Abstract
Hedgehog-regulated processing of the transcription factor cubitus interruptus (Ci) in Drosophila depends on phosphorylation of the C-terminal region of Ci by cAMP-dependent protein kinase and subsequently by casein kinase 1 and glycogen synthase kinase 3. Ci processing also requires Slimb, an F-box protein of SCF (Skp1/Cullin/F-box proteins) complex, and the proteasome, but the interplay between phosphorylation and the activity of Slimb and the proteasome remains unclear. Here we show that processing of the Gli3 protein, a homolog of Ci, also depends on phosphorylation of a set of four cAMP-dependent protein kinase sites that primes subsequent phosphorylation of adjacent casein kinase 1 and glycogen synthase kinase 3. Our gain- and loss-of-function analyses in cultured cells further reveal that βTrCP, the vertebrate homolog of Slimb, is required for Gli3 processing, and we demonstrate that βTrCP can bind phosphorylated Gli3 both in vitro and in vivo. We also find that the Gli3 protein is polyubiquitinated in the cell and that its processing depends on proteasome activity. Our findings provide evidence for a direct link between phosphorylation of Gli3/Ci proteins and βTrCP/Slimb action, thus supporting the hypothesis that the processing of Gli3/Ci is affected by the proteasome.
Keywords: casein kinase 1, Gli3, hedgehog, cAMP-dependent protein kinase
Secreted hedgehog (Hh) signaling proteins play fundamental roles in the embryonic patterning of multicellular organisms ranging from insects to humans (1, 2). In Drosophila, Hh signaling is mediated by the zinc-finger-containing transcription factor cubitus interruptus (Ci). In the absence of Hh signaling, a significant fraction of full-length Ci protein (Ci155) is proteolytically processed from its C terminus to the zinc finger DNA-binding domain to generate a transcriptional repressor, Ci75 (3). Hh signaling blocks Ci155 processing and also induces the translocation of Ci155 into the nucleus (4, 5). Ci protein thus acts both positively and negatively in executing the transcriptional response to Hh signaling, and the regulation of Ci protein processing represents a key step in the transduction of the Hh signal.
Although the mechanism of Ci processing reaction is not completely understood, Ci processing has been shown to require cAMP-dependent protein kinase (PKA) activity and five PKA sites within the C-terminal region of the Ci protein (6–8). The phosphorylation of the first three PKA sites appears to prime further phosphorylation at adjacent glycogen synthase kinase 3 (GSK3) and casein kinase 1 (CK1) sites, which are also essential for Ci155 processing (9, 10). Consistent with these observations, Hh pathway activity can be triggered by mutation of the shaggy gene, the fly homolog of GSK3 (9, 10), or by RNA interference (RNAi)-mediated knockdown of CK1α mRNA in cultured fly cells (11).
In addition to the three kinases mentioned above, loss of Slimb function in Drosophila also results in failure of Ci processing and activation of the Hh signaling pathway (12). Slimb encodes an F-box/WD40-repeat-containing protein of the SCF complex (12, 13). Studies of βTrCP, the vertebrate homolog of fly Slimb, have shown that βTrCP specifically binds its phosphorylated substrates through the DSpGX2-4Sp binding motif, where Sp refers to phosphoserine and X refers to any residue (14–21). βTrCP binding recruits the ubiquitination machinery to its substrates and mediates conjugation of multiple ubiquitins, resulting in either degradation or processing (22). The involvement of Slimb in Ci processing raises the possibility that Ci processing may depend on the proteasome activity. Indeed, Ci processing can be inhibited by specific proteasome inhibitors (4, 23). Based on these observations, it would be tempting to speculate that Ci processing is mediated by the proteasome. But it remains unclear whether Slimb is directly or indirectly involved in Ci processing because it has not been shown whether it is able to bind phosphorylated Ci protein. Because it is difficult to detect polyubiquitinated forms of Ci protein, it has been proposed that Slimb may act indirectly through an unidentified factor(s) (4, 11). To understand the molecular mechanism of Hh signaling, it is therefore important to distinguish direct vs. indirect modes of Slimb action in Ci processing.
The vertebrate homologs of Drosophila Ci comprise Gli1, Gli2, and Gli3, of which only Gli3 has been shown to be processed into the Gli3–83 repressor in vivo (24). Like Ci processing, Gli3 processing is also inhibited by Shh signaling (24–26) and requires the activity of PKA and six PKA sites within its C-terminal region. It is not clear, however, whether βTrCP, CK1, and GSK3 are involved in Gli3 processing, or whether the proteasome mediates Gli3 processing. In the present study we show that Gli3 phosphorylation by PKA primes further phosphorylation by CK1 and GSK3 and that PKA-primed phosphorylation is required for Gli3 processing. Similar to the role of Slimb in Ci processing, βTrCP is also required for Gli3 processing. Most importantly, we demonstrate that βTrCP can directly bind phosphorylated Gli3 protein both in vitro and in vivo, that Gli3 is polyubiquitinated in the cell, and that Gli3 processing requires proteasome activity. Our findings strongly support the hypothesis that βTrCP acts directly on the Gli3 protein and that Gli3 processing is affected by the proteasome.
Results
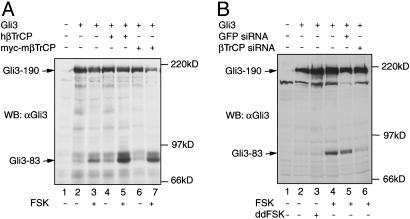
The Role of βTrCP in Gli3 Protein Processing. Because Slimb is required for Ci155 processing in Drosophila (12), we asked whether βTrCP was also involved in Gli3 protein processing. The simple coexpression of Gli3 with either human βTrCP (hβTrCP) or myc-tagged mouse βTrCP (myc-mβTrCP) in HEK293 cells did not result in an increase in Gli3–83 levels (Fig. 1A, lanes 2, 4, and 6). However, treatment with forskolin (FSK), which activates PKA, significantly enhanced Gli3 processing as measured by an increase in the levels of Gli3–83 protein (Fig. 1 A, lanes 3, 5, and 7). A similar enhancement of Gli3 processing by βTrCP was also observed when a constitutively active PKA was coexpressed or when the primary culture of chicken limb bud cells was transfected (data not shown). These data suggest that βTrCP is also involved in Gli3 processing in vertebrates and that it acts downstream of PKA with respect to Gli3 processing.
Fig. 1.
Gli3 processing requires βTrCP. HEK293 cells were transfected with the expression constructs or together with either βTrCP siRNA or GFP siRNA control as shown above the blots and treated overnight with FSK as indicated. Gli3 protein was detected by immunoblotting with an anti-Gli3 antibody. Overexpression of βTrCP resulted in an increase in the Gli3–83 levels (A, compare lanes 5 and 7 with lane 3), whereas βTrCP RNAi blocks Gli3 processing (B, compare lane 6 with lanes 4 and 5).
To determine whether βTrCP is required for Gli3 processing, RNAi was used to knock down the endogenous βTrCP gene product. Cotransfection of a Gli3 expression construct with a double-stranded βTrCP small interfering RNA (βTrCP siRNA), which has been previously shown to effectively knock down βTrCP RNA in HEK293 cells (27), dramatically inhibited the production of Gli3–83 (Fig. 1B, compare lane 6 with lanes 4 and 5). However, RNAi using control GFPsiRNA had little effect on Gli3 processing (Fig. 1B, compare lane 5 with lane 4). The effect of βTrCP RNAi was confirmed by quantitative RT-PCR analysis of βTrCP RNA (Fig. 6, which is published as supporting information on the PNAS web site). These results indicate that βTrCP is required for Gli3 processing.
PKA-Primed Gli3 Phosphorylation by GSK3 and CK1. We have previously shown that PKA stimulation induces hyperphosphorylation of Gli3 protein as measured by the accumulation of slowly migrating Gli3 species in SDS/PAGE. The hyperphosphorylation of Gli3 protein depended on six PKA sites (24) (Fig. 2B, compare lane 13 with lane 4), but it cannot be explained by phosphorylation of the PKA sites alone because the in vitro phosphorylation of immunoprecipitated Gli3 protein by PKA caused no noticeable shift in migration of the protein (data not shown). Two recent studies have shown that phosphorylation of first three PKA sites in the Ci C-terminal region primed the further phosphorylation of adjacent GSK3 and CK1 sites, which appear to be required for Ci155 processing (9, 10). Because these sites are conserved in Gli3, we tested their role in Gli3 protein hyperphosphorylation.
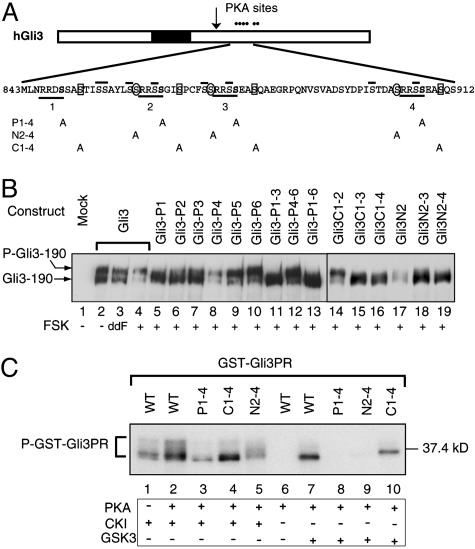
Fig. 2.
PKA-primed phosphorylation of Gli3 protein by CK1 and GSK3. (A)A schematic drawing of hGli3 protein. Six dots stand for PKA sites, and a filled box represents the zinc finger DNA-binding domain. An arrow indicates the approximate cleavage site. Underlined is the consensus sequence for PKA phosphorylation with S residues shown in bold. Numbers below the lines correspond to the designated names of the Gli3 mutant constructs. PKA-primed phosphorylation sites for GSK3 and CK1 are circled and boxed, respectively. Overlined are the secondary S residues that may potentially become GSK3 and CK1 phosphorylation sites. (B) Immunoblot analysis revealed phosphorylation of Gli3 and its mutants as measured by mobility shift of the proteins in transfected chicken limb bud cells after treatment with dideoxy FSK (ddFSK) and FSK. Proteins were separated by 5% SDS/PAGE. (C) PKA-primed phosphorylation of GST-Gli3PR (839–920 aa) and its mutants by CK1 and GSK3 in vitro. Affinity-purified GST-Gli3PR and its mutant proteins were first incubated with PKA catalytic subunit in the presence of nonradioactive ATP and subsequently with CK1 or GSK3 in the presence of [γ-32P]ATP. The phosphorylated proteins were resolved by SDS/PAGE and detected by autoradiography.
The Gli3 C-terminal region contains six instead of five PKA sites; the first one of the six PKA sites is flanked by a CK1 site alone, and the second, third, and fourth sites are flanked by both GSK3 and CK1 sites (Fig. 2 A). Point mutations were introduced into some of the three GSK3 sites or four CK1 sites either individually or in combinations, including GSK3 sites 2, 2–3, and 2–4 or CK1 sites 1–2, 1–3, and 1–4 (designated Gli3N2, Gli3N2–3, and Gli3N2–4, or Gli3C1–2, Gli3C1–3, and Gli3C1–4, respectively). The phosphorylation status of the Gli3 mutant proteins was examined in transfected primary chicken limb bud cells in response to PKA stimulation. Whereas mutations in single GSK3 or CK1 sites had little effect on phosphorylation of the Gli3 protein as measured by protein mobility shift, mutations in two or more GSK3 or CK1 sites significantly reduced the mobility shift in Gli3 protein (Fig. 2B, compare lanes 14–19 with lane 4, and data not shown). The extent of reduction appeared to be directly correlated with the number of sites mutated. Nevertheless, Gli3N2–4 and Gli3C1–4 mutant proteins did not migrate as fast as Gli3P1–3 and Gli3P1–6 (Fig. 2B, compare lanes 16 and 19 with lanes 11 and 13). This finding is consistent with the idea that in vivo Gli3 phosphorylation by GSK3 and CK1 occurs only after the protein is phosphorylated by PKA.
To directly examine PKA-primed Gli3 phosphorylation by GSK3 and CK1, we performed an in vitro sequential kinase reaction (see Materials and Methods for details) using a human Gli3 fragment (839–920 residues) fused with GST. The fusion protein, GST-Gli3PR (Gli3 PKA region), which contains the first four PKA sites, three GSK3 sites, and four CK1 sites, was weakly phosphorylated by CK1 even without preincubation with PKA (Fig. 2C, lane 1), indicating that CK1 can phosphorylate the fusion protein without priming the PKA sites in vitro. Nevertheless, the fusion protein did exhibit greater phosphorylation by CK1 after preincubation with PKA (Fig. 2, compare lane 2 with lane 1). Phosphorylation by PKA also induced phosphorylation of the fusion protein by GSK3 (Fig. 2C, compare lane 7 with lane 6). To further confirm the PKA-primed Gli3 phosphorylation by CK1 and GSK3 in vitro, the same kinase assay was performed by using phosphorylation site GST-Gli3PR mutants. The alteration of either PKA or CK1 sites (P1–4 and C1–4, respectively) significantly reduced phosphorylation of fusion proteins by CK1, although not completely (Fig. 2C, compare lanes 3 and 4 with lane 2). Similarly, mutations at either PKA or GSK3 sites (N2–4) completely eliminated the phosphorylation of fusion proteins by GSK3 (Fig. 2C, compare lanes 8 and 9 with lane 7). From these results we conclude that phosphorylation of the first four PKA sites of the Gli3 protein primes the further phosphorylation of the protein by CK1 and GSK3 in vitro. The same synergistic phosphorylation phenomenon is also likely to occur in vivo because Gli3, when expressed in chick limb bud cells, was hyperphosphorylated, whereas mutations in the PKA sites of the proteins abolished the hyperphosphorylation (Fig. 2B) (24).
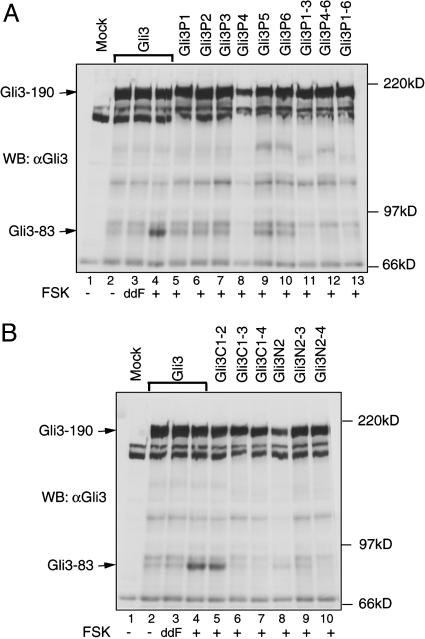
The Role of GSK3 and CK1 Sites in Gli3 Processing. To determine the significance of Gli3 phosphorylation in Gli3 processing, we asked whether mutations in GSK3 or CK1 sites affected Gli3 processing. We also examined the Gli3PKA (renamed Gli3P) mutant for processing in our new transfection conditions using primary cultures of chicken limb bud cells. Mutations in combinations of six PKA sites, including Gli3P1–3, Gli3P4–6, and Gli3P1–6, completely inhibited Gli3 processing, whereas single mutations in each of six PKA sites resulted in the significantly reduced levels of Gli3–83 protein (Fig. 3A), indicating that all six PKA sites contribute to Gli3 processing and confirming our previous findings (24). Similarly, the extent of Gli3 protein processing for Gli3-C and Gli3-N mutants appeared to correlate directly with the number of CK1 or GSK3 sites mutated. Whereas mutations in the first two CK1 sites or the first GSK3 site had little effect on Gli3 processing (Fig. 3B, lanes 5 and 8; note that Gli3N2 expression was lower in this particular experiment), mutations in either all four CK1 sites or all three GSK3 sites completely blocked Gli3 processing (Fig. 3B, lanes 7 and 10). These results indicate that, like PKA sites, CK1 and GSK3 sites are also required for Gli3 processing. Mutations in the PKA, CK1, or GSK3 sites also prevented the ability of βTrCP to promote Gli3 processing (Fig. 7, which is published as supporting information on the PNAS web site), indicating that the PKA, CK1, and GSK3 phosphorylation sites are required for βTrCP action and that βTrCP acts downstream of those kinases.
Fig. 3.
Putative PKA, CK1, and GSK3 phosphorylation sites in Gli3 protein are required for Gli3 processing. (A and B) Primary chick limb bud monolayer cultures were transfected with wild-type Gli3 or its mutant constructs with point mutations at PKA sites (A) or CK1 or GSK3 sites (B) (see Fig. 2 A for sites mutated). Cells were treated with FSK, dideoxy FSK (ddFSK), or DMSO vehicle for 16–18 h, and Gli3 processing was examined by immunoblotting with the anti-Gli3 antibody. Gli3–83 protein was not detected for Gli3P1–3, Gli3P4–6, and Gli3P1–6 (A, lanes 11–13) and for Gli3C1–4 and Gli3N2–4 (B, lanes 7 and 10).
βTrCP Interacts Directly with Gli3. Studies of several βTrCP substrates have shown that βTrCP only recognizes substrates that have been phosphorylated (22). The binding of βTrCP to its substrates recruits E1, E2, and the SCF complex so that polyubiquitin chains can be conjugated to these substrates. Although the vast majority of polyubiquitinated substrates then undergo complete degradation by the proteasome, a few ubiquitinated proteins have been shown to undergo limited site-specific proteasome-mediated processing. The best example is proteasome-mediated processing of NF-κBp105 to generate functional NF-κBp50 (28–33).
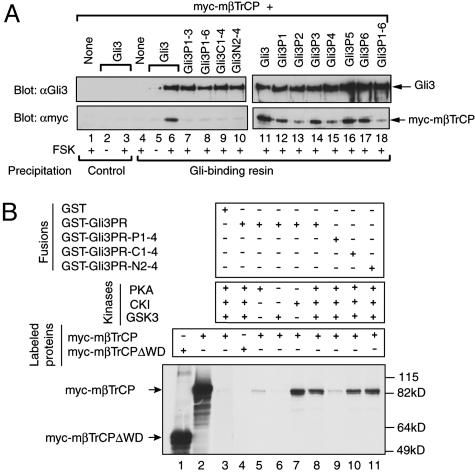
Because βTrCP is required for Gli3 processing, we next asked whether βTrCP was directly involved in Gli3 processing by examining the physical interaction between βTrCP and Gli3. As shown in Fig. 4A, βTrCP was coprecipitated by the Gli-specific DNA beads only when it was coexpressed with wild-type Gli3 protein, not Gli3P1–3, Gli3P1–6, Gli3C1–4, or Gli3N2–4 mutant proteins (compare lane 6 with lanes 7–10). The coprecipitation of βTrCP must be due to its specific interaction with Gli3 protein because the same beads failed to precipitate βTrCP when it was expressed either alone or together with wild-type Gli3 protein in the absence of FSK (Fig. 4A, lanes 4 and 5). In addition, the Gli-binding beads used must specifically recognize Gli3 protein because the nonspecific Gli-binding beads precipitated neither Gli3 protein nor βTrCP (Fig. 4A, lanes 1–3). We also tried to perform a reciprocal experiment to see whether Gli3 protein can be coimmunoprecipitated by anti-myc antibody, but attempts failed, most likely because of the inefficiency of precipitating myc-mβTrCP. From these results, we conclude that βTrCP interacts with phosphorylated Gli3 protein in the cell and that PKA, GSK3, and CK1 sites in Gli3 are required for βTrCP binding.
Fig. 4.
Direct interaction of βTrCP with Gli3 protein. (A) Interaction between βTrCP and Gli3 in the cell. HEK293 cells were transfected with myc-mβTrCP alone or together with Gli3 or its mutant constructs and treated with FSK overnight as shown below the blots. The protein lysates from the cells were incubated with DNA-conjugated Sepharose beads containing specific Gli-binding consensus sequence (Gli-binding resin) or nonspecific sequence (control). The precipitated proteins were then resolved by 7% SDS/PAGE and transferred to nitrocellulose filters. The filters were horizontally cut into two halves. The upper half was immunoblotted with anti-Gli3 antibody (Upper), and the lower half was immunoblotted with anti-myc antibody to detect myc-mβTrCP (Lower). (B) Interaction between a phosphorylated Gli3 fragment and βTrCP. GST, GST-Gli3PR, or its mutants were first phosphorylated with the indicated kinases and were then used to pull down 35S-labeled myc-mβTrCP or myc-mβTrCPΔWD proteins, which were detected by autoradiography.
The existence of multiple PKA, GSK3, and CK1 sites in the Gli3 C-terminal region raises the possibility that the Gli3 protein may contain more than one βTrCP-binding site. To test this, we examined how well PKA site Gli3 mutants were able to bind βTrCP in the cell. Mutating the first or third PKA site (Gli3P1 and Gli3P3) in Gli3 significantly reduced its ability to interact with βTrCP (Fig. 4A, lanes 12 and 14). Surprisingly, Gli3 protein with mutations in either the second or fourth PKA site consistently showed extremely weak binding to βTrCP as compared with the Gli3P1–6 negative control (Fig. 4A, compare lanes 13 and 15 with lane 18), whereas mutations in either the fifth or sixth PKA site, both of which lack adjacent GSK3 and CK1 phosphorylation sites, had little effect on its binding to βTrCP (Fig. 4A, lanes 16–17). We also examined some CK1 or GSK3 site Gli3 mutants for βTrCP binding. The amount of βTrCP pulled down by the specific Gli-binding beads was proportionally correlated with the number of phosphorylation sites mutated in Gli3 (Fig. 4A, lanes 9–10, and data not shown). Taken together, these data suggest that Gli3 has more than one βTrCP binding site and that each of the first four PKA sites contributes to βTrCP binding.
To determine whether βTrCP directly binds Gli3 protein, we examined the ability of the phosphorylated GST-Gli3PR fusion protein (Fig. 2 A) to pull down βTrCP protein in vitro. Myc-mβTrCP was brought down only by GST-Gli3PR that had been phosphorylated by CK1 alone, or all three kinases, but not by PKA or GSK3 alone (Fig. 4B, lanes 5–8), indicating that βTrCP binds only the phosphorylated fusion protein. It should be noted that CK1 phosphorylation in vitro is not very specific (Fig. 2C, lanes 3 and 4, and data not shown), which explains why GST-Gli3PR phosphorylated by CK1 alone can pull down βTrCP. Interaction between βTrCP and the fusion protein must occur between the Gli3PR sequence and WD40 repeats of βTrCP because GST alone failed to precipitate βTrCP (Fig. 4B, lane 3), and a deletion of WD40 repeats in βTrCP abolished the binding (Fig. 4B, lane 4). We then used the phosphorylation site mutant fusion proteins to test the binding specificity. Although mutations in the primary CK1 and GSK3 sites did not abrogate the binding to βTrCP, which is most likely because of the phosphorylation of the secondary CK1 and/or GSK3 sites by CK1 in vitro, PKA site mutant GST-Gli3PR-P1–4 failed to bind βTrCP (Fig. 4B, compare lane 9 with lanes 10 and 11), indicating that PKA sites are required for βTrCP binding. Taken together, our results indicate that βTrCP interacts directly with phosphorylated Gli3.
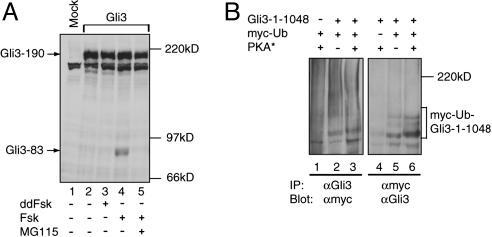
The Role of the Proteasome in Gli3 Processing. The phosphorylation-dependent direct interaction between βTrCP and Gli3 protein supports the hypothesis that Gli3 processing is mediated by the proteasome. If that were the case, Gli3 would be polyubiquitinated and inhibition of proteasome activity would block Gli3 processing. Indeed, whereas treatment of transfected HEK293 cells with FSK alone induced Gli3 processing, treatment of the same transfected cells with both FSK and MG115, a specific proteasome inhibitor, blocked Gli3 processing (Fig. 5A, compare lane 5 with lane 4).
Fig. 5.
Proteasome-dependent Gli3 processing. (A) An immunoblot showing that Gli3 processing was blocked by treatment of transfected HEK293 cells with the proteasome inhibitor MG115. (B) Gli3 is polyubiquitinated. HEK293 cells were transfected with expression constructs shown above the blots. After they were denatured, protein lysates from the cells were subjected to immunoprecipitation with either anti-Gli3 or anti-myc antibodies, followed by immunoblotting with reciprocal antibodies as indicated below the blots.
To determine whether Gli3 is polyubiquitinated, Gli3–1-1048, a construct truncated after residue 1048, was chosen to facilitate the separation and detection of polyubiquitinated forms of Gli3 because its size is smaller and yet it remains efficiently processed (Fig. 8, which is published as supporting information on the PNAS web site). HEK293 cells were cotransfected with myc-Ub (myc-tagged ubiquitin), Gli3–1-1048, and PKA* (a constitutively active PKA) (34) constructs, or with two of the three constructs in combinations as controls. The ubiquitination of Gli3 protein was examined by coimmunoprecipitation. As shown in Fig. 5B, several slowly migrating protein species were precipitated by the myc antibody only when Gli3–1-1048 and myc-Ub were coexpressed, but not when myc-Ub was expressed alone, indicating that those are ubiquitinated forms of the Gli3 protein. The ubiquitinated forms of Gli3–1-1048 were also detected in the absence of PKA*, even though their levels were lower than those in the presence of PKA* (Fig. 5B, compare lane 5 with lane 6), suggesting that the endogenous PKA activity is adequate to induce certain levels of ubiquitination. This finding is consistent with our observation that Gli3 protein can be processed without PKA stimulation (Fig. 1 A, lane 2). Similarly, ubiquitinated forms of the Gli3 protein were also precipitated by anti-Gli3 antibody, albeit less efficiently (Fig. 5B, lanes 2 and 3), probably because of the fact that the antibody has a lower affinity than the myc antibody. Taken together, these data indicate that Gli3 is indeed polyubiquitinated in transfected cells.
Discussion
We have previously shown that PKA stimulation caused hyperphosphorylation of Gli3, as measured by reduced gel mobility (24). Like Ci in the fly (9, 10), our present study indicates that such hyperphosphorylation is mostly due to the PKA-primed phosphorylation of GSK3 and CK1 sites (the primary sites) adjacent to the PKA sites and subsequent phosphorylation of the secondary GSK3 and CK1 sites that are primed by phosphorylation of the primary GSK3 and CK1 sites. In principle, PKA, CK1, and GSK3 can phosphorylate as many as 19 serine residues in the Gli3PR region: four PKA sites, three primary GSK3 sites, four primary CK1 sites, and eight secondary GSK3 and CK1 sites together (Fig. 2 A). The secondary CK1 and GSK3 sites are most likely phosphorylated in the cell because mutations in the primary CK1 sites or GSK3 sites significantly reduce the hyperphosphorylated Gli3 species (Fig. 2B).
Like PKA sites, the primary GSK3 and CK1 sites in Gli3 protein are required for Gli3 processing. Mutations in any set of PKA sites, primary GSK3, or CK1 sites also inhibit the ability of βTrCP to promote Gli3 processing, indicating that phosphorylation is a prerequisite for βTrCP action. Indeed, here we demonstrate that βTrCP directly binds phosphorylated Gli3 protein both in the cell and in vitro, indicating that βTrCP acts directly on Gli3 instead of another unidentified factor(s). In addition, because PKA, CK1, and GSK3 sites are conserved between Gli3 and Ci, it is most likely that Slimb acts directly on Ci too.
The βTrCP recognition sequence has been defined from its interaction with many other substrates. Earlier studies identified DSpGX2Sp as the binding motif for βTrCP (14–20), but two recent studies have shown that βTrCP may bind a broad range of sequences. It is able to bind the DSpGX4Sp motif of Cdc25A (21), which has five residues between two phosphoserines. It has also been suggested that it binds EEGFGSp and DSpAFQE sequences in Wee1 protein, where E residues are believed to mimic one of the two phosphoserines in the DSpGXXSp motif (35). Thus, the βTrCP binding consensus sequence seems more diverse than it was originally thought to be, which allows βTrCP to target a broader range of proteins.
Inspection of the Gli3PR amino acid sequence revealed no known consensus sequence for βTrCP binding. This finding raised the question of which sequence motifs in Gli3 are bound by βTrCP. Our protein–protein binding analysis using Gli3 mutants indicates that the phosphorylation of any set of PKA, primary CK1, or GSK3 sites is required for βTrCP binding in the cell (Fig. 4), but the results do not necessarily indicate that these phosphorylated sites are actually involved in direct contact with βTrCP. In fact, the distance between a PKA site and a primary CK1 site in Gli3 is only two residues, and between a PKA site and a primary GSK3 site there are three residues, two of which are basic R residues that have never been found in the known βTrCP-binding motifs. Such a short spacer between the two phosphoserines makes it unlikely that the first four PKA sites and the primary GSK3 and CK1 sites are all involved in direct binding with βTrCP. In support of this view, we found that mutations in all four primary CK1 or GSK3 sites did not affect the ability of phosphorylated GST-Gli3PR fusion protein to bind βTrCP in vitro, whereas mutations in the PKA sites did eliminate βTrCP binding (Fig. 4B). These results can be interpreted if PKA sites and secondary instead of primary CK1 and/or GSK3 sites are the sites that are actually involved in βTrCP binding. A PKA site and a secondary GSK3 and/or CK1 site are five or six residues apart (Fig. 2 A), thus resembling the βTrCP-binding motif in Cdc25A. The difference between in vitro and in vivo phosphorylation is that CK1 can phosphorylate nonprimed serine residues in Gli3 in vitro such as the secondary CK1 sites and probably even secondary GSK3 sites when the primary CK1 and GSK3 sites are mutated, whereas PKA, CK1, and GSK3 activities inside the cell are well regulated so that the phosphorylation of PKA sites primes the primary GSK3 and CK1 sites, which in turn prime the secondary GSK3 and CK1 sites. Mutations in the primary CK1 and GSK3 sites thus block the phosphorylation of secondary CK1 and GSK3 sites, consequently preventing Gli3 from binding to βTrCP. There are eight secondary CK1 and GSK3 sites, including those immediately next to the second, third, and fourth PKA sites in Gli3 (Fig. 2 A). These S residues, once phosphorylated, become negatively charged and may mimic D residues in the βTrCP binding motif: DSpGX2-4Sp. It may be difficult to test this, because mutating altered secondary CK1 and GSK3 sites might interfere with the phosphorylation of the Gli3 by PKA, given that three of the eight sites are within the PKA consensus sequences. Our in vitro and in vivo binding analyses support the notion that phosphorylated PKA sites and secondary CK1 and/or GSK3 sites are directly involved in βTrCP binding. Analysis of PKA site Gli3 mutants also reveals the presence of more than one βTrCP-binding site in the Gli3PR region. This kind of redundant sequence arrangement may add robustness to Gli3 protein processing, which is a critical event for normal embryonic development.
Although the vast majority of βTrCP substrates undergo complete degradation after their polyubiquitination, there is at least one exception. The site-specific processing of NF-κBp105 to form the functional NF-κBp50 has been shown to be mediated by the proteasome (28–33). Several features of Gli3/Ci processing are reminiscent of NF-κBp105 processing. First, like NF-κBp105, both Ci and Gli3 are processed in a site-specific manner. Second, both Gli3 and NF-κB105 are polyubiquitinated in the cell, and the processing of both Gli3/Ci and NF-κB105 depends on proteasome activity (Fig. 5) (4, 23). Third, consistent with the idea that the C-terminal fragments of Gli3/Ci and NF-κB105 are degraded by the proteasome, they have never been detected after the proteins are processed. Fourth, processing of both NF-κB and Gli3/Ci requires phosphorylation of their C termini, and both phosphorylated Gli3 and NF-κBp105 proteins can be bound by βTrCP (Fig. 4) (36). The only difference between Gli3 and NF-κBp105 phosphorylation is that the former is phosphorylated at numerous sites by at least three kinases, whereas the latter is phosphorylated at fewer sites by only IκB kinase (36). These observations strongly suggest that, like NF-κB processing, Gli3 processing is mediated by the proteasome. However, the possibility that Gli3 is cleaved by a site-specific protease cannot be ruled out because it is possible that the proteasome may simply degrade the polyubiquitinated Gli3 C-terminal region after it is cleaved. If this proves to be true, then one would predict that deleting the cleavage site would inhibit processing because the amino acid sequence around the cleavage site is usually essential for proper cleavage of the protein. Contrary to this prediction, we have found that a Gli3 mutant whose cleavage site has been deleted was still efficiently processed (data not shown). A similar observation has also been reported for Ci155 (37). Taken together, these observations support the hypothesis that Gli3 processing is directly affected by the proteasome, although further studies will be needed to prove this conclusively.
Materials and Methods
DNA Constructs. Expression constructs for human Gli3 (hGli3) and its PKA site mutants were described in ref. 24, but the mutants were renamed as Gli3P followed by a number for the site(s). PCR-based mutagenesis was used to alter serine residues to alanine at first one, two, three, and all four putative CK1 phosphorylation sites (residues 852, 868, 880, and 910) and first one, two, and all three GSK3 putative phosphorylation sites (residues 861, 873, and 903) in hGli3, designated Gli3C1, Gli3C1–2, Gli3C1–3, Gli3C1–4, Gli3N2, Gli3N2–3, and Gli3N2–4, respectively. The Gli3–1-1048 (1–1048 aa) construct was generated by a combination of restriction digestion and PCR strategies. Gex2T-Gli3PR (839–920 aa), Gex2T-Gli3PR-P1–4 (first four PKA sites mutated), Gex2T-Gli3PR-C1–4, and Gex2T-Gli3PR-N2–4 were generated by PCR and insertion in-frame into BamHI and EcoRI sites of the pGex2T vector (Amersham Pharmacia). A myc-tagged human ubiquitin construct, myc-Ub, was cloned into the pRK expression vector by RT-PCR. Myc-mβTrCP, hβTrCP, and PKA* (constitutively active PKA) (34) constructs were described in refs. 13 and 18. Myc-mβTrCPΔWD was generated by a deletion from the BglII site to the stop codon of mβTrCP.
Cell Culture, Transfection, and Analysis. Cell culture, transfection, pharmacological treatment, immunoprecipitation, and immunoblot analysis were mostly performed as described in ref. 24. To determine proteasome-dependent Gli3 processing, cells were treated with FSK (40 μM) and MG115 (50 μM) for 6 h. Cells used for the detection of ubiquitinated Gli3 protein were first lysed in a well of a six-well plate with 100 μl of denaturing buffer (1% SDS/50 mM Tris, pH 7.5/0.5 mM EDTA/1 mM DTT). After incubation for 5 min at 100°C, the lysates were diluted 10× with lysis buffer and then subjected to coimmunoprecipitation and immunoblot analysis. Coprecipitation of βTrCP with phosphorylated Gli3 protein was performed by using affinity Sepharose beads conjugated with double-stranded oligonucleotide containing a Gli-binding site (38). The affinity Gli-binding Sepharose beads were prepared as described in ref. 39 and consisted of the following oligonucleotide sequences: A1 Gli-binding site, 5′-TGG GCG AAG ACC ACC CAC AAT GA (sense) and 5′-ACC ATC ATT GTG GGT GGT CTT CG (antisense); B1 Gli-binding site, 5′-GAT CCG TGG ACC ACC CAA GAC GAA ATT (sense) and 5′-GAT CAA TTT CGT CTT GGG TGG TCC ACG (antisense); nonspecific Gli-binding site, 5′-GAT CAC AGA TAC ATC TCT CAG ACT GC (sense) and 5′-GAT CGC AGT CTG AGA GAT GTA TCT GT (antisense). RNAi of βTrCP was mostly performed as described in ref. 27.
In Vitro Phosphorylation and Binding Assays. Affinity-purified GST-Gli3PR and its mutants were incubated with PKA catalytic subunit (Sigma) in the presence of ATP at 30°C for 30 min. After PKA and ATP were washed off, the fusion proteins were incubated with CK1 and GSK3 (NEB, Beverly, MA) alone or both in the presence of [γ-32P]ATP at 30°C for 30 min. The phosphorylated proteins were detected by autoradiography. For binding assay, ≈5 μg of freshly made GST fusion proteins were phosphorylated on glutathione beads and incubated with [35S]methionine-labeled myc-mβTrCP or myc-mβTrCPΔWD, which was prepared by using the TNT system (Promega), in 150 μl of lysis buffer containing 0.1% Triton X-100 for 1 h at 4°C with rotation. After glutathione beads were extensively washed with lysis buffer, proteins were separated by SDS/PAGE. The bound βTrCP was detected by fluorography followed by autoradiography.
Supplementary Material
Acknowledgments
We thank Drs. Zhijian J. Chen (University of Texas Southwestern Medical Center, Dallas) and Tian Xu (Yale University School of Medicine, New Haven, CT) for mouse and human βTrCP expression constructs. We also thank Anna Rodina for assistance in the in vitro interaction between Gli3 and βTrCP. This work was initiated in the laboratory of Dr. Philip Beachy and was supported by the start-up fund from Weill Medical College and National Institutes of Health Grant R01 CA111673 (to B.W.).
Author contributions: B.W. designed research; B.W. and Y.L. performed research; B.W. analyzed data; and B.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: Hh, hedgehog; Ci, cubitus interruptus; PKA, cAMP-dependent protein kinase; CK1, casein kinase 1; GSK3, glycogen synthase kinase 3; RNAi, RNA interference; siRNA, small interfering RNA; myc-Ub, myc-tagged ubiquitin; FSK, forskolin.
References
- 1.Ingham, P. W. & McMahon, A. P. (2001) Genes Dev. 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- 2.McMahon, A. P., Ingham, P. W. & Tabin, C. J. (2003) Curr. Top. Dev. Biol. 53, 1–114. [DOI] [PubMed] [Google Scholar]
- 3.Aza-Blanc, P., Ramirez-Weber, F. A., Laget, M. P., Schwartz, C. & Kornberg, T. B. (1997) Cell 89, 1043–1053. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. H., von Kessler, D. P., Park, W., Wang, B., Ma, Y. & Beachy, P. A. (1999) Cell 98, 305–316. [DOI] [PubMed] [Google Scholar]
- 5.Wang, Q. T. & Holmgren, R. A. (2000) Development (Cambridge, U.K.) 127, 3131–3139. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., Gallaher, N., Goodman, R. H. & Smolik, S. M. (1998) Proc. Natl. Acad. Sci. USA 95, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., Cardinaux, J. R., Goodman, R. H. & Smolik, S. M. (1999) Development (Cambridge, U.K.) 126, 3607–3616. [DOI] [PubMed] [Google Scholar]
- 8.Price, M. A. & Kalderon, D. (1999) Development (Cambridge, U.K.) 126, 4331–4339. [DOI] [PubMed] [Google Scholar]
- 9.Price, M. A. & Kalderon, D. (2002) Cell 108, 823–835. [DOI] [PubMed] [Google Scholar]
- 10.Jia, J., Amanai, K., Wang, G., Tang, J., Wang, B. & Jiang, J. (2002) Nature 416, 548–552. [DOI] [PubMed] [Google Scholar]
- 11.Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. & Beachy, P. A. (2003) Science 299, 2039–2045. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, J. & Struhl, G. (1998) Nature 391, 493–496. [DOI] [PubMed] [Google Scholar]
- 13.Theodosiou, N. A., Zhang, S., Wang, W. Y. & Xu, T. (1998) Development (Cambridge, U.K.) 125, 3411–3416. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, S. Y., Chen, A., Xiong, Y., Pan, Z. Q. & Ronai, Z. (1999) Oncogene 18, 2039–2046. [DOI] [PubMed] [Google Scholar]
- 15.Hart, M., Concordet, J. P., Lassot, I., Albert, I., del los Santos, R., Durand, H., Perret, C., Rubinfeld, B., Margottin, F., Benarous, R. & Polakis, P. (1999) Curr. Biol. 9, 207–210. [DOI] [PubMed] [Google Scholar]
- 16.Latres, E., Chiaur, D. S. & Pagano, M. (1999) Oncogene 18, 849–854. [DOI] [PubMed] [Google Scholar]
- 17.Margottin, F., Benichou, S., Durand, H., Richard, V., Liu, L. X., Gomas, E. & Benarous, R. (1996) Virology 223, 381–386. [DOI] [PubMed] [Google Scholar]
- 18.Spencer, E., Jiang, J. & Chen, Z. J. (1999) Genes Dev. 13, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winston, J. T., Strack, P., Beer-Romero, P., Chu, C. Y., Elledge, S. J. & Harper, J. W. (1999) Genes Dev. 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., Andersen, J. S., Mann, M., Mercurio, F. & Ben-Neriah, Y. (1998) Nature 396, 590–594. [DOI] [PubMed] [Google Scholar]
- 21.Busino, L., Donzelli, M., Chiesa, M., Guardavaccaro, D., Ganoth, D., Dorrello, N. V., Hershko, A., Pagano, M. & Draetta, G. F. (2003) Nature 426, 87–91. [DOI] [PubMed] [Google Scholar]
- 22.Cardozo, T. & Pagano, M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Q. T. & Holmgren, R. A. (1999) Development (Cambridge, U.K.) 126, 5097–5106. [DOI] [PubMed] [Google Scholar]
- 24.Wang, B., Fallon, J. F. & Beachy, P. A. (2000) Cell 100, 423–434. [DOI] [PubMed] [Google Scholar]
- 25.Litingtung, Y., Dahn, R. D., Li, Y., Fallon, J. F. & Chiang, C. (2002) Nature 418, 979–983. [DOI] [PubMed] [Google Scholar]
- 26.Huangfu, D. & Anderson, K. V. (2005) Proc. Natl. Acad. Sci. USA 102, 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong, A. & Sun, S. C. (2002) J. Biol. Chem. 277, 22111–22114. [DOI] [PubMed] [Google Scholar]
- 28.Fan, C. M. & Maniatis, T. (1991) Nature 354, 395–398. [DOI] [PubMed] [Google Scholar]
- 29.Orian, A., Whiteside, S., Israel, A., Stancovski, I., Schwartz, A. L. & Ciechanover, A. (1995) J. Biol. Chem. 270, 21707–21714. [DOI] [PubMed] [Google Scholar]
- 30.Coux, O. & Goldberg, A. L. (1998) J. Biol. Chem. 273, 8820–8828. [DOI] [PubMed] [Google Scholar]
- 31.Heusch, M., Lin, L., Geleziunas, R. & Greene, W. C. (1999) Oncogene 18, 6201–6208. [DOI] [PubMed] [Google Scholar]
- 32.Palombella, V. J., Rando, O. J., Goldberg, A. L. & Maniatis, T. (1994) Cell 78, 773–785. [DOI] [PubMed] [Google Scholar]
- 33.Lin, L., DeMartino, G. N. & Greene, W. C. (1998) Cell 92, 819–828. [DOI] [PubMed] [Google Scholar]
- 34.Orellana, S. A. & McKnight, G. S. (1992) Proc. Natl. Acad. Sci. USA 89, 4726–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, N., Arai, H., Nishihara, Y., Taniguchi, M., Hunter, T. & Osada, H. (2004) Proc. Natl. Acad. Sci. USA 101, 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orian, A., Gonen, H., Bercovich, B., Fajerman, I., Eytan, E., Israel, A., Mercurio, F., Iwai, K., Schwartz, A. L. & Ciechanover, A. (2000) EMBO J. 19, 2580–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Methot, N. & Basler, K. (1999) Cell 96, 819–831. [DOI] [PubMed] [Google Scholar]
- 38.Kinzler, K. W. & Vogelstein, B. (1990) Mol. Cell. Biol. 10, 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerrigan, L. A. & Kadonaga, J. T. (1988) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. A., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 12.10.1–12.10.18.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.