Abstract
The prokaryotic β recombinase catalyzes site-specific recombination between two directly oriented minimal six sites in chromatin-integrated substrates. Here, we demonstrate that an enhanced green fluorescent protein (EGFP)-fused version of β recombinase (β-EGFP) is fully active, retaining most specific activity. It is used to develop a recombination-dependent activatable gene expression (RAGE) system based on the androgen receptor (AR) ligand-binding domain (LBD). Two hybrid molecules, a direct fusion of the LBD-AR to the C-terminus of β recombinase (β-AR) and a triple fusion of β-EGFP to the same ligand-binding domain (β-EGFP-AR), were engineered and their subcellular behavior, stability and catalytic activity were evaluated. Both chimeric β recombinase proteins showed in vivo inducible recombinogenic activity dependent on addition of an androgen receptor agonist, although the β-AR fusion protein demonstrated more accurate ligand-dependent translocation from cytoplasm to nucleus.
INTRODUCTION
Site-specific recombinases have become a standard tool for conditional gene modifications, as an alternative to classical gene targeting technologies (1–3). These systems allow programmed inter- and intramolecular recombination that overcome some limitations of classical knockout systems, such as embryonic lethality or generation of compensation mechanisms. Cre-loxP and Flp-FRT are currently the systems of choice, due to their ability to induce targeted changes in animal models and in plants (4). Using these systems, tissue-specific, conditional and inducible gene targeting events have been reported in a wide variety of tissues and organs (5–8); in addition, the Int-attP site-specific recombination (SSR) system from lambda phage recently proved to be successful in plants (9). There are few reports on the combined use of these systems (10); it is thus necessary to characterize other SSR systems that could be used as an alternative to or in combination with existing models.
β Recombinase from the Gram-positive plasmid pSM19035 induces specific intramolecular recombination in mammalian cells, in both episomal plasmids and chromatin-associated substrate structures (11,12). Plasmid pSM19035 has an unusual structure, as almost 80% of the molecule consists of a repeated sequence and two replication origins (13). Since replication of this plasmid follows the classical theta model, a mechanism must exist to ensure its complete replication (inversion) and maximization of plasmid segregation (resolution). Development of an in vitro recombination system based on purified β recombinase showed both inversion and deletion activities associated to the protein, delimited the sequences required for directing the SSR reactions and characterized the requirements for both reactions (13,14). Unlike Cre and Flp, which belong to the Int recombinase family, β recombinase is included in the resolvase/invertase family and catalyzes exclusively intramolecular recombination events (14–16). At difference from Cre and Flp SSR, which do not require additional factors [reviewed in 17,18), for deletion, β recombinase requires a supercoiled substrate and a chromatin-associated protein (e.g. bacterial Hbsu or eukaryotic HMG1 proteins) (16). The mammalian cell environment can provide such a host factor (12), and nuclear genomic DNA supercoiling seems to be sufficient for β recombinase function (11). The β recombinase-dependent SSR that uses the minimal defined recognition sequences (six) was denominated β-six (12). Owing to its specific characteristics, particularly to its exclusively intramolecular recombination capacity, the β-six SSR system was proposed as the ideal choice when several independently controlled recombination events are needed in the same animal or cell (12). This would prevent intermolecular recombination events between remaining recombination sites, which can occur when integrase family recombinases are used. Combined use of the β-six system with the Cre- and/or Flp-based SSR systems would expand current possibilities for programmed modification of eukaryotic genomes in complex spatio-temporal combinations.
Controlled induction of recombinogenic activity of SSR systems is a highly desirable feature. A first level of control is obtained by lineage or cell type selective expression of the recombinase activity. The second is achieved using an inducible promoter or by including a protein module in the recombinase activity that is able to maintain the recombinase in the cytoplasmic compartment; following interaction with inductor molecules, it promotes importation of the recombinase activity to the nuclear compartment [reviewed in (19)]. This latter method has been developed successfully in the Cre-loxP and Flp-FRT systems, using the ligand-binding domain (LBD) of the estrogen (ER), androgen (AR) or the progesterone (PR) nuclear receptors. The combined use of tissue- or cell type-specific promoters with fusion proteins of recombinase with nuclear receptor LBD allows spatio-temporal control of the targeted genomic modification (20).
Here, we explored the development of such an inducible SSR system using the β-six model and the androgen receptor LBD (AR-LBD). The AR is located primarily in cytoplasm; after binding to an AR agonist it translocates to the nucleus; after dimerization, it binds specific DNA regions and activates transcription of its target genes (21). The AR is composed of 917 amino acids, with the C-terminal portion forming the LBD, although functional regulatory interactions are described between the N- and C-termini (21). Modulation of FLP (22) and Cre (23) recombinogenic activity by fusion of the LBD-AR has been demonstrated. Our results indicate that β recombinase fused in its C-terminus to LBD-AR or a triple fusion of β recombinase in the same way could be controlled by addition of selective AR agonists.
MATERIALS AND METHODS
Plasmids
Plasmids pBT233, pRecombiner and pβ-EGFP have been described previously (12,24). Plasmid pEGFP-N1 (Clontech, Palo Alto, CA) and pTA-cloning vector (Promega, Madison, WI) were from commercial sources.
Generation of pLuc and psps-Luc
For recombination-activated gene expression, we designed and obtained a set of two vectors. The luciferase gene was obtained from the pRVSINloxPTP-Luc-IRES-ΔNGFR HindIII/XbaI and ligated in the same restriction sites to pCDNA3.1 Hygro. The resulting plasmid, p-Luc-Hygro, was digested with MluI/NheI to eliminate the CMV promoter, which was replaced with the EF-1α promoter from the pEF-4aMyc-His after MluI and SpeI digestion. We thus obtained the pLuc positive control for luciferase expression in our experiments. The recombination substrate was obtained by cloning the six-pur-six (sps) cassette from pRecombiner (12) in pLuc using HindIII. Resulting plasmids were purified for transfection using Qiagen columns (Qiagen, Hilden, Germany), following the manufacturer's protocols.
Retroviral vector construction
pLZR-ires-EGFP and pLZRβ-EGFP vector development has been described previously (12). To generate pLZR-β-IRES-EGFP, we introduced the β recombinase gene in the pLZR-ires-EGFP vector, and appropriate 5′ BamHI and 3′ EcoRI sites on the β gene from pBT233. Using primers beta(RES)-5 (5′-GAGAGAGGATCCATGGCTAAAATTGGT TATG-3′) and beta(RES)-3 (5′-GTGTGTGAATTCTTAACTATCCCTCTTTCC-3′), we performed PCR amplification with Taq Gold polymerase (Roche) using the supplier's standard conditions (one denaturation cycle at 94°C for 2 min, followed by 35 cycles at 94°C for 15 s; 59°C for 30 s; 72°C for 1 min and a final elongation cycle at 72°C for 5 min). The amplification product was ligated to BamHI/EcoRI-digested pLZR-ires-EGFP following the standard cloning procedures, resulting in pLZR-β-ires-EGFP.
To generate pLZR-β-AR(LBD)-ires-EGFP, the LBD-AR was obtained by PCR amplification of the pAdapt.hrtTA-VP16-AR-ires-Egfp plasmid (received from Dr K. Anastassiadis) with the primers SAR (5′-CAGCCGCGGATGCATGTGTCACACATTG AAGG-3′) and ARNN (5′-CAGGCGGCCGCAGGCGCCTCACTGGGTGTGGAAATAG ATGG-3′). The amplified product was purified and cloned using the pTA-Cloning vector system, to give the pTA-S/NN plasmid. This was digested with SacII and NotI and the resulting LBD-containing fragment cloned in the same pβ-EGFP restriction sites, resulting in the pβ-LBD vector containing the fusion open reading frame (ORF) in a correct translation phase. The BglII, PvuII and NarI fragment containing the β-LBD ORF from this vector was cloned in pLZR-ires-EGFP vector, digested with BamHI/SfuI, resulting in the pLZR-β-LBD-ires-EGFP retroviral vector.
For the pLZR-β-EGFP-AR(LBD), the LBD-AR was obtained by PCR amplification of the same parental plasmid with the primers BAR (5′-CAGAGATCTATGCATGTGTCACACA TTGAAGG-3′) and ARNH (5′-CAGAAGCTTGCGGCCGCTCACTGGGTGTGGAAATAG ATGG-3′). The amplification product was purified and cloned using the pTA-Cloning vector system, to give the pTA-B/NH plasmid. This was digested with BglII and HindIII, and the resulting LBD-containing fragment was cloned in the same restriction sites of pEGFP-N1 (Clontech), to give the pEGFP-LBD vector containing the fusion ORF in a correct translation phase. After AgeI/NotI digestion, we obtained the LBD linked to a 3′-EGFP fragment that was ligated to the 5′-EGFP fragment of pβ-EGFP, which had been digested as above. The resulting plasmid, pβ-EGFP-LBD, was XhoI/NotI-digested and ligated to pLZR-IRES-EGFP, giving rise to the pLZR-β-EGFP-LBD retroviral vector.
Plasmids pLZR-β-ires-EGFP, pLZR-β-AR(LBD)-ires-EGFP, pLZR-β-EGFP-AR(LBD) and pLZR-β-EGFP were purified for transduction experiments with Qiagen columns and further packaged in 293T cells as described previously (12).
Cell lines and culture
NIH-3T3 and 293T cell lines were obtained from the American Type Culture Collection (ATCC CRL-1658; Manassas, VA). All cell lines were cultured in DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Cultek, Madrid, Spain), 10 U/l penicillin, 10 µg/ml streptomycin and 2 mM l-glutamine (Merck, Darmstadt, Germany) (5% CO2, 37°C).
Cell transfection
To obtain stable transformed cell lines of the recombination psps-Luc construct and its control pLuc, plasmid DNA (20 µg) was introduced in NIH-3T3 cells (2 × 106) by electroporation at a concentration of 107 cells/ml in supplemented DMEM, pulsed at 220 V, 950 µF (Gene Pulser; Bio-Rad, Hercules, CA). Cells were replated and cultured (48 h), after which selection antibiotics (200 µg/ml hygromycin B, 2 µg/ml puromycin) were added to medium. Individual clones (sps-Luc), obtained by limiting dilution, were collected and transduced 20–25 days after electroporation and selection.
Retroviral transduction
pLZR-β-ires-EGFP, pLZR-β-EGFP, pLZR-β-AR(LBD)-ires-EGFP and pLZR-β-EGFP-AR(LBD) retroviral vectors were packaged transiently in 293T as described previously (12). Viral supernatants were titrated on NIH-3T3 cells on the basis of EGFP fluorescence and used to infect sps-Luc clones. Transduction was performed using 1 ml of viral supernatant (plus 8 µg/ml polybrene) added to the culture for 8 h, after which medium was renewed. After 48 h, genomic DNA was purified for PCR analysis using a miniprep purification kit (Qiagen)
Cell purification
Transduced EGFP-expressing cells were purified by cell sorting. Cells were trypsin-treated, centrifuged, resuspended in 1 ml of phosphate-buffered saline (PBS) and FACS purified using an EPICS Elite sorter (Coulter). Once isolated, each EGFP+ population was cultured without antibiotics, and 20–25 days later genomic DNA was purified by PCR for recombination analysis.
Isolation and analysis of subclones
Subclones of one sps-Luc clone infected with the pLZR-β-EGFP retroviral vector were obtained by limiting dilution, replated and their recombination rates analyzed for 7 weeks (samples were prepared after 5, 6 and 7 culture weeks). Cells were washed, trypsinized and collected; genomic DNA was purified as above and analyzed in PCR.
Androgen treatment
To determine the functionality of the inducible proteins, mibolerone (DuPont, Boston, MA) dissolved in absolute ethanol was added to the culture during exponential growth, in a concentration range from 10−6 to 10−8 M. Control cultures received similar amounts of ethanol alone; final ethanol concentrations did not exceed 0.01% (v/v). Luciferase assays were performed 24, 48, 72 h or 6 days post-induction. For 6-day inductions, culture medium with ligand was replaced after 72 h.
Analysis of recombination products
Transduced and non-transduced pools, isolated clones and subclones were harvested from 6-well plates when they reached confluence. Genomic DNA was obtained by the Easy-DNA method (Invitrogen) and 50 ng were used for recombined substrate detection by PCR in a Perkin Elmer thermocycler. We used the Expand Long Template PCR system (Roche) as follows: 93°C for 2 min; 35 cycles of 94°C for 10 s; 49°C for 30 s; 68°C for 90 s and final elongation at 68°C for 7 min. Primers for PCR detection were EF-1α-s (5′-GCACTTGA TGTAATTCTCC-3′) and lucif-as (5′-GTAAGTGATGTCCACCTCG-3′). The PCR protocol was optimized for amplification not only of the recombined product (0.6 kb band), but also of the non-recombined substrate (1.7 kb band), which permitted us to measure the relative recombination rates between samples and to quantify the recombination event.
Immunoblot
For immunoblot analysis, total proteins were extracted from pLZRβ-ires-EGFP, pLZRβ-EGFP, pLZRβ-AR(LBD)-ires-EGFP and pLZRβ-EGFP-AR(LBD)-transduced cells, separated by SDS–PAGE and transferred to polyvinylidene fluoride membranes (Immunobilon-P, Millipore, Bedford, MA) using a semidry blotting apparatus (Transblot-SD, Bio-Rad). Antibodies used were anti-EGFP (mouse mAb 8362-1, 1:3000 dilution; Clontech, Palo Alto, CA), anti-β recombinase [rabbit pAb, 1:500 (11)], anti-AR(LBD) (rabbit pAb sc-815, 1:1000: Santa Cruz, Santa Cruz, CA), anti-tubulin (mouse mAb T-9026, 1:500; Sigma, St Louis, MO) and anti-histone H1 (mouse mAb sc-8030, 1:1000; Santa Cruz). After immunobloting with secondary antibody, proteins were visualized by chemiluminescent detection (ECL; Amersham Pharmacia Biotech, Bucks., UK).
Confocal laser scanning
Transduced cells (103) were plated on a 35 mm glass coverslip in 6-well plates and cultured for 24 h (when assaying inducible recombination activity, ligand was added at this time). Cells were fixed (10 min) with 2% paraformaldehyde and 0.05% Triton X-100 in PBS, and stained with rabbit anti-β recombinase [1:500 (11)] and anti-AR(LBD) (SC-815, 1:1000; Santa Cruz Biotechnology). Secondary antibody was Cy3-labeled goat anti-rabbit IgG (H+L) (111-166-008, 1:400; Amersham). For Hoechst staining, cells were incubated with 0.8 µg/ml Hoechst 33258 (Molecular Probes, Eugene, OR) for 5 min, immediately after second antibody incubation. Cells were mounted in Vectashield (Vector, Burlingame, CA) and observed with a 63 × 1.4 oil-immersion lens. Images were collected by confocal laser microscopy (Leica TCSNT), noise-filtered, corrected for background and processed using Adobe Photoshop.
Luciferase assay
Infected cells were seeded into 6-well plates and harvested at confluence. Proteins were extracted and analyzed using the Luciferase reporter assay (Promega) according to the manufacturer's protocols. Cells were washed in 1× PBS and 250 µl lysis reagent was added per well. Lysate was collected after 10 min and cleared by centrifugation (13000 r.p.m., 15 min). Luciferase assays were performed using 20 µl of lysate and a luminometer (Berthold Technologies, Vienna, Austria). Total protein was quantified to normalize results. Data shown are representative of at least three independent experiments.
RESULTS
C-terminal fusion proteins of β recombinase maintain full activity
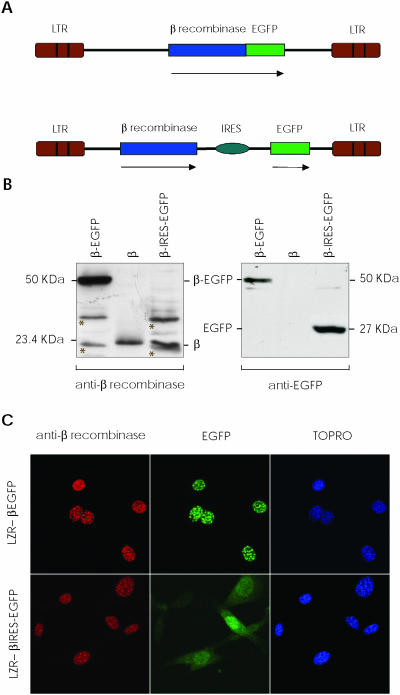
Two different vectors expressing β recombinase were constructed using pLZR (12) as retroviral backbone. One expressed a direct fusion protein between the β recombinase C-terminus and the EGFP N-terminus; the other is a bicistronic vector that expresses both proteins as individual ORFs separated by the encephalomyocarditis virus RNA internal ribosome entry site (IRES) (Figure 1A). These plasmids were used to obtain high-titer supernatants by transient transfection in 293T cells (see Materials and Methods). To verify the β recombinase expression pattern using integrative retroviral vectors context, we transduced NIH-3T3 cells with both vectors. About 2–7 days later, proteins were extracted from transduced cells and used to detect β recombinase and EGFP by immunoblotting (Figure 1B). The autoradiograph shows 23.4 and 27 kDa bands that correspond to β recombinase and EGFP, respectively, with a 50 kDa band representing the β-EGFP fusion protein.
Figure 1.
β Recombinase expression using retroviral vectors. (A) Scheme of LZR-β-EGFP and LZR-β-ires-EGFP retroviral vectors used. Expression of a direct fusion protein between β recombinase and EGFP (upper); independent expression of both proteins using an IRES (lower). Arrows indicate the transcribed sequence. (B) NIH-3T3 cells were transduced with both vectors; after 48 h, total proteins were extracted. Western blot autoradiograph shows expression of the fusion protein (β-EGFP, 50 kDa, left lane), individual proteins (β, 23.4 kDa; EGFP, 27 kDa, right) and 5 ng of purified β recombinase (center). Non-specific bands are marked with an asterisk on the membranes. (C) Immunofluorescence of NIH-3T3 cells transduced with LZR-β-EGFP (upper) and LZR-β-ires-EGFP (lower), showing nuclear localization of β recombinase, fused or alone. EGFP is detected in the same nuclear dots when LZR-β-EGFP is used, but not when β recombinase and EGFP are expressed independently (LZR-β-ires-EGFP). TOPRO stains nuclei.
Similar results were obtained when expression in transduced cell was tested in immunofluorescence with anti-β recombinase antibody and EGFP autofluorescence (Figure 1C). Fluorescence microscopy of β-EGFP-expressing cells showed exclusively nuclear localization of both proteins, indicating that spontaneous subcellular localization of β recombinase is not modified by fusion or the retroviral dependent-context. A diffuse distribution pattern was detected for EGFP in β-ires-EGFP cells, whereas β recombinase maintained its nuclear localization. These results concurred with previous data for β-EGFP episomal expression plasmids (12), demonstrating the feasibility of obtaining high, stable, uniform β recombinase expression, as an independent or a fusion protein in integrative retroviral vectors.
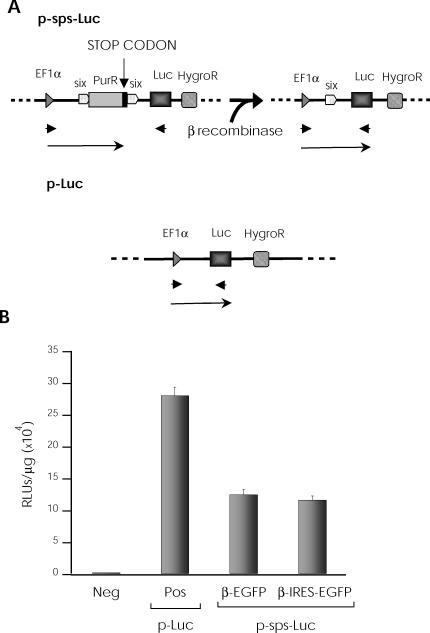
Experiments were developed to validate the potential of these vectors in regulated expression systems. The psps-Luc plasmid, in which gene expression is driven by the EF-1α promoter, was developed as a recombination target (Figure 2A). We inserted a puromycin resistance gene between two directed six sites; the luciferase gene was located downstream of the recombinant cassette, followed by a hygromycin resistance gene. In this construct, luciferase expression is impaired by the upstream puromycin stop codon; following β recombinase expression, one six site and the entire puromycin gene are deleted, including the stop codon; reporter gene (luciferase) expression is thus allowed (Figure 2A). As a positive control, we used the same plasmid without the puromycin gene or the six sites (pLuc).
Figure 2.
Retroviral vector potential in regulated expression systems. (A) Scheme for target plasmids used. psps-Luc plasmid, in which a puromycin gene is located between two directed six sites under the control of an EF-1α promoter (upper). A luciferase gene is inserted downstream, followed by a hygromycin resistance gene. β Recombinase expression causes deletion of the puromycin gene including its stop codon and one six site, allowing luciferase reporter gene expression. Positive control plasmid (pLuc) with direct luciferase expression, under the same promoter (lower). Arrows indicate transcription units and arrowheads indicate specific primers used to detect recombination. (B) NIH-3T3 cells were electroporated with psps-Luc and pLuc, and hygromycin-resistant cells were selected. psps-Luc NIH-3T3-resistant cells were transduced with β-EGFP or β-ires-EGFP retroviral vectors; luciferase expression was measured after 48 h. The negative sample (neg) involves transduction of psps-Luc NIH-3T3-resistant cells with a control vector.
NIH-3T3 cells were electroporated with these plasmids (psps-Luc and pLuc) and hygromycin-resistant cells were selected. The pool of resistant cells was transduced with the β-ires-EGFP or β-EGFP retroviral vectors and luciferase expression was measured after 48 h. We detected no luciferase activity in the absence of β recombinase expression, but luciferase expression levels were greatly increased in its presence, alone or as a fusion protein (Figure 2B). Equivalent recombination rates were obtained with both retroviral vectors, indicating that the β-EGFP fusion protein retains most of the catalytic activity shown by native β recombinase. This result enables the design of novel β recombinase fusion proteins using the C-terminus as the fusion point. All further experiments were performed with the β-EGFP fusion protein.
β recombinase-mediated RAGE is genomic context-dependent
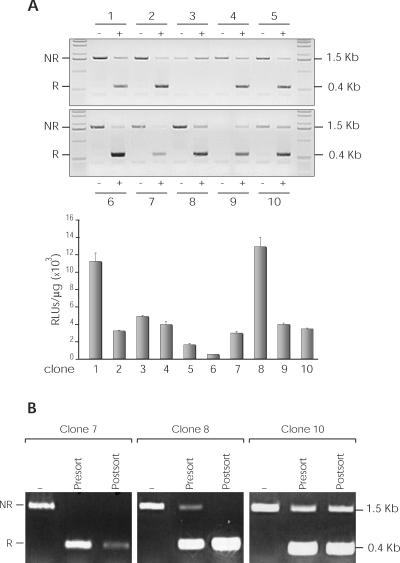
To determine whether the genome integration sites of responder plasmids influenced recombination effectiveness, we obtained stable puromycin resistant psps-Luc-transfected NIH-3T3 clones, each of which was transduced with the β-EGFP retroviral vector; SSR was analyzed by PCR and luciferase expression. Genomic DNA was isolated from 10 clones and amplified with specific primers of the EF-1α promoter and the luciferase gene (Figure 2A), which allowed us to estimate the relative recombination rates (Figure 3A, upper panel). Ninety percent of clones showed substantial recombination of the substrate structure; the recombination level was also determined by quantification of luciferase activity (Figure 3A, lower panel). To determine whether these results were affected by differences in the percentage of β-EGFP transduction, we purified the EGFP+ population in three clones (C7, C8 and C10) by cell sorting and analyzed recombination by PCR (Figure 3B). This analysis showed an improved net recombination in C7 and C8; C10 showed roughly similar proportions for recombined and unrecombined pools, indicating partial inhibition of β recombinase activity.
Figure 3.
Analysis of recombination in psps-Luc-transduced NIH-3T3 clones. Ten clones (1–10) were isolated and SSR analyzed by PCR and luciferase expression. (A) Genomic DNA of each clone (−, untransduced cells; +, transduced cells) was amplified using specific primers of the EF-1α promoter and luciferase gene (see Figure 2A). Non-recombined DNA (NR; 1.5 kb band), recombined DNA (R; 0.4 kb band) (upper panel). The recombination level was determined by luciferase activity quantification (lower panel). (B) EGFP-positive subpopulations were purified by cell sorting of three different clones (7, 8 and 10). Recombination level in untransduced (−) and transduced (pre-sorting and post-sorting) cells was analyzed by PCR.
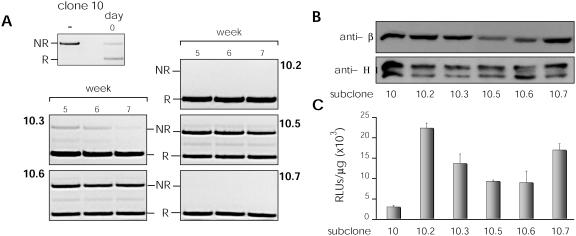
To analyze whether the results for C10 were due to chromatin inaccessibility, insufficient recombinase expression, or a reversible recombination process (absent in in vitro experiments), we studied β-EGFP-transduced C10 subclones. Several C10 subclones were obtained by limiting dilution and their recombination rates analyzed for 7 weeks (samples were prepared after 5, 6 and 7 culture weeks). Some subclones showed complete recombination from the start, whereas for others, recombination rates improved with time (Figure 4A). These differences appeared to be related to β recombinase levels, as western blot analysis showed reduced expression in the less efficient clones (Figure 4B); β recombinase expression correlated with molecular recombination rates as well as with relative luciferase activity (Figure 4C).
Figure 4.
Analysis of recombination time-course. Several C10 subclones were obtained and their recombination rates analyzed over a 7-week period. The recombination rate obtained for five of them (10.2, 10.3, 10.5, 10.6 and 10.7) was analyzed by several techniques. (A) Non-recombined (NR) and recombined (R) DNA fragments amplified by PCR. (B) Detection of β recombinase expression and histone H1, as control, by western blot. (C) Luciferase activity. Western blot and luciferase activity were measured in the fifth week.
β recombinase-mediated RAGE regulation using inducible systems
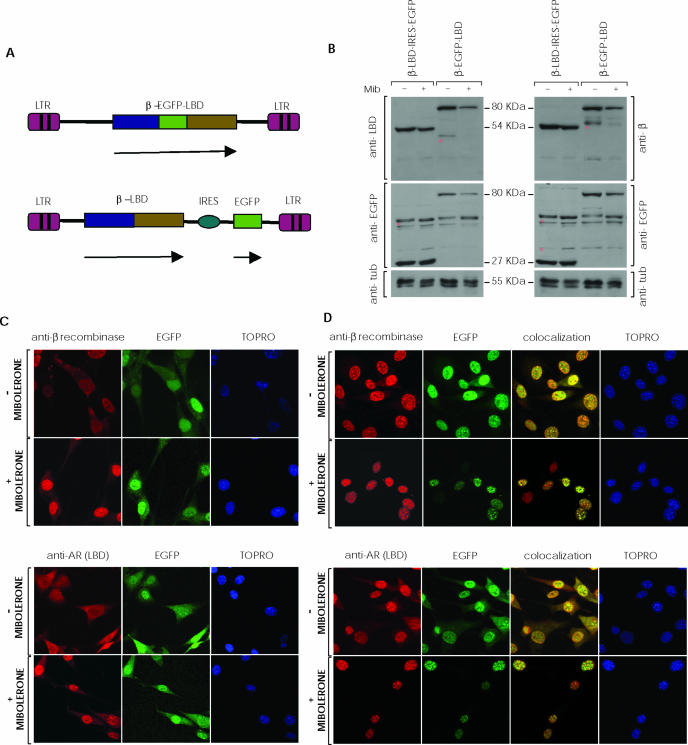
The previous result showed that β-EGFP retains full enzymatic activity; we thus used this strategy to test β recombinase potential in an AR-based inducible system. We constructed two retroviral vectors containing β recombinase and the AR-LBD as fusion proteins (Figure 5A). In the first, we generated a bicistronic vector that fused β recombinase to the LBD (β-AR), permitting independent IRES-driven EGFP expression; in the second, we fused β recombinase to EGFP and LBD in a triple fusion protein (β-EGFP-AR). As recombination substrate, we used the previously tested clones (C7, C8 and C10).
Figure 5.
SSR using an inducible system based on fusion of β recombinase and the androgen receptor. (A) Scheme showing the retroviral vectors used in the inducible system. LZR-β-EGFP-(AR)LBD vector expresses the triple fusion protein (β-EGFP-LBD) of β recombinase, EGFP and the androgen receptor LBD (upper); the lower panel shows LZR-β-(AR)LBD-ires-EGFP vector which allows independent expression of the β-LBD fusion protein and EGFP by an IRES. Arrows indicate transcription units. (B) C7, C8 and C10 clones were transduced with the retroviral vectors and the EGFP-positive subpopulation selected by cell sorting. Western blot shows expression of β-EGFP-LBD (80 kDa), β-LBD (54 kDa), EGFP (27 kDa), and tubulin (control, 55 kDa) in transduced cells, untreated (−) or treated with (+) mibolerone (Mib; 10−8 M, 48 h). Non-specific bands are marked with an asterisk on the membranes. (C) Immunocytochemistry of β-LBD-ires-EGFP-expressing cells, alone or mibolerone-treated. (D) Immunocytochemistry of β-EGFP-LBD-transduced cells, alone or with mibolerone. Only data obtained in clone C8 have been included in (B–D). Clones C7 and C10 rendered quite similar results.
Based on previous results using the androgen receptor LBD (AR-LBD) to control FLP and Cre recombinase enzyme activity (22,23), LBD inclusion would retain the β recombinase fusion proteins in cytoplasm, impeding access to their genomic target recombination sequences. Following ligand administration (mibolerone), AR-LBD would prevent further cytoplasmic interactions; β-AR and β-EGFP-AR would subsequently be able to translocate to the nucleus via the β recombinase nuclear localization signal. Once in the nucleus, the β recombinase domain would gain access to its targets.
C7, C8 and C10 cells were transduced with both retroviral vectors and selected by cell sorting. Luciferase activity was analyzed in the resulting EGFP+ fraction, for different mibolerone concentrations and induction times.
To quantify induced and uninduced fusion protein levels, we developed a western blot using antibodies to β recombinase, EGFP and AR-LBD. The bands for mibolerone-treated cells showed a lesser intensity than those of controls. EGFP levels were similar in cells transduced with bicistronic vectors, in which EGFP is expressed independently (Figure 5B). To confirm this effect and protein loading equivalence, blots were stained with anti-histone1 and anti-tubulin control antibodies; intensity of all samples was measured and normalized with controls. We calculated a 30 and 50% decrease in fusion protein expression in mibolerone-treated cells for double- and triple-fusion proteins, respectively.
In addition, we studied fusion protein subcellular localization by immunocytochemistry, before and after mibolerone treatment. Transduced cells were fixed and stained with anti-β recombinase or anti-LBD antibody; EGFP was visualized directly in a fluorescence microscope. As predicted, the EGFP signal was throughout-diffused in cells transduced with the bicistronic vector; the fusion protein localized to cytoplasm before and primarily to the nucleus after mibolerone treatment (Figure 5C). Following mibolerone treatment, the triple fusion protein localized exclusively in the nucleus; in untreated cells, this protein was found in the cytoplasm as well as in nuclear aggregates (Figure 5D).
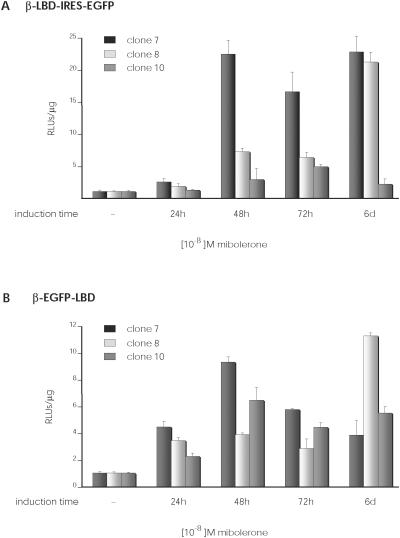
In both groups of cells, mibolerone treatment upregulated luciferase expression; the treated triple fusion protein cells had the highest luciferase levels, but also their untreated control cells showed more background values than untreated double fusion protein cells (data not shown). In this regard, normalized luciferase activity in mibolerone-treated cells was higher in cells transduced with the double fusion protein than with the triple fusion protein, indicating better regulation of the β recombinase-dependent RAGE system (Figure 6).
Figure 6.
Analysis of luciferase expression using the inducible system. C7, C8 and C10 clones were transduced with these retroviral vectors and the EGFP-positive subpopulation selected by cell sorting. Graphs show luciferase expression in function of induction time in culture with 10−8 M mibolerone. (A) Luciferase expression of β-LBD-ires-EGFP-transduced clones. (B) Luciferase expression of β-EGFP-LBD transduced clones.
DISCUSSION
Site-specific recombinases have been established as powerful tools for gene manipulation and the generation of genetically modified living organisms (3). Two classes of these enzymes have been studied in detail, both of which store the energy of a broken DNA phosphodiester bond in covalent protein–DNA intermediates; depending on the choice of nucleophile, they are referred to as serine or tyrosine recombinases [reviewed in (25)].
The most widely used SSR models are the Cre-loxP system derived from bacteriophage P1, and Flp-FRT from the yeast Saccharomyces cerevisae. Both have demonstrated versatility for strict spatio-temporal control of the genomic modification induced, eliminating the consequences of undesired effects due to systemic lack of an essential gene (26,27) or bypass effects due to unforeseeable redundancy that produces no detectable phenotypic changes (28,29).
Cre and Flp belong to one of the major families of site-specific recombinases, the tyrosine recombinases. These enzymes cleave, exchange and rejoin one pair of DNA strands, generating a Holliday junction as a recombination intermediate before initiating these reactions on the other pair of DNA strands. The reaction output (integration, deletion or inversion) depends on the existence of recombination sites in the same or in distinct molecules and their relative orientation [reviewed in (25,30)].
The other major SSR family is that of the serine recombinases, which comprises two subfamilies; the small recombinases (<250 amino acids), which catalyze mainly intramolecular recombinations, and the large recombinases (>450 amino acids), which catalyze both inter- and intramolecular reactions [reviewed in (30)]. The small recombinases can again be subdivided into three groups, which catalyze resolution (resolvases such as γδ and Tn3), inversion (invertases such as Hin and Gin) or both (resolvase-invertases such as β and Sin). All catalyze recombination via a concerted four-strand cleavage and rejoining mechanism [reviewed in (30)], best explained by a recombinase subunit rotation model during DNA synapsis [reviewed in (25)]. Resolvase-invertases are peculiar, as they do not have the same high selectivity as resolvases and invertases for resolution or inversion [reviewed in (30)]; for example, β recombinase can catalyze resolution between two directly oriented recombination sites, and both resolution and inversion between two inversely oriented sites (31). It was proposed that the distinctive properties of β recombinase are more likely due to a degree of flexibility in the structure and assembly of the synaptic complex, than to any difference in the strand exchange mechanism (30).
We recently described the potential application of a new SSR system based on β recombinase for manipulation of the mammalian genome (11,12). In contrast to Cre-loxP and Flp-FRT systems, the in vitro requirements of the β-six model include a host protein factor and a supercoiled substrate to catalyze resolution reactions (32); those requirements seem to be provided by the mammalian chromatin environment (11,12).
The versatility of SSR systems is based on their potential to be controlled in an inducible manner, which was shown to be feasible and efficient for the Cre-loxP and Flp-FRT systems (33–35) using fusions of recombinase activity to receptor LBD in several hormone systems. Studies also indicated that EGFP fusion to the β recombinase C-terminus does not abolish its recombinogenic capacity, opening the way for evaluation of an inducible β-six model (12).
Using improved retroviral expression vectors, we show here that the specific catalytic activity of the β-EGFP protein is fully retained and is comparable with native β recombinase in eukaryotic environments. We thus used this fusion scheme to evaluate a potential β-six-dependent inducible recombination system (β-SSR). To test the influence of the chromosomal integration environment on the effectiveness of the β-SSR system, we analyzed several parameters and characterized the responder clones (NIH-3T3-sps-Luc). We found that 90% of the integration sites are accessible to the β recombinase activity, although there were notable differences in efficiency. We obtained several subclones of a selected cloned cell line, which harbored the same recombination substrate but different β recombinase expression levels due to random retroviral expression vector integration. This allowed us to analyze the influence of β recombinase expression levels on the recombination rate, independently of substrate positional constraints. We found strong correlation between recombination effectiveness and β recombinase expression, suggesting that an undetermined minimal amount of β recombinase is necessary for SSR, probably in a genomic integration site-dependent manner.
The β-SSR was obtained by replacing the EGFP module of β-EGFP protein with the human AR-LBD (β-AR) or by adding AR-LBD to the β-EGFP protein (β-EGFP-AR). Based on earlier models (36,37), these two fusion proteins would remain inactive in cytoplasm until stimulated by a receptor agonist (mibolerone) that promotes nuclear translocation and recombination of the responder substrate.
In the β-AR-expressing cells, β-AR accumulated in cytoplasm, with partial translocation to the nucleus only after mibolerone treatment. Mibolerone treatment of β-EGFP-AR-expressing cells promoted complete translocation to the nucleus. When these cells were not treated, however, the β-EGFP-AR fusion protein was distributed throughout the cell, with nuclear foci, anticipating a probable higher basal background. We confirmed the expression levels of both fusion proteins in untreated and mibolerone-treated cells in western blot, using anti-β recombinase, anti-EGFP and anti-LBD antibodies. Total fusion protein decreased after mibolerone administration (30 and 50% decrease in β-AR and β-EGFP-AR expression, respectively); control anti-EGFP antibody in β-AR-expressing cells showed no significant decrease. The data thus strongly suggest that the presence of the AR-LBD appears to decrease fusion protein half-life after mibolerone treatment. Previous studies indicated that the AR N-terminal region is necessary for receptor stabilization (38). Stabilization is agonist-induced and prevents ubiquitin-mediated degradation by the proteosome. Deletion or mutation of this region accelerates normal degradation when the AR is bound to the agonist (39). Our data using an N-terminal-deficient AR-LBD concur with these results.
Evaluation of the inducible luciferase reporter levels corroborated that it increased after mibolerone treatment of transduced cells expressing both fusions. Following induction, absolute luciferase levels were higher for cells expressing β-EGFP-AR protein than β-AR, although untreated β-EGFP-AR control cells also showed a higher luciferase background. The diminished protein amount in treated triple fusion protein cells, in addition to its higher background in control cells, could explain the lower up-regulation shown for this fusion in data from Figure 6.
These functional results concur with the subcellular localization described for these fusion proteins. The literature shows contradictory findings on basal AR localization, depending on the cell line and immunocytochemical procedure used, expression level and the fusion partner (40–42); probably the same undefined factors could be affecting our experimental design explaining, at least in part, the higher luciferase background founded in untreated cells.
These results demonstrate the feasibility of combining the requirements for an accurate inducible system with the SSR mediated by this β recombinase. Further work would provide a well-regulated system for controlled deletion of selected target sequences, which could be used in combination with Cre-loxP and Flp/FRT, the systems of choice for conditional targeting.
Acknowledgments
The authors would like to thank Catherine Mark for editorial assistance. This work was partially supported by the Spanish Ministry of Science and Technology, CICYT, (SAF2001-2262 and GEN2001-4856-C13-02) to A.B. V.D. and J.G.-C. received a fellowship from the Spanish Ministry of Science and Technology, and M.A.G. was fellow of the Comunidad Autónoma de Madrid. The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and by Pfizer. Fund-ing to pay the Open Access publication charges for this article was provided by Spanish Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Metzger D., Feil R. Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol. 1999;10:470–476. doi: 10.1016/s0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 2.Sauer B. Site-specific recombination: developments and applications. Curr. Opin. Biotechnol. 1994;5:521–527. doi: 10.1016/0958-1669(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 3.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 4.Rossant J., McMahon A. ‘Cre’-ating mouse mutants- a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 5.Agah R., Frenkel P.A., French B.A., Michael L.H., Overbeek P.A., Schneider M.D. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow C., Schroeder M., Lekstrom-Himes J., Kylefjord H., Deng C.X., Wynshaw-Boris A., Spiegelman B.M., Xanthopoulos K.G. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 1997;25:2543–2545. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang B.H., Liao W., Li L., Nakamuta M., Mack D., Chan L. Liver-specific inactivation of the abetalipoproteinemia gene completely abrogates very low density lipoprotein/low density lipoprotein production in a viable conditional knockout mouse. J. Biol. Chem. 1999;274:6051–6055. doi: 10.1074/jbc.274.10.6051. [DOI] [PubMed] [Google Scholar]
- 8.Utomo A.R., Nikitin A.Y., Lee W.H. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat. Biotechnol. 1999;17:1091–1096. doi: 10.1038/15073. [DOI] [PubMed] [Google Scholar]
- 9.Zubko E., Scutt C., Meyer P. Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol. 2000;18:442–445. doi: 10.1038/74515. [DOI] [PubMed] [Google Scholar]
- 10.Meyers E.N., Lewandoski M., Martin G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nature Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 11.Diaz V., Rojo F., Martinez A.C., Alonso J.C., Bernad A. The prokaryotic beta-recombinase catalyzes site-specific recombination in mammalian cells. J. Biol. Chem. 1999;274:6634–6640. doi: 10.1074/jbc.274.10.6634. [DOI] [PubMed] [Google Scholar]
- 12.Diaz V., Servert P., Prieto I., Gonzalez M.A., Martinez-A C., Alonso J.C., Bernad A. New insights into host factor requirements for prokaryotic beta-recombinase-mediated reactions in mammalian cells. J. Biol. Chem. 2001;276:16257–16264. doi: 10.1074/jbc.M011725200. [DOI] [PubMed] [Google Scholar]
- 13.Rojo F., Weise F., Alonso J.C. Purification of the beta product encoded by the Streptococcus pyogenes plasmid pSM19035. A putative DNA recombinase required to resolve plasmid oligomers. FEBS Lett. 1993;328:169–173. doi: 10.1016/0014-5793(93)80987-6. [DOI] [PubMed] [Google Scholar]
- 14.Rojo F., Alonso J.C. A novel site-specific recombinase encoded by the Streptococcus pyogenes plasmid pSM19035. J. Mol. Biol. 1994;238:159–172. doi: 10.1006/jmbi.1994.1278. [DOI] [PubMed] [Google Scholar]
- 15.Rojo F., Alonso J.C. The beta recombinase of plasmid pSM19035 binds to two adjacent sites, making different contacts at each of them. Nucleic Acids Res. 1995;23:3181–3188. doi: 10.1093/nar/23.16.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso J.C., Weise F., Rojo F. The Bacillus subtilis histone-like protein Hbsu is required for DNA resolution and DNA inversion mediated by the beta recombinase of plasmid pSM19035. J. Biol. Chem. 1995;270:2938–2945. doi: 10.1074/jbc.270.7.2938. [DOI] [PubMed] [Google Scholar]
- 17.Kilby N.J., Snaith M.R., Murray J.A. Site-specific recombinases: tools for genome engineering. Trends Genet. 1993;9:413–421. doi: 10.1016/0168-9525(93)90104-p. [DOI] [PubMed] [Google Scholar]
- 18.Landy A. Mechanistic and structural complexity in the site-specific recombination pathways of Int and FLP. Curr. Opin. Genet. Dev. 1993;3:699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- 19.Lewandoski M. Conditional control of gene expression in the mouse. Nature Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 20.Tronche F., Casanova E., Turiault M., Sahly I., Kellendonk C. When reverse genetics meets physiology: the use of site-specific recombinases in mice. FEBS Lett. 2002;529:116–121. doi: 10.1016/s0014-5793(02)03266-0. [DOI] [PubMed] [Google Scholar]
- 21.Wong C.I., Zhou Z.X., Sar M., Wilson E.M. Steroid requirement for androgen receptor dimerization and DNA binding. Modulation by intramolecular interactions between the NH2-terminal and steroid-binding domains. J. Biol. Chem. 1993;268:19004–19012. [PubMed] [Google Scholar]
- 22.Logie C., Stewart A.F. Ligand-regulated site-specific recombination. Proc. Natl Acad. Sci. USA. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaczmarczyk S.J., Green J.E. Induction of cre recombinase activity using modified androgen receptor ligand binding domains: a sensitive assay for ligand-receptor interactions. Nucleic Acids Res. 2003;31:86. doi: 10.1093/nar/gng087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canosa I., Rojo F., Alonso J.C. Site-specific recombination by the beta protein from the streptococcal plasmid pSM19035: minimal recombination sequences and crossing over site. Nucleic Acids Res. 1996;24:2712–2717. doi: 10.1093/nar/24.14.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice P.A. Resolving integral questions in site-specific recombination. Nature Struct. Mol. Biol. 2005;12:641–643. doi: 10.1038/nsmb0805-641. [DOI] [PubMed] [Google Scholar]
- 26.Gu H., Marth J.D., Orban P.C., Mossmann H., Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 27.Ioffe E., Stanley P. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl Acad. Sci. USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schorle H., Holtschke T., Hunig T., Schimpl A., Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature. 1991;352:621–624. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn R., Rajewsky K., Muller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 30.Canosa I., Lopez G., Rojo F., Boocock M.R., Alonso J.C. Synapsis and strand exchange in the resolution and DNA inversion reactions catalysed by the beta recombinase. Nucleic Acids Res. 2003;31:1038–1044. doi: 10.1093/nar/gkg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canosa I., Lurz R., Rojo F., Alonso J.C. Beta Recombinase catalyzes inversion and resolution between two inversely oriented six sites on a supercoiled DNA substrate and only inversion on relaxed or linear substrates. J. Biol. Chem. 1998;273:13886–13891. doi: 10.1074/jbc.273.22.13886. [DOI] [PubMed] [Google Scholar]
- 32.Alonso J.C., Gutierrez C., Rojo F. The role of chromatin-associated protein Hbsu in beta-mediated DNA recombination is to facilitate the joining of distant recombination sites. Mol. Microbiol. 1995;18:471–478. doi: 10.1111/j.1365-2958.1995.mmi_18030471.x. [DOI] [PubMed] [Google Scholar]
- 33.Brocard J., Warot X., Wendling O., Messaddeq N., Vonesch J.L., Chambon P., Metzger D. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl Acad. Sci. USA. 1997;94:14559–14563. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brocard J., Feil R., Chambon P., Metzger D. A chimeric Cre recombinase inducible by synthetic,but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 1998;26:4086–4090. doi: 10.1093/nar/26.17.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D., Chambon P. Ligand-activated site-specific recombination in mice. Proc. Natl Acad. Sci. USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anastassiadis K., Kim J., Daigle N., Sprengel R., Scholer H.R., Stewart A.F. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene. 2002;298:159–172. doi: 10.1016/s0378-1119(02)00979-4. [DOI] [PubMed] [Google Scholar]
- 37.Hartig P.C., Bobseine K.L., Britt B.H., Cardon M.C., Lambright C.R., Wilson V.S., Gray L.E., Jr Development of two androgen receptor assays using adenoviral transduction of MMTV-luc reporter and/or hAR for endocrine screening. Toxicol. Sci. 2002;66:82–90. doi: 10.1093/toxsci/66.1.82. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S., Liang X., Danielsen M. Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Mol. Endocrinol. 1996;10:24–34. doi: 10.1210/mend.10.1.8838142. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z.X., Lane M.V., Kemppainen J.A., French F.S., Wilson E.M. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol. Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 40.Jenster G., Trapman J., Brinkmann A.O. Nuclear import of the human androgen receptor. Biochem. J. 1993;293:761–768. doi: 10.1042/bj2930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao T., McPhaul M.J. Functional activities of the A and B forms of the human androgen receptor in response to androgen receptor agonists and antagonists. Mol. Endocrinol. 1998;12:654–663. doi: 10.1210/mend.12.5.0112. [DOI] [PubMed] [Google Scholar]
- 42.Poukka H., Karvonen U., Yoshikawa N., Tanaka H., Palvimo J.J., Jänne O.A. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J. Cell Sci. 2000;113:2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]