Abstract
Understanding the specificity of cell-surface carbohydrates interaction with antibodies and receptors is important for the development of new therapeutics and high-sensitivity diagnostics. This approach is, however, limited to the availability of natural and truncated sequences of the oligosaccharides and the sensitivity of the assay system. Reported here is the synthesis of the cancer antigen Globo H hexasaccharide, an epitope found on the cell surface of breast, prostate, and ovarian cancers, and its truncated sequences by using the programmable one-pot synthesis strategy. The saccharides were then arrayed covalently on glass slides with different density and used for the fluorencense-based binding analysis of two monoclonal antibodies against Globo H and the serum from breast cancer patients, to define the specificity of these antibodies. It was shown that the terminal tetrasaccharide binds the monoclonal antibodies equally well as does the hexasaccharide and the fucose residue is required for effective binding. The serum binds both the defucosylated pentasaccharide and the fucosylated hexasaccharide without a significant difference, perhaps because of the polyclonal nature of the serum or the presence of diverse immune responses to different sugar epitopes at various stages. This method requires very small amounts of materials and is more effective and sensitive than the traditional ELISA method, and thus provides another platform to monitor the immune response to carbohydrate epitopes at different stages during differentiation, metastasis, or treatment.
Keywords: programmable one-pot synthesis, glycoarray, glycan epitope, Globo H-truncated sequences
The cell-surface glycosphingolipid Globo H is a member of a family of antigenic carbohydrates that are highly expressed on a range of cancer cell lines, especially breast cancer cells (1–4). Furthermore, it has been established that the serum of breast cancer patients contains high levels of antibodies against the Globo H epitope, and this epitope is also targeted by the monoclonal antibodies MBr1 (5–7) and VK-9 (8). As a result, this hexasaccharide has been the focus of studies aimed at anticancer vaccine development (9–15). Many elegant syntheses of Globo H have been reported (11, 16–24), including one approach that uses the one-pot programmable oligosaccharide synthesis developed in our laboratory (25). Previously, it has been reported that certain truncated Globo H derivatives can still be effective in binding MBr1 and VK-9 antibodies, which could increase the efficiency of immunogen development for vaccine therapy (8, 26–30). We set out to further characterize the binding specificities of these and cancer patient antibodies by using carbohydrate microarray analysis.
Carbohydrate microarrays allow for the direct characterization of carbohydrate–protein interactions. In addition, the attachment of sugars to surfaces can effectively mimic the presentation of these compounds on the cell membrane. A large factor which is present in this system is the formation of multivalent interactions, which have been shown to yield high affinity and specificity interactions in natural systems (31). In addition, only a very small amount of material is required for arraying (≈0.1–0.5 fmol per spot). Thus, the microarray provides a more appropriate and more sensitive model system for studying cellular events than traditional solution-phase ELISA analysis. Successful glycoarray systems have followed advancements in carbohydrate synthesis and have implemented both covalent (32–41) and noncovalent (42–46) strategies for sugar immobilization.
In a previous study, we performed the microarray analysis of an undecasaccharide presented on the surface of the HIV-1 envelope glycoprotein gp120 (47) by using 2G12, a broadly neutralizing anti-HIV-1 antibody. The results indicated that truncated sugars competed more strongly against the natural ligand. This array method provides a different direction for the specificity of protein–carbohydrate interaction and is thus a useful tool for glycobiology research.
Inspired by these results, we looked to evaluate the recruitment of monoclonal antibodies MBr1 and VK-9 to the surface by Globo H analogs 1–6 within the microarray platform. Compounds 1–6 were synthesized chemically by our programmable one-pot methodology, which also provided access to biotin- and fluorescence-labeled Globo H derivatives 7–9. We also tested cancer patient serum for the presence of antibodies against analogs 1–6 by using the microarray approach. In addition, we implemented fluorescence-tagged analytical sequencing to provide structural confirmation of synthetic oligosaccharides. This method requires picomole amounts of material and is complementary to traditional methods for the determination of the structure of synthetic sugars. The combination of these microarray and sequencing tools allows for a thorough characterization of synthetic Globo H, its interactions with corresponding monoclonal antibody binding partners, MBr1 and VK-9, and the presence of antibodies against it in cancer patient serum.
Materials and Methods
General. Primary antibodies used were mouse anti-Globo H monoclonal antibodies MBr1 (IgM, Alexis Biochemicals, Lausen, Switzerland) and VK-9 (IgG, gift from Philip Livingston, Memorial Sloan-Kettering Cancer Center, New York). Secondary antibodies were FITC-tagged goat anti-mouse IgM, goat anti-mouse IgG, goat anti-human IgM, and goat anti-human IgG (Calbiochem). The cancer patient serum was also a gift from Philip Livingston.
General Procedure for One-Pot Synthesis of Protected Globo H 13 and Analogs 15 and 16. The first donor (1.2 eq), reducing end acceptor (1 eq), and molecular sieves were stirred in CH2Cl2 for 1 h at room temperature. The reaction was cooled to –50°C, and N-iodosuccinimide (NIS) (1.2 eq) was added, followed by trifluoromethanesulfonic acid (TfOH) (1 M solution in ether, 0.3 eq). The mixture was stirred for 2 h at –40°C to –50°C and followed by TLC until completion. The nonreducing end second donor (1 eq) was dissolved in CH2Cl2 and added to the reaction mixture. NIS (1.2 eq) was added, followed by TfOH (1 M solution in ether, 0.16 eq). The reaction was stirred at –30°C to –20°C for 2 h, then diluted with CH2Cl2 and quenched with a few drops of Et3N. Next, the reaction mixture was washed with saturated aqueous (sat. aq.) NaHCO3 and sat. aq. Na2S2O3 and dried over Na2SO4. Purification by column chromatography provided the protected saccharide.
General Procedure for the Diazotransfer Reaction (1–4b). Sodium azide (20 eq) was dissolved in a minimum volume of water and cooled to 0°C. An equal volume of dichloromethane was added, and trifluoromethanesulfonic anhydride (10 eq) was added slowly to the vigorously stirring solution. The reaction was stirred at 0°C for 2 h. Saturated aqueous (sat. aq.) NaHCO3 was added to quench the reaction. The mixture was extracted twice with CH2Cl2, and the organic layers were washed once with sat. aq. NaHCO3 and used for the next reaction without further purification. The substrate and CuSO4 (0.1 eq) were dissolved in H2O with the same volume as the triflyl azide solution to be added. Triethylamine (3 eq) was added to the mixture. The freshly prepared CH2Cl2 solution of triflyl azide was added at once with vigorous stirring. MeOH was added to obtain the desired 3:10:3 ratio of H2O/MeOH/CH2Cl2. The solution was stirred overnight, then evaporated and purified by column chromatography.
Microarray Analysis of Globo H Derivatives of 1–4a, 5, and 6. NHS-coated glass slides were spotted with solutions of sugars 1–4a, 5, and 6 with concentrations of 1, 2, 5, 7.5, 10, 12.5, 15, 17.5, 20, 30, 40, 50, 60, 80, and 100 μM from bottom to top with 15 replicates horizontally placed in each grid, then blocked with ethanol amine. The slides were dried and stored at room temperature in a desiccator. The slide was washed with PBS buffer (pH 7.4) before use. Next, a 50 μg/ml solution in 0.05% Tween 20/PBS buffer (pH 7.4) of MBr1 anti-Globo H monoclonal antibody (IgM) from mouse or VK-9 anti-Globo H monoclonal antibody (IgG) from mouse was added and spread throughout the grid by application of a coverslip. Incubation in a glass humidifying chamber was performed with shaking for 1 h. Then the slide was washed three times with 0.05% Tween 20/PBS buffer (pH 7.4), three times with PBS buffer (pH 7.4), and three times with water. Next, a 50 μg/ml solution of FITC-tagged goat anti-mouse IgM (for MBr1) or IgG (for VK-9) antibody was added to the slide as before. Humidifying chamber incubation with shaking was performed under foil for 1 h. The slide was washed five times with 0.05% Tween 20/PBS buffer (pH 7.4), five times with PBS buffer (pH 7.4), and five times with H2O and dried. The slide was scanned with a microarray fluorescence scanner. The resulting image was analyzed with the program imagene (BioDiscovery, El Segundo, CA) to locate and quantify the fluorescence of all of the spots within the grid. These data were plotted against the concentration of the solution used for sugar printing to obtain carbohydrate-antibody binding curves.
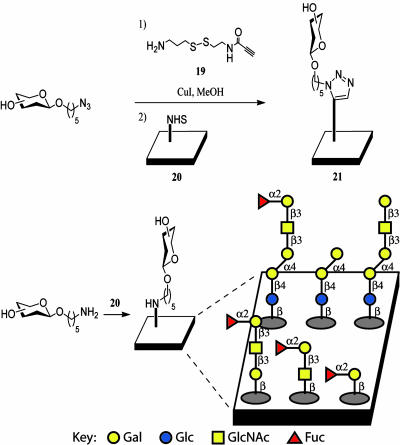
Disulfide Linker Immobilization (of Type 21). The N-t-butoxycarbonyl-protected derivative of disulfide linker 19 (3.9 mg, 13.5 μmol) was dissolved in 1 ml of CH2Cl2, and 1 ml of trifluoroacetic acid was added. The reaction was stirred for 1 h, then quenched by solvent removal. The remaining trifluoroacetic acid was removed by azeotroping twice with MeOH and benzene. A 1-mM solution of the deprotected linker (19) was prepared. Samples of the azide-modified sugars were weighed (1 mg for 1b), and the linker solutions were added to each sugar (2.29 ml for 1b) such that 1 eq of the two starting materials was present. A spatula tip of copper iodide was added, and the reactions were stirred overnight. The MeOH was removed, and 1-mM stock solutions were formed by adding water. These solutions were spotted and analyzed alongside 1–4a, 5, and 6.
Microarray Analysis of Cancer Patient Serum for Antibodies that Bind Globo H Derivatives 1–6. N-hydroxysuccinimide (NHS)-coated glass slides (spotted the same as those used in the antibody binding assays) were washed with PBS buffer. Breast cancer patient serum was diluted 1:100 with 0.05% Tween 20/3% BSA/PBS buffer (pH 7.4), applied to the grid, and incubated in a glass humidifying chamber with shaking for 1 h. Then the slide was washed three times with 0.05% Tween 20/PBS buffer (pH 7.4), three times with PBS buffer (pH 7.4), and three times with water. Next, a 50 μg/ml solution of FITC-tagged goat anti-human IgM or IgG was added to the slide, and it was incubated in a humidifying chamber with shaking under foil for 1 h. The slide was washed five times with 0.05% Tween 20/PBS buffer (pH 7.4), five times with PBS buffer (pH 7.4), and five times with water and then dried with air. The slide was then scanned with a microarray fluorescence scanner, and the data were analyzed with imagene software.
Globo H Structure Confirmation by Analytical Sequencing. Globo H with a free hydroxyl group at the reducing terminus was labeled by reductive amination with 2-aminobenzamide according to published procedures (48). This fluorescently labeled Globo H was subjected to arrays of exoglycosidases [α-fucosidase (bovine kidney), Sigma], β-1,3-galactosidase (recombinant from Escherichia coli, Calbiochem), β-N-acetylglucosaminidase (recombinant from Streptococcus pseumoniae, Prozyme, San Leandro, CA), and β-N-acetylhexosaminidase (from Jack Bean, Prozyme) according to the manufacturer's recommended buffers. Exoglycosidase reactions were then desalted on C4 ZipTips (Millipore) before analysis by normal phase-HPLC (Glycosep N HPLC column, Prozyme) to determine the sequence, monosaccharide type, and linkage of sugar residues. HPLC conditions used were described fully by Rudd and coworkers (48).
For more details see Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
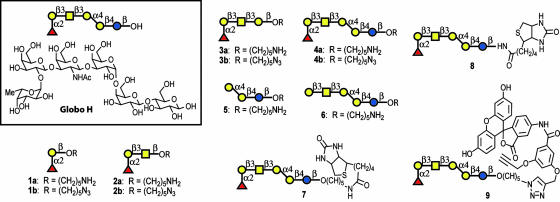
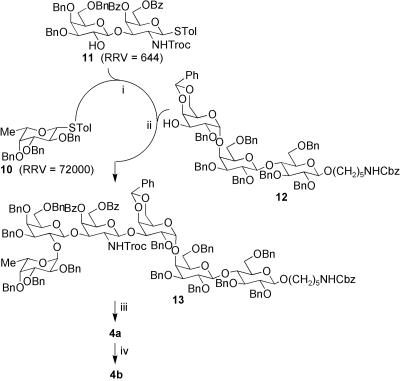
Truncated Globo H analogs 1–6 (Fig. 1) were prepared by using the one-pot programmable protocol for oligosaccharide synthesis such that binding to monoclonal antibodies MBr1 and VK-9 could be evaluated by using microarray analysis. Analogs 1–4 contain the saccharide domain of the natural glycolipid with sequentially clipped sugars, and compounds 5 and 6 are similar to 3 and 4, respectively, without the terminal fucose moiety. Furthermore, 1-aminopentyl or 1-azidopentyl linkers were included at the reducing ends for immobilization via covalent linkage to NHS-coated glass slides, allowing analysis with two different immobilization methods. Although our laboratory had previously reported the one-pot programmable synthesis of Globo H (25), we implemented a different synthetic strategy for this study (Fig. 2). Instead of using two one-pot reactions, we constructed the entire hexasaccharide in a single one-pot reaction by using a [1 + 2 + 3] approach. Formation of the most difficult Galα1–4Gal bond in advance improved the yield of the one-pot reaction (83% versus 62% for the previous strategy). The trisaccharide building block 12 is also valuable in the synthesis of all Globo family oligosaccharides.
Fig. 1.
Structure of Globo H, truncated derivatives, and conjugates.
Fig. 2.
One-pot synthesis of Globo H hexasaccharide analogs 4a and 4b. (i) NIS, TfOH, –40°C. (ii) NIS, TfOH, –30°C, 83% from 10. (iii) a) Zn, AcOH; b) Ac2O, Py; c) NaOMe; d) H2, Pd-black, HCOOH, 70% over four steps. (iv) TfN3, CuSO4, 70%.
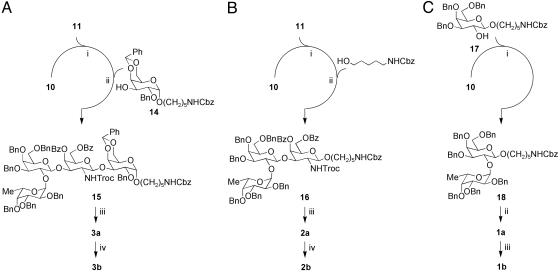
The synthesis of the tetrasaccharide, trisaccharide, and disaccharide analogs implemented the same building blocks as the full Globo H hexamer. The coupling of building blocks 10 and 11 to galactose building block 14 or the linker N-Cbz-5-hydroxylpentamine gave tetrasaccharide 15 (Fig. 3A) or trisaccharide 16 (Fig. 3B), respectively. The coupling of galactose building block 17 and fucose building block 10 gave disaccharide 18 (Fig. 3C). Deprotection of Globo H hexasaccharide 13, tetrasaccharide 15, and trisaccharide 16 began with the removal of the trichloroethoxycarbonyl group by using activated Zn particles and acetic acid. This deprotection was followed by acetylation of the amine group with acetic anhydride and pyridine. The benzoate groups were removed with sodium methoxide in methanol. Final deprotection of the benzyl ether, benzylidene acetal, and N-Cbz groups was accomplished by using Pd-black in 5% formic acid/methanol under 1 atm (1 atm = 101.3 kPa) hydrogen, which yielded the fully deprotected Globo H hexasaccharide 4a, tetrasaccharide 3a, and trisaccharide 2a. Deprotection of disaccharide 18 through treatment with Pd-black in 5% formic acid in methanol as described above gave compound 1a. The diazotransfer reaction (triflyl azide with copper sulfate catalyst) was used to convert the amine groups of deprotected oligosaccharides 1a–4a to the corresponding azido-sugars 1b–4b, respectively. Similar methods were used to synthesize 5 from disaccharide 11, galactose building block 14, and the N-Cbz-5-hydroxylpentamine linker. Compound 6 was made by treatment of 4a with α-fucosidase from bovine kidney.
Fig. 3.
Synthesis of truncated Globo H sequences. (A) One-pot synthesis of Globo H tetrasaccharide analogs 3a and 3b. (i) NIS, TfOH, –50°C. (ii) NIS, TfOH, –20°C, 42% from 10. (iii) a) Zn, AcOH; b) Ac2O, Py; c) NaOMe; d) H2, Pd-black, 69% over four steps. (iv) TfN3, CuSO4, 64%. (B) One-pot synthesis of Globo H trisaccharide analogs 2a and 2b. (i) NIS, TfOH, –50°C. (ii) NIS, TfOH, –20°C, 54% from 10. (iii) a) Zn, AcOH; b) Ac2O, Py; c) NaOMe; d) H2, Pd-black, 66% over four steps. (iv) TfN3, CuSO4, 89%. (C) Synthesis of Globo H disaccharide analogs 1a and 1b. (i) NIS, TfOH, –20°C, 60% (α:β = 2:1). (ii) H2, Pd-black, 59%. (iii) TfN3, CuSO4, 52%.
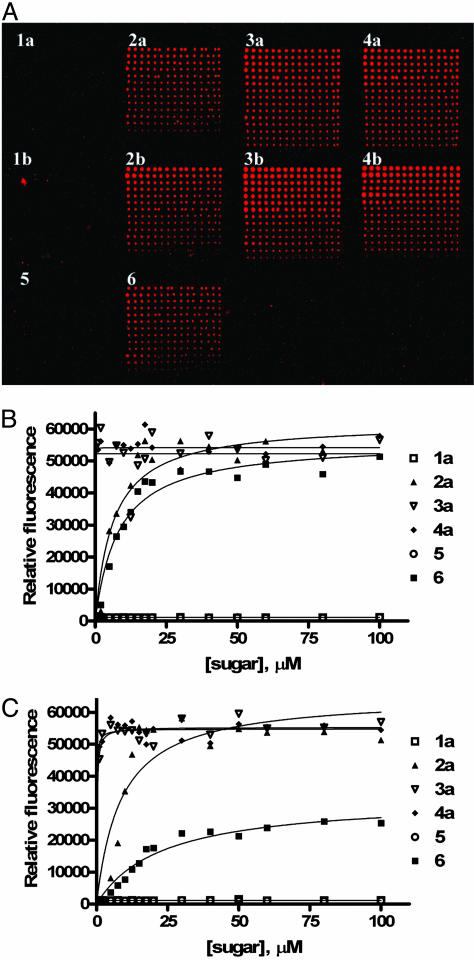
After their synthesis, the antibody binding abilities of Globo H analogs 1–6 were studied within the microarray platform. Amino-functionalized Globo H analogs 1a–4a, 5, and 6 were directly immobilized onto NHS-coated glass slides as reported (40). For azido-analogs 1b–4b, we implemented a previously developed disulfide linker strategy for surface attachment (37). The azides were combined with disulfide linker 19 via the 1,3-dipolarcycloaddition reaction followed by spotting onto the NHS microplate (20) for immobilization to 21 (Fig. 4). Sugars were spotted in a range of concentrations to allow for antibody binding curve generation. The assay involved initial treatment with either monoclonal mouse IgM antibody MBr1 or monoclonal mouse IgG VK-9, followed by incubation with a fluorescein-tagged secondary antibody, either goat anti-mouse IgM or IgG, respectively, for detection. Scanning the slide for fluorescence yielded images (Fig. 5A), in which the binding of the antibody to printed oligosaccharide spots could be directly observed. The slides contained sugars printed in grids with 1–4a in the top row from left to right, 1–4b in the middle row, and 5–6 in the bottom row (Fig. 5A). Initial visual analysis indicated that the shorter oligosaccharides show weaker recruitment of the antibody to the plate surface.
Fig. 4.
Methods for immobilization of sugars on NHS-coated glass slides.
Fig. 5.
Binding of monoclonal antibodies to Globo H and truncated sequences. (A) Slide image obtained from fluorescence scan after antibody incubation assay with VK-9. The grids contain sugars 1–4a printed in the top row from left to right, 1–4b in the middle row, and 5–6 in the bottom row. Each grid has decreasing concentrations of the glycan from 100 to 1 μM from top to bottom with each row containing 15 replicates. (B and C) Carbohydrate-antibody binding curves for Globo H analogs 1–4a, 5, and 6 with MBr1 (B) and VK-9 (C).
The fluorescence scans were processed to quantify the intensity of each spot. The resulting data are shown as binding curves for the different carbohydrate–antibody interactions with 1–4a, 5, and 6 (Fig. 5). Amino-derivatized oligosaccharide analogs 1–4a yielded higher antibody-recruitment properties than the azide containing moieties (1–4b) immobilized via the disulfide linker. This observation could be caused by poor solubility of linker containing sugars, lack of full conversion in linker attachment, or, simply, that the shorter linker is more suited to binding. The assay results showed that antibody binding generally increases with the complexity of the oligosaccharide structure. Disaccharide Globo H fragments 1a and 1b produced no recruitment of antibody to the surface. Trisaccharides 2a and 2b bound antibody, but not to the point where they could compete with the full natural hexasaccharides 4a and 4b. Tetramers 3a and 3b, however, displayed similar binding on the surface to the full natural hexamers, indicating that the tetrasaccharide core structure is fully effective for binding the antibody and the inner disaccharide moiety has little effect on binding. Also, the presence of the fucose moiety is required for binding of 3a and 3b to MBr1 or VK-9 (compare 3a and 3b with 5). In agreement with previous solution-phase assays (8), 6 shows some binding to MBr1, although we also observed similar low affinity binding between 6 and VK-9 (Fig. 5C).
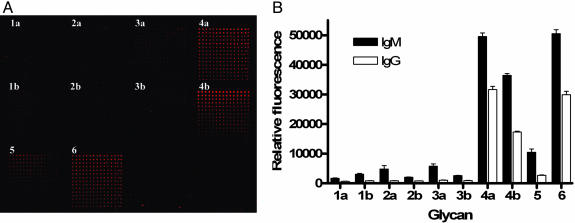
In addition to characterizing the specificities of Globo H antibodies MBr1 and VK-9 in a microarray format, we tested cancer patient serum for antibodies that bind to Globo H or Globo H fragments. Glass slides printed in microarray format with compounds 1–6 were treated with patient serum, and bound antibodies were detected with fluorescein-labeled goat anti-human IgM or IgG secondary antibody (Fig. 6). These experiments revealed the presence of antibodies that bind Globo H hexasaccharides 4a and 4b and pentasaccharide 6, along with less binding to trisaccharide 5. This result is perhaps caused by the polyclonal nature of the antibodies from patients against Globo H, or the antibodies are generated at different stages to recognize the pentasaccharide and the fucosylated hexasaccharide.
Fig. 6.
Detection of antibodies in breast cancer patient serum. (A) Slide image obtained from fluorescence scan after cancer patient serum assay with IgM detection. The grids contain sugars 1–4a printed in the top row from left to right, 1–4b in the middle row, and 5 and 6 in the bottom row, similar to Fig. 5A (B) Binding specificities of antibodies in cancer patient serum to Globo H analogs 1–4a, 5, and 6. Relative fluorescence intensities observed in spots containing 100 μM of glycan is plotted for IgM (filled bars) and IgG (empty bars) binding.
Globo H conjugates 7–9 were also synthesized. These derivatives are valuable for biological screening assays, fluorescence-based binding assays, and the generation of Globo H-carrier protein conjugates.
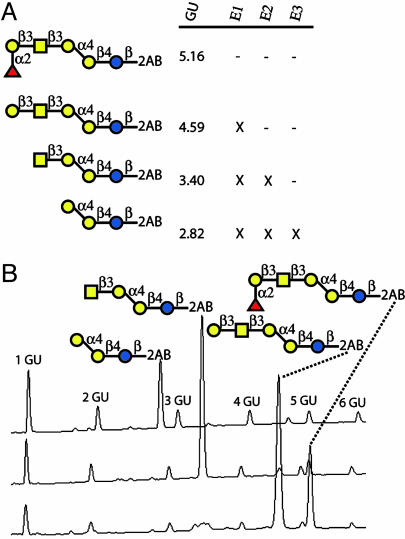
In further characterization of the Globo H oligosaccharide epitope, we sought to implement analytical sequencing for structure confirmation (48–51). For this purpose, a Globo H derivative containing a fluorescent tag was prepared by reductive amination of the reducing free terminus of Globo H with 2-aminobenzamide. This labeled Globo H was then subjected to various digestions by the exoglycosidases (Fig. 7A) and subjected to normal phase-HPLC analysis with fluorescence detection (Fig. 7B). The sensitivity of this sequencing methodology is very appropriate for structure determination of limited quantities of oligosaccharides, as 0.1 pmol is reported to have a signal-to-noise ratio of 19:1 (49). The glycan fragments obtained from digestion were as expected and act to confirm the structure of the Globo H antigen. The Globo H structure was assigned based on glycosidase specificities and change in retention time on HPLC, thus illustrating the potential of this approach for obtaining structural information of a minute amount of oligosaccharides prepared by the one-pot synthesis.
Fig. 7.
Globo H structural confirmation by analytical sequence analysis. (A) Shown are the glycans obtained by exoglycosidase cleavage with the indicated enzymes (E1 = α-fucosidase, E2 = β1,3-galactosidase, E3 = β-N-acetylhexosaminidase), marked by an X, and the glucose unit (GU) value relative to fluorescently labeled dextran standard. (B) Sample chromatograms from normal phase-HPLC with fluorescence detection (excitation = 330 nm, emission = 420 nm) highlighting glycans obtained during sequence analysis.
Conclusion
Overall, these studies expand on understanding the oligosaccharide epitope found on the crucial glycosphingolipid Globo H and its interactions with antibodies MBr1 and VK-9 and cancer patient serum. One-pot chemical synthesis of carbohydrates, along with sensitive techniques for sequence analysis and carbohydrate–protein interactions, should assist in the advancement of anticancer vaccine development. One aspect of this pursuit has involved the display of Globo H on a scaffold for multivalent presentation to yield an innate immune response in patients. Toward this end, it is beneficial to facilitate the synthesis of the immunogen such that the cost and efficiency of production are optimal. Simplified Globo H tetrasaccharide 3 shows similar binding affinity to 4 in multivalent format, whereas the synthetic route to this compound is shorter. As a result, this derivative shows great promise for the efficient development of an anticancer vaccine and for diagnostic methods. In addition, the presence of antibodies against full-length Globo H and pentasaccharide 6 were observed in cancer patient serum. The result is, however, different from that based on the analysis of monoclonal antibodies, probably because of the polyclonal nature of antibodies, or the antibodies are generated at different stages to recognize different epitopes. The advancement of cancer therapy will require an arsenal of tools for understanding and treating the disease, which has become more vital because of the recent reports of cancer stem cells and the promise and challenges exhibited by this field. The sequencing and microarray techniques presented herein represent effective and sensitive methods for rapid analysis of the specificity of protein–carbohydrate interactions and characterization of differentiation processes pertaining to cancer onset at the molecular level.
Supplementary Material
Author contributions: C.-H.W. designed research; C.-Y.H., D.A.T., A.Y.C., M.D.B., J.H., and S.H. performed research; and D.A.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: NIS, N-iodosuccinimide; TfOH, trifluoromethanesulfonic acid; NHS, N-hydroxysuccinimide.
References
- 1.Kannagi, R., Levery, S. B., Ishigami, F., Hakomori, S. I., Shevinsky, L. H., Knowles, B. B. & Solter, D. (1983) J. Biol. Chem. 258, 8934–8942. [PubMed] [Google Scholar]
- 2.Zhang, S. L., Cordon-Cardo, C., Zhang, H. S., Reuter, V. E., Adluri, S., Hamilton, W. B., Lloyd, K. O. & Livingston, P. O. (1997) Int. J. Cancer 73, 42–49. [DOI] [PubMed] [Google Scholar]
- 3.Hakomori, S. & Zhang, Y. M. (1997) Chem. Biol. 4, 97–104. [DOI] [PubMed] [Google Scholar]
- 4.Dube, D. H. & Bertozzi, C. R. (2005) Nat. Rev. Drug Discov. 4, 477–488. [DOI] [PubMed] [Google Scholar]
- 5.Menard, S., Tagliabue, E., Canevari, S., Fossati, G. & Colnaghi, M. I. (1983) Cancer Res. 43, 1295–1300. [PubMed] [Google Scholar]
- 6.Canevari, S., Fossati, G., Balsari, A., Sonnino, S. & Colnaghi, M. I. (1983) Cancer Res. 43, 1301–1305. [PubMed] [Google Scholar]
- 7.Bremer, E. G., Levery, S. B., Sonnino, S., Ghidoni, R., Canevari, S., Kannagi, R. & Hakomori, S. I. (1984) J. Biol. Chem. 259, 4773–4777. [PubMed] [Google Scholar]
- 8.Kudryashov, V., Ragupathi, G., Kim, I. J., Breimer, M. E., Danishefsky, S. J., Livingston, P. O. & Lloyd, K. O. (1998) Glycoconjugate J. 15, 243–249. [DOI] [PubMed] [Google Scholar]
- 9.Ragupathi, G., Slovin, S. F., Adluri, S., Sames, D., Kim, I. J., Kim, H. M., Spassova, M., Bornmann, W. G., Lloyd, K. O., Scher, H. I., et al. (1999) Angew. Chem. Int. Ed. 38, 563–566. [DOI] [PubMed] [Google Scholar]
- 10.Slovin, S. F., Ragupathi, G., Adluri, S., Ungers, G., Terry, K., Kim, S., Spassova, M., Bornmann, W. G., Fazzari, M., Dantis, L., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 5710–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danishefsky, S. J. & Allen, J. R. (2000) Angew. Chem. Int. Ed. 39, 836–863. [DOI] [PubMed] [Google Scholar]
- 12.Wang, Z. G., Williams, L. J., Zhang, X. F., Zatorski, A., Kudryashov, V., Ragupathi, G., Spassova, M., Bornmann, W., Slovin, S. F., Scher, H. I., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 2719–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilewski, T., Ragupathi, G., Bhuta, S., Williams, L. J., Musselli, C., Zhang, X. F., Bencsath, K. P., Panageas, K. S., Chin, J., Hudis, C. A., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragupathi, G., Coltart, D. M., Williams, L. J., Koide, F., Kagan, E., Allen, J., Harris, C., Glunz, P. W., Livingston, P. O. & Danishefsky, S. J. (2002) Proc. Natl. Acad. Sci. USA 99, 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slovin, S. F., Ragupathi, G., Fernandez, C., Jefferson, M. P., Diani, M., Wilton, A. S., Powell, S., Spassova, M., Reis, C., Clausen, H., et al. (2005) Vaccine 23, 3114–3122. [DOI] [PubMed] [Google Scholar]
- 16.Bilodeau, M. T., Park, T. K., Hu, S. H., Randolph, J. T., Danishefsky, S. J., Livingston, P. O. & Zhang, S. L. (1995) J. Am. Chem. Soc. 117, 7840–7841. [Google Scholar]
- 17.Park, T. K., Kim, I. J., Hu, S. H., Bilodeau, M. T., Randolph, J. T., Kwon, O. & Danishefsky, S. J. (1996) J. Am. Chem. Soc. 118, 11488–11500. [Google Scholar]
- 18.Lassaletta, J. M. & Schmidt, R. R. (1996) Liebigs Ann. 1417–1423.
- 19.Zhu, T. & Boons, G. J. (1999) Angew. Chem. Int. Ed. 38, 3495–3497. [DOI] [PubMed] [Google Scholar]
- 20.Allen, J. R., Allen, J. G., Zhang, X. F., Williams, L. J., Zatorski, A., Ragupathi, G., Livingston, P. O. & Danishefsky, S. J. (2000) Chem. Eur. J. 6, 1366–1375. [DOI] [PubMed] [Google Scholar]
- 21.Allen, J. R., Harris, C. R. & Danishefsky, S. J. (2001) J. Am. Chem. Soc. 123, 1890–1897. [DOI] [PubMed] [Google Scholar]
- 22.Bosse, F., Marcaurelle, L. A. & Seeberger, P. H. (2002) J. Org. Chem. 67, 6659–6670. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekaran, E. V., Chawda, R., Locke, R. D., Piskorz, C. F. & Matta, K. L. (2002) Glycobiology 12, 153–162. [DOI] [PubMed] [Google Scholar]
- 24.Keding, S. J., Endo, A. & Danishefsky, S. J. (2003) Tetrahedron 59, 7023–7031. [Google Scholar]
- 25.Burkhart, F., Zhang, Z. Y., Wacowich-Sgarbi, S. & Wong, C. H. (2001) Angew. Chem. Int. Ed. 40, 1274–1277. [DOI] [PubMed] [Google Scholar]
- 26.Lay, L., Nicotra, F., Panza, L., Russo, G. & Adobati, E. (1994) Helv. Chim. Acta 77, 509–514. [Google Scholar]
- 27.Lay, L., Panza, L., Russo, G., Colombo, D., Ronchetti, F., Adobati, E. & Canevari, S. (1995) Helv. Chim. Acta 78, 533–538. [Google Scholar]
- 28.Toma, L., Ciuffreda, P., Colombo, D., Ronchetti, F., Lay, L. & Panza, L. (1994) Helv. Chim. Acta 77, 668–678. [Google Scholar]
- 29.Toma, L., Colombo, D., Ronchetti, F., Panza, L. & Russo, G. (1995) Helv. Chim. Acta 78, 636–646. [Google Scholar]
- 30.Adobati, E., Panza, L., Russo, G., Colnaghi, M. I. & Canevari, S. (1997) Glycobiology 7, 173–178. [DOI] [PubMed] [Google Scholar]
- 31.Mammen, M., Choi, S. K. & Whitesides, G. M. (1998) Angew. Chem. Int. Ed. 37, 2755–2794. [DOI] [PubMed] [Google Scholar]
- 32.Park, S. & Shin, I. (2002) Angew. Chem. Int. Ed. 41, 3180–3182. [DOI] [PubMed] [Google Scholar]
- 33.Houseman, B. T. & Mrksich, M. (2002) Chem. Biol. 9, 443–454. [DOI] [PubMed] [Google Scholar]
- 34.Feizi, T., Fazio, F., Chai, W. & Wong, C.-H. (2003) Curr. Opin. Struct. Biol. 13, 637–645. [DOI] [PubMed] [Google Scholar]
- 35.Disney, M. D., Magnet, S., Blanchard, J. S. & Seeberger, P. H. (2004) Angew. Chem. Int. Ed. 43, 1591–1594. [DOI] [PubMed] [Google Scholar]
- 36.Ratner, D. M., Adams, E. W., Su, J., O'Keefe, B. R., Mrksich, M. & Seeberger, P. H. (2004) ChemBioChem 5, 379–383. [DOI] [PubMed] [Google Scholar]
- 37.Bryan, M. C., Fazio, F., Lee, H.-K., Huang, C.-Y., Chang, A., Best, M. D., Calarese, D. A., Blixt, O., Paulson, J. C., Burton, D., et al. (2004) J. Am. Chem. Soc. 126, 8640–8641. [DOI] [PubMed] [Google Scholar]
- 38.Disney, M. D. & Seeberger, P. H. (2004) Chem. Eur. J. 10, 3308–3314. [DOI] [PubMed] [Google Scholar]
- 39.Adams, E. W., Ratner, D. M., Bokesch, H. R., McMahon, J. B., O'Keefe, B. R. & Seeberger, P. H. (2004) Chem. Biol. 11, 875–881. [DOI] [PubMed] [Google Scholar]
- 40.Blixt, O., Head, S., Mondala, T., Scanlan, C. N., Huflejt, M. E., Alvarez, R., Bryan, M. C., Fazio, F., Calarese, D. A., Stevens, J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 17033–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, I., Park, S. & Lee, M.-R. (2005) Chem. Eur. J. 11, 2894–2901. [DOI] [PubMed] [Google Scholar]
- 42.Fazio, F., Bryan, M. C., Blixt, O., Paulson, J. C. & Wong, C. H. (2002) J. Am. Chem. Soc. 124, 14397–14402. [DOI] [PubMed] [Google Scholar]
- 43.Bryan, M. C., Plettenburg, O., Sears, P., Rabuka, D., Wacowich-Sgarbi, S. & Wong, C. H. (2002) Chem. Biol. 9, 713–720. [DOI] [PubMed] [Google Scholar]
- 44.Fukui, S., Feizi, T., Galustian, C., Lawson, A. M. & Chai, W. G. (2002) Nat. Biotechnol. 20, 1011–1017. [DOI] [PubMed] [Google Scholar]
- 45.Wang, D. N., Liu, S. Y., Trummer, B. J., Deng, C. & Wang, A. L. (2002) Nat. Biotechnol. 20, 275–281. [DOI] [PubMed] [Google Scholar]
- 46.Bryan, M. C. & Wong, C.-H. (2004) Tetrahedron Lett. 45, 3639–3642. [Google Scholar]
- 47.Calarese, D. A., Lee, H.-K., Huang, C.-Y., Best, M. D., Astronomo, R. D., Stanfield, R. L., Katinger, H., Burton, D. R., Wong, C.-H. & Wilson, I. A. (2005) Proc. Natl. Acad. Sci. USA 102, 13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Royle, L., Mattu, T. S., Hart, E., Langridge, J. I., Merry, A. H., Murphy, N., Harvey, D. J., Dwek, R. A. & Rudd, P. M. (2002) Anal. Biochem. 304, 70–90. [DOI] [PubMed] [Google Scholar]
- 49.Guile, G. R., Rudd, P. M., Wing, D. R., Prime, S. B. & Dwek, R. A. (1996) Anal. Biochem. 240, 210–226. [DOI] [PubMed] [Google Scholar]
- 50.Rudd, P. M., Guile, G. R., Kuster, B., Harvey, D. J., Opdenakker, G. & Dwek, R. A. (1997) Nature 388, 205–207. [DOI] [PubMed] [Google Scholar]
- 51.Rudd, P. M. & Dwek, R. A. (1997) Curr. Opin. Biotechnol. 3, 488–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.