identification at atherosclerosis QTLs.
Supplementary Material
Acknowledgments
We thank Helen Yu and Suey Lee for technical assistance. We thank Dr. Connie Zhao, Sigal Lifshitz-Arama, and Marcia Sullivan (The Rockefeller University Genomics Resource Center) for running the genotyping samples. This work was supported by National Institutes of Health Grants HL-54591-08 and HL70524-02. D.T. was a fellow of the Emmy Noether Program of the Deutsche Forschungsgemeinschaft (DFG Te 342/1-1).
Author contributions: D.T., M.T., and J.L.B. designed research; D.T., M.T., A.D.P., and J.L.B. performed research; D.T., M.T., and J.L.B. contributed new reagents/analytic tools; D.T., M.T., A.D.P., and J.L.B. analyzed data; and D.T., M.T., and J.L.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: LDL, low-density lipoprotein; LDLR–/–, LDL receptor-deficient; ApoE–/–, apolipoprotein E-deficient; QTL, quantitative trait locus; VLDL, very low-density lipoprotein; LOD, maximal logarithm of odds.
References
- 1.Breslow, J. L. (2001) Clin. Cardiol. 24, Suppl. II, 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lusis, A. J., Fogelman, A. M. & Fonarow, G. C. (2004) Circulation 110, 1868–1873. [DOI] [PubMed] [Google Scholar]
- 3.Lusis, A. J., Mar, R. & Pajukanta, P. (2004) Annu. Rev. Genomics Hum. Genet. 5, 189–218. [DOI] [PubMed] [Google Scholar]
- 4.Zdravkovic, S., Wienke, A., Pedersen, N. L., Marenberg, M. E., Yashin, A. I. & De Faire, U. (2002) J. Intern. Med. 252, 247–254. [DOI] [PubMed] [Google Scholar]
- 5.Fox, C. S., Polak, J. F., Chazaro, I., Cupples, A., Wolf, P. A., D'Agostino, R. A. & O'Donnell, C. J. (2003) Stroke 34, 397–401. [DOI] [PubMed] [Google Scholar]
- 6.Yancey, P. G. & St Clair, R. W. (1992) Arterioscler. Thromb. 12, 1291–1304. [DOI] [PubMed] [Google Scholar]
- 7.Thiery, J., Nebendahl, K., Rapp, K., Kluge, R., Teupser, D. & Seidel, D. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 1181–1188. [DOI] [PubMed] [Google Scholar]
- 8.Roberts, A. & Thompson, J. S. (1977) Prog. Biochem. Pharmacol. 13, 298–305. [PubMed] [Google Scholar]
- 9.Allayee, H., Ghazalpour, A. & Lusis, A. J. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 1501–1509. [DOI] [PubMed] [Google Scholar]
- 10.Paigen, B., Morrow, A., Brandon, C., Mitchell, D. & Holmes, P. (1985) Atherosclerosis 57, 65–73. [DOI] [PubMed] [Google Scholar]
- 11.Colinayo, V. V., Qiao, J. H., Wang, X., Krass, K. L., Schadt, E., Lusis, A. J. & Drake, T. A. (2003) Mamm. Genome 14, 464–471. [DOI] [PubMed] [Google Scholar]
- 12.Ishimori, N., Li, R., Kelmenson, P. M., Korstanje, R., Walsh, K. A., Churchill, G. A., Forsman-Semb, K. & Paigen, B. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 161–166. [DOI] [PubMed] [Google Scholar]
- 13.Mehrabian, M., Wong, J., Wang, X., Jiang, Z., Shi, W., Fogelman, A. M. & Lusis, A. J. (2001) Circ. Res. 89, 125–130. [DOI] [PubMed] [Google Scholar]
- 14.Mu, J. L., Naggert, J. K., Svenson, K. L., Collin, G. B., Kim, J. H., McFarland, C., Nishina, P. M., Levine, D. M., Williams, K. J. & Paigen, B. (1999) J. Lipid Res. 40, 1328–1335. [PubMed] [Google Scholar]
- 15.Paigen, B. (1995) Am. J. Clin. Nutr. 62, 458S–462S. [DOI] [PubMed] [Google Scholar]
- 16.Paigen, B., Mitchell, D., Reue, K., Morrow, A., Lusis, A. J. & LeBoeuf, R. C. (1987) Proc. Natl. Acad. Sci. USA 84, 3763–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart-Phillips, J. L., Lough, J. & Skamene, E. (1989) Clin. Invest. Med. 12, 121–126. [PubMed] [Google Scholar]
- 18.Zhang, S. H., Reddick, R. L., Piedrahita, J. A. & Maeda, N. (1992) Science 258, 468–471. [DOI] [PubMed] [Google Scholar]
- 19.Plump, A. S., Smith, J. D., Hayek, T., Aalto-Setala, K., Walsh, A., Verstuyft, J. G., Rubin, E. M. & Breslow, J. L. (1992) Cell 71, 343–353. [DOI] [PubMed] [Google Scholar]
- 20.Ishibashi, S., Brown, M. S., Goldstein, J. L., Gerard, R. D., Hammer, R. E. & Herz, J. (1993) J. Clin. Invest. 92, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, H., Johnson, J. L., Carson, K. G. & Jackson, C. L. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 788–792. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima, Y., Plump, A. S., Raines, E. W., Breslow, J. L. & Ross, R. (1994) Arterioscler. Thromb. 14, 133–140. [DOI] [PubMed] [Google Scholar]
- 23.Dansky, H. M., Charlton, S. A., Sikes, J. L., Heath, S. C., Simantov, R., Levin, L. F., Shu, P., Moore, K. J., Breslow, J. L. & Smith, J. D. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1960–1968. [DOI] [PubMed] [Google Scholar]
- 24.Teupser, D., Persky, A. D. & Breslow, J. L. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 1907–1913. [DOI] [PubMed] [Google Scholar]
- 25.Dansky, H. M., Shu, P., Donavan, M., Montagno, J., Nagle, D. L., Smutko, J. S., Roy, N., Whiteing, S., Barrios, J., McBride, T. J., et al. (2002) Genetics 160, 1599–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch, C. L., Bretschger, S., Latib, N., Bezouevski, M., Guo, Y., Pleskac, N., Liang, C. P., Barlow, C., Dansky, H., Breslow, J. L. & Tall, A. R. (2001) Proc. Natl. Acad. Sci. USA 98, 7946–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidelmann, S. B., De Luca, C., Leibel, R. L., Breslow, J. L., Tall, A. R. & Welch, C. L. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teupser, D., Pavlides, S., Tan, M., Gutierrez-Ramos, J. C., Kolbeck, R. & Breslow, J. L. (2004) Proc. Natl. Acad. Sci. USA 101, 17795–17800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brownstein, M. J., Carpten, J. D. & Smith, J. R. (1996) Biotechniques 20, 1004.-1006, 1008–1010. [DOI] [PubMed] [Google Scholar]
- 30.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930–932. [DOI] [PubMed] [Google Scholar]
- 31.Ahmadiyeh, N., Churchill, G. A., Solberg, L. C., Baum, A. E., Shimomura, K., Takahashi, J. S. & Redei, E. E. (2005) Behav. Genet. 35, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solberg, L. C., Baum, A. E., Ahmadiyeh, N., Shimomura, K., Li, R., Turek, F. W., Churchill, G. A., Takahashi, J. S. & Redei, E. E. (2004) Mamm. Genome 15, 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan, H. D., Santos, F., Green, K., Dean, W. & Reik, W. (2005) Hum. Mol. Genet. 14, R47–R58. [DOI] [PubMed] [Google Scholar]
- 34.Purcell, M. K., Mu, J. L., Higgins, D. C., Elango, R., Whitmore, H., Harris, S. & Paigen, B. (2001) Mamm. Genome 12, 495–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.