Abstract
Background
Members of the M2 family of peptidases, related to mammalian angiotensin converting enzyme (ACE), play important roles in regulating a number of physiological processes. As more invertebrate genomes are sequenced, there is increasing evidence of a variety of M2 peptidase genes, even within a single species. The function of these ACE-like proteins is largely unknown. Sequencing of the A. gambiae genome has revealed a number of ACE-like genes but probable errors in the Ensembl annotation have left the number of ACE-like genes, and their structure, unclear.
Results
TBLASTN and sequence analysis of cDNAs revealed that the A. gambiae genome contains nine genes (AnoACE genes) which code for proteins with similarity to mammalian ACE. Eight of these genes code for putative single domain enzymes similar to other insect ACEs described so far. AnoACE9, however, has several features in common with mammalian somatic ACE such as a two domain structure and a hydrophobic C terminus. Four of the AnoACE genes (2, 3, 7 and 9) were shown to be expressed at a variety of developmental stages. Expression of AnoACE3, AnoACE7 and AnoACE9 is induced by a blood meal, with AnoACE7 showing the largest (approximately 10-fold) induction.
Conclusion
Genes coding for two-domain ACEs have arisen several times during the course of evolution suggesting a common selective advantage to having an ACE with two active-sites in tandem in a single protein. AnoACE7 belongs to a sub-group of insect ACEs which are likely to be membrane-bound and which have an unusual, conserved gene structure.
Background
In mammals, angiotensin-converting enzyme (EC 3.4.15.1, ACE, peptidyl-dipeptidase A), an important member of the M2 peptidase family, is found on the surface of endothelial cells and is best known for its role in the biosynthesis of angiotensin II, as well as the degradation of circulating bradykinin and the haemoregulatory peptide, N-acetyl SDKP [1-3]. Mammalian ACE exists as two isoforms, a somatic form (sACE, 150–180 kDa) and a smaller protein (germinal ACE, 90–110 kDa) found exclusively in adult testes. The ACE gene comprises twenty-six exons and two promoters, and has clearly arisen by gene duplication [4]. The somatic promoter drives expression of the larger protein (exons 1–12 and exons 14–26), which consists of two very similar domains connected in tandem by a short inter-domain peptide. Each domain possesses a functional peptidase active site and they are commonly called the N- and C-domains after their relative position to the amino and carboxy termini. The bulk of the protein including the amino terminus is extracellular and is linked by a hydrophobic transmembrane sequence to a short intracellular peptide. The biological rationale for two catalytic units linked in tandem is not known, although there is experimental evidence to suggest that co-operativity exists between the two halves of the protein [5-7].
In developing spermatids, an alternative intragenic promoter utilises exons 13–26 and generates a single domain ACE (gACE) which is largely identical to the C-terminal domain of somatic ACE [4,8]. Male mice lacking gACE are infertile as a result of sperm that are defective in migration in the oviduct and in binding to the zona pellucida [9].
Insect ACE has broad substrate specificity and its presence in the haemolymph of insects raises the possibility that, like mammalian sACE, it is required for extracellular metabolism of peptide hormones [10,11]. Drosophila melanogaster has six ACE-like genes (Ance, Acer, Ance-2, Ance-3, Ance-4 and Ance-5), all coding for single domain proteins. The ANCE protein has been relatively well characterised and has been shown to have several biochemical similarities to the mammalian enzyme. Its activity is developmentally regulated, with highest levels during metamorphosis [12,13]. ANCE is also expressed in spermatocytes and appears to have an important role in spermiogenesis [14]. Of the other Drosophila ACEs, only the Acer gene product has been studied biochemically and it is likely that ANCE-2, ANCE-3, ANCE-4 and ANCE-5 do not function as peptidases since they lack one or more of the residues that are essential for peptidase activity [15]. ACER, like ANCE and human ACE, is a peptidyl dipeptidase, but is generally less efficient than ANCE at cleaving dipeptides from many oligopeptide substrates [16]. Acer is expressed in the embryonic heart [17] and in both the male and female gonads and brain of adult flies (A. Carhan, R. E. Isaac and A. D. Shirras, unpublished results) where it is assumed to have a role in the metabolism of, as yet unidentified, biologically active peptides involved in neuroendocrine signalling and reproduction.
ACE activity has been found in the gonads and accessory glands of several insect species in addition to D. melanogaster, suggesting a conserved role for this enzyme in insect reproduction [10,11]. Recently, we have provided evidence for a role for ACE in reproduction in the mosquito, A. stephensi. ACE activity in the adult female mosquito increases by 260% following a blood meal [10,18] and it has been proposed that the induced ACE has a role in regulating peptide signalling in response to a blood meal. We have shown that feeding two different ACE inhibitors to male A. stephensi in their glucose diet, resulted in approx. 80% reduction in the number of eggs laid [10]. In addition, ACE inhibitors introduced in the blood meal, resulted in a dose-dependent effect on brood-size, but had no effect on oocyte development, nor the rate of digestion of the blood. The only observable difference between inhibitor-fed and control insects was that the inhibitor-fed females could not lay eggs, even 8 days after the blood meal, suggesting that the blood-induced ACE is involved in the control of egg-laying [19].
To understand the mechanism of ACE induction in blood-fed mosquitoes and the role of the enzyme in mosquito reproductive physiology, we have characterised the ACE gene family in Anopheles gambiae, a mosquito responsible for the transmission of the human malaria parasite and whose genome has recently been sequenced [20]. The partially annotated A gambiae genome [21] reveals between eight and twelve potential ACE-like genes. We now report the correct number of A. gambiae ACE (AnoACE) genes, confirm their corresponding protein sequences and establish their expression patterns during development and in response to a blood meal. We show that several of the AnoACEs are similar to other insect ACEs, but that AnoACE9 is a novel two-domain ACE whose structure resembles that of human somatic ACE.
Results
ACE-like genes in the A. gambiae genome
The Ensembl annotation of the A. gambiae genome (version 34.2 g) [21] suggests that there are eight genes encoding proteins belonging to the M2 (ACE) family of peptidases. BLAST searching of the genome and an examination of the predicted genes, however, shows that not all ACE-like proteins have been predicted, while several of those that have been identified have missing exons, or are hybrids, composed of exons from more than one ACE-like gene. These problems are particularly acute for a cluster of ACE-like sequences at 35C on chromosome 3R (Fig. 1). In order to confirm the number and organisation of ACE-like genes in the A. gambiae genome a combination of TBLASTN analysis [21], using Drosophila melanogaster ACE proteins as query sequences, and cDNA sequence analysis was carried out. Results are summarised in Tables 2 and 3 and may be viewed as a custom track in Ensembl Mosquito [37].
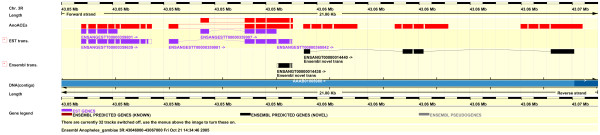
Figure 1.
Annotation of ACE-like genes in the 35C cluster. Ensembl ContigView display of the 43.05–43.07 Mb region of chromosome 3R showing AnoACE custom track annotation (red bars), Ensembl EST transcripts (ENSANGESTT) and Ensembl predicted transcripts (ENSANGT). From left to right the AnoACE genes are numbered 2, 3 (with two alternative 5' exons), 4, 5 and 6.
Table 2.
ACE-like genes in the A. gambiae genome. Genes were identified by a combination of TBLASTN using Drosophila genes as query sequences and cDNA sequence analysis. Corresponding Ensembl (version 34.2 g) transcripts and ESTs (where known) are indicated.
| AnoACE | ENSANGT | ENSANGESTT | Chromosome | Contig | Coordinates |
| 1 | 0000010993 | - | 2L, 28C | AAAB01008807_41 | 47,788,338–47,790,069 |
| 2 | - | 00000359639 00000359801 |
3R, 35C | AAAB01008980_249 | 43,046,775 – 43,049,518 |
| 3 | 00000014438 (part) |
0000035981* 00000359997* 00000360042 |
3R, 35C | AAAB01008980_249 | 43,050,199 – 43,055,041 |
| 4 | 00000014440 (part) |
- | 3R, 35C | AAAB01008980_249 | 43,055,473 – 43,057,570 |
| 5 | 00000014440 (part) |
- | 3R, 35C | AAAB01008980_249 | 43,059,057 – 43,061,130 |
| 6 | 00000014440 (part) |
- | 3R, 35C | AAAB01008980_249 | 43,063,988 – 43,066,391 |
| 7 | 00000022325 00000014398† (part) |
00000360081 | 3R, 35C | AAAB01008980_250 AAAB01008980_252† |
43,068,658 – 43,072,250 43,136,048 – 43,139,362† |
| 8 | 00000007115 | - | 3R, 29C | AAAB01008964_349 | 3,659,796 – 3,661,800 |
| 9 | 00000028617 00000011721 |
00000365570 00000365630 |
2R, 19C | AAAB01008898_151 | 59,413,237 – 59,418,650 |
*alternative transcripts with different 5' exons
† 3' exons of AnoACE7, 69 kb downstream of the rest of the gene
Table 3.
Exon coordinates of AnoACE genes. Coordinates are as in Ensembl Mosquito version 34.2 g. Exon annotations may be viewed as a custom track in Ensembl Mosquito [37].
| AnoACE, Chr. | Exon | Start | End | Length | Start codon | Stop codon |
| 1, 2L | 1 | 47788338 | 47790069 | 1731 | 47788338 | 47790069 |
| 2, 3R | 1 | 43046775 | 43046948 | 173 | 43047058 | 43049381 |
| 2 | 43047050 | 43047327 | 277 | |||
| 3 | 43047407 | 43047771 | 364 | |||
| 4 | 43047832 | 43048053 | 221 | |||
| 5 | 43048118 | 43048261 | 143 | |||
| 6 | 43048324 | 43049032 | 708 | |||
| 7 | 43049101 | 43049274 | 173 | |||
| 8 | 43049343 | 43049518 | 175 | |||
| 3A, 3R | 1 | 43050199 | 43050555 | 356 | 43050288 | 43054998 |
| 2 | 43053178 | 43053542 | 364 | |||
| 3 | 43053612 | 43053976 | 364 | |||
| 4 | 43054035 | 43054916 | 881 | |||
| 5 | 43054981 | 43055041 | 60 | |||
| 3B, 3R | 1 | 43051447 | 43051757 | 310 | 43051523 | 43054998 |
| 2 | 43053178 | 43053542 | 364 | |||
| 3 | 43053612 | 43053976 | 364 | |||
| 4 | 43054035 | 43054916 | 881 | |||
| 5 | 43054981 | 43055041 | 60 | |||
| 4, 3R | 1 | 43055473 | 43055722 | 249 | 43055473 | 43057570 |
| 2 | 43055795 | 43056159 | 364 | |||
| 3 | 43056227 | 43056590 | 363 | |||
| 4 | 43056672 | 43057570 | 898 | |||
| 5, 3R | 1 | 43059057 | 43059300 | 243 | 43059057 | 43061130 |
| 2 | 43059368 | 43059732 | 364 | |||
| 3 | 43059801 | 43060165 | 364 | |||
| 4 | 43060238 | 43061130 | 892 | |||
| 6, 3R | 1 | 43063988 | 43064231 | 243 | 43063988 | 43066391 |
| 2 | 43064299 | 43064663 | 364 | |||
| 3 | 43064732 | 43065096 | 364 | |||
| 4 | 43065169 | 43066391 | 1222 | |||
| 7, 3R | 1 | 43068658 | 43069121 | 463 | 43068773 | 43139362 |
| 2 | 43069190 | 43069805 | 615 | |||
| 3 | 43070472 | 43070631 | 159 | |||
| 4 | 43070704 | 43070827 | 123 | |||
| 5 | 43070892 | 43071025 | 133 | |||
| 6 | 43071102 | 43071687 | 585 | |||
| 7 | 43071770 | 43072014 | 244 | |||
| 8 | 43072151 | 43072250 | 99 | |||
| 9 | 43136048 | 43136189 | 141 | |||
| 10 | 43139040 | 43139362 | 322 | |||
| 8, 3R | 1 | 3659796 | 3661191 | 1395 | 3659796* | 3661800 |
| 2 | 3661258 | 3661542 | 284 | |||
| 3 | 3661607 | 3661800 | 193 | |||
| 9, 2R | 1 | 59413237 | 59413683 | 446 | 59413677 | 59418197 |
| 2 | 59414399 | 59414615 | 216 | |||
| 3 | 59414676 | 59416593 | 1917 | |||
| 4 | 59416658 | 59418650 | 1992 |
*Start of open reading frame, no start methionine identified
TBLASTN analysis revealed 11 possible ACE-like coding sequences. Six of these sequences are arranged in a tightly linked tandem cluster at 35C (Figs. 1 &2). These were numbered (from 5' to 3') AnoACE 2,3,4,5,6 and 7. Several EST clones exist for AnoACE2 and AnoACE3, allowing the exons of these genes, and their coding regions to be unambiguously assigned. AnoACE3 has two alternative transcripts which differ in their first exon. No cDNA clones are available for AnoACEs 4, 5 and 6. The exons of these genes were identified by BLAST similarity and the SNAP exon predictions of Ensembl [21]. The nucleotide sequences of AnoACEs 5 and 6 are 97% identical, including the introns, which raised the possibility that these two genes are the result of a sequence assembly error caused by nucleotide polymorphisms in the PEST strain. This high level of identity is only observed within the putative transcribed region, however. The upstream flanking sequence of AnoACE6 shares only around 75% identity with the AnoACE5 upstream DNA and there is no significant similarity between the downstream flanking sequences of the two genes. An assembly error can therefore be ruled out.
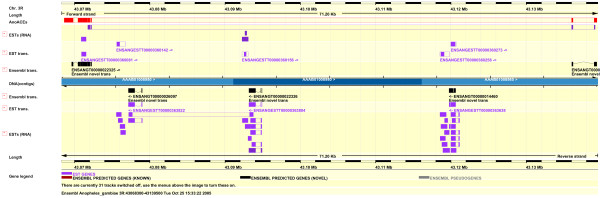
Figure 2.
Annotation of AnoACE7. Ensembl ContigView display of the 43.07–43.13 Mb region of chromosome 3R showing AnoACE custom track annotation (red bar), Ensembl ESTs (purple bars), EST transcripts (ENSANGESTT) and Ensembl predicted transcripts (ENSANGT). Eight upstream exons, encoding the bulk of the protein, are situated at the 3' end of the 35C cluster, downstream of AnoACE6 (Fig. 1). Two exons encoding the C terminus of the protein are situated 69 kb downstream. The sequence of EST clone P141-G-08-5 spans upstream and downstream exons of AnoACE7 and is shown below the gene annotation. The large intron contains several potential genes. Not all introns are shown at this scale.
The AnoACE7 gene has an unusual organisation; there are eight upstream exons situated at the 3' end of the 35C cluster which encode the bulk of the protein and there are two exons encoding the C terminus of the protein 69 kb downstream (Fig. 2). These exons were shown to belong to the AnoACE7 gene by the sequence of NAP1 cDNA P141-G-08-5 which spans exon 8 and the two downstream exons (Fig. 2., Ensembl annotation). The gene structure was further confirmed by TBLASTN alignment with the Drosophila ANCE-3 protein. The 5' end of AnoACE7 was determined by RT-PCR analysis using nested primers designed against the genomic sequence. A 1.1 kb RT-PCR product was cloned, sequenced and found to contain a long open reading frame coding for a protein which overlapped existing predicted AnoACE7 protein sequence and which had a putative signal peptide. This was assumed to correspond to the N terminus of the AnoACE7 precursor.
The AnoACE9 gene contains two ACE-like coding regions. The sequence of cDNA 19600449703686 (MRA-468-36) spans both coding regions (Ensembl annotation), indicating that this is a single gene coding for a double-domain protein, rather than two separate genes (Fig. 3).
Figure 3.
Annotation of AnoACE9. Ensembl ContigView display of the 59.413–59.419 Mb region of chromosome 2R showing AnoACE custom track annotation (red bar), Ensembl EST transcripts (ENSANGESTT) and Ensembl predicted transcripts (ENSANGT). The AnoACE9 gene consists of 4 exons. Exons 3 and 4 both contain ACE active site coding regions and are spanned by cDNA 19600449703686 (ENSANGESTT00000365630) indicating a double-domain enzyme.
AnoACE proteins
Protein sequences were predicted from cDNAs for AnoACEs 2, 3, 7 and 9. AnoACEs 1, 4, 5, 6 and 8 were predicted from genomic sequence and alignment with Drosophila ACE proteins. Predicted properties of the proteins are summarised in Table 4. Signal peptides were identified for all proteins except AnoACEs 1 and 8. All of the AnoACE genes, except for AnoACE9, code for single domain proteins similar to those previously found in insects. AnoACE9 is a putative double domain enzyme, similar to mammalian somatic ACE. Two of the AnoAces (7 and 9) have hydrophobic C termini which might serve as membrane anchors or be lost as a result of post-translational modification with a GPI anchor [22,23]. AnoACE7 has an extended N terminus, compared to the other proteins. All proteins have a full complement of required active site amino acids in the HExxH...EAV/I/L motif (Fig. 4). Predicted protein sequences are available as additional data file 1.
Table 4.
Predicted AnoACE proteins. Conceptual proteins were translated from cDNA sequences for AnoACEs 2, 3, 7 and 9 and from genomic sequence for AnoACEs 1, 4, 5, 6, and 8.
| AnoACE | Length (aa) | Active sites | Hydrophobic C terminus | Predicted GPI anchor |
| 1‡ | 576 | 1 | No | No |
| 2 | 638 | 1 | No | No |
| 3A* | 630 | 1 | No | No |
| 3B* | 619 | 1 | No | No |
| 4 | 625 | 1 | No | No |
| 5† | 619 | 1 | No | No |
| 6† | 730 | 1 | No | No |
| 7 | 917 | 1 | Yes | No/marginal |
| 8‡ | 623 | 1 | No | No |
| 9 | 1225 | 2 | Yes | Yes |
*AnoACE3A and AnoACE3B are derived from alternative transcripts and differ at their N termini
†AnoACE5 and AnoACE6 are 97% identical, except for an extended C terminus in AnoACE6
‡N-terminus probably missing, no signal peptide
Figure 4.
Active site region of ACE-like proteins. The active site regions of AnoACEs, the N and C domains of human ACE, D. melanogaster ANCE, ACER and ANCE-3, and Apis 3929 were aligned using CLUSTALX [36]. Amino acids shown to be required for activity in mammalian ACE are highlighted. The histidines in the HExxH motif are required for zinc coordination, whereas the glutamate is required for catalysis. The downstream glutamate is required for zinc coordination. This is replaced by glutamine in ANCE-3.
Comparative genomics of ACE-like genes
A phylogram showing the relationships of the AnoACEs to other insect and chordate ACEs is shown in Fig. 5. AnoACEs 2, 3, 4, 5 and 6 cluster with Drosophila ANCE and ACER, and ACE-like proteins from Apis mellifera, Haematobia irritans, Lutzomyia longipalpis and Bombyx mori. These proteins are all single domain proteins which lack a hydrophobic C terminus. A second cluster of insect proteins contains AnoACEs 7 and 8, the two remaining Apis ACEs and Drosophila ANCE-3. AnoACE7 is a one-to-one orthologue of ANCE-3 and of Apis 3929. AnoACE8 is a one-to-one orthologue of Apis 10228. There appears to be no Drosophila orthologue of AnoACE8. The similarity of the AnoACE7 group extends to the genomic organisation of the genes. In each case the bulk of the protein is encoded by a set of upstream exons and these are separated from the downstream exons, encoding the C terminus of the protein by a large intron. In the case of Anopheles the intron is 69 kb in length, in Drosophila 35 kb and in Apis 21 kb. The proteins in this group are characterised by an extended N terminus, rich in proline and charged amino acids. AnoACE7 and Drosophila ANCE-3 have a hydrophobic C terminus but this is absent from the predicted Apis protein. GPI anchor prediction software [23] predicts no GPI anchor for AnoACE7 whereas Drosophila ANCE-3 has a strong GPI anchor prediction.
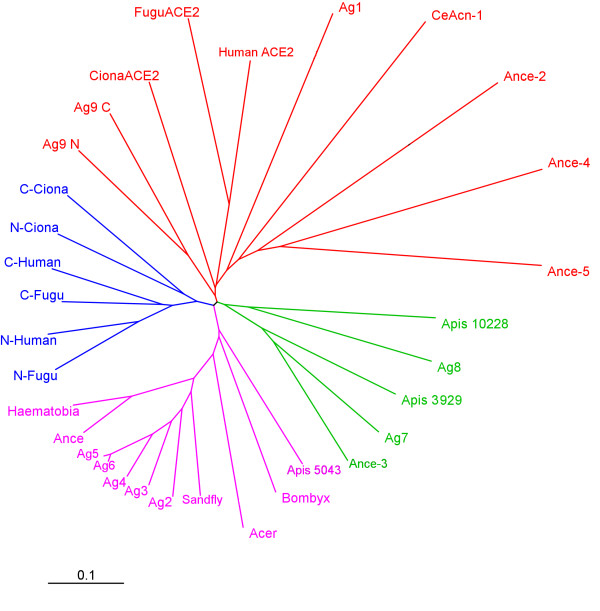
Figure 5.
Evolutionary relationships of invertebrate and chordate ACE-like proteins. Core sequences (corresponding to amino acids161 to 569 of the AnoACE2 pre-protein) of each protein were aligned and a neighbour-joining tree calculated using CLUSTALX [36]. TreeView was used to produce a radial tree. Sequences used were AnoACEs (Ag)1, 2, 3, 4, 5, 6, 7, 8, and 9 (N and C domains); D. melanogaster ANCE, ACER, ANCE-2, -3, -4 and -5; B. mori ACE (accession no. BAA97657); H. irritans ACE (accession no. Q10715); C. elegans ACN-1; N and C domains of Human ACE, Fugu rubripes (Ensembl SINFRUP00000174092) ACE; A. mellifera Ensembl proteins ENSAPMP00000005043, ENSAPMP00000003929 and ENSAPMP000000010228; Sandfly (Lutzomyia longipalpis) ACE (accession no. AAS16911); Human and F. rubripes (Ensembl SINFRUP00000161972) ACE2. N and C domains of Ciona intestinalis ACE, were assembled from genomic sequence in contig AABS01000302.1. Scale bar shows number of amino acid substitutions per site.
AnoACEs 1 and 9 are placed in a cluster with a diverse group of ACE-like proteins that includes Drosophila ANCEs-2, -4 and -5, the C. elegans ACE-like protein ACN-1 and vertebrate ACE2. The latter protein, while having sequence similarity to ACEs, has a different enzymatic activity and acts as a carboxypeptidase rather than a dipeptidase. Several members of this group (Drosophila ANCEs-2, -4 and -5, ACN-1) lack one or more essential active site amino acids and are therefore unlikely to be active peptidases. AnoACE9 has several structural similarities with chordate ACEs. It has two active site domains and a hydrophobic C terminus with a strong GPI anchor prediction (Fig. 6). The N and C domains of this protein are more similar to each other than to other ACEs, unlike the vertebrate N and C domains which cluster together.
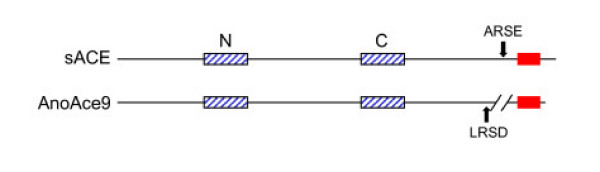
Figure 6.
Diagrammatic alignment of human somatic ACE (1306 amino acids) and AnoACE9 (1225 amino acids). The hatched boxes represent the N and C domain active site regions. Filled boxes are the C-terminal hydrophobic potential membrane anchor regions. AnoACE9 has a very short potential cytoplasmic region and a sequence gap relative to the human enzyme on the N terminal side of the putative transmembrane region. Arrows indicate potential secretase recognition sequences.
Expression of AnoACE genes
Primers for all AnoACE genes were validated by PCR amplification of genomic DNA or cDNA. Each primer pair produced a product of the expected size (Fig. 7).
Figure 7.
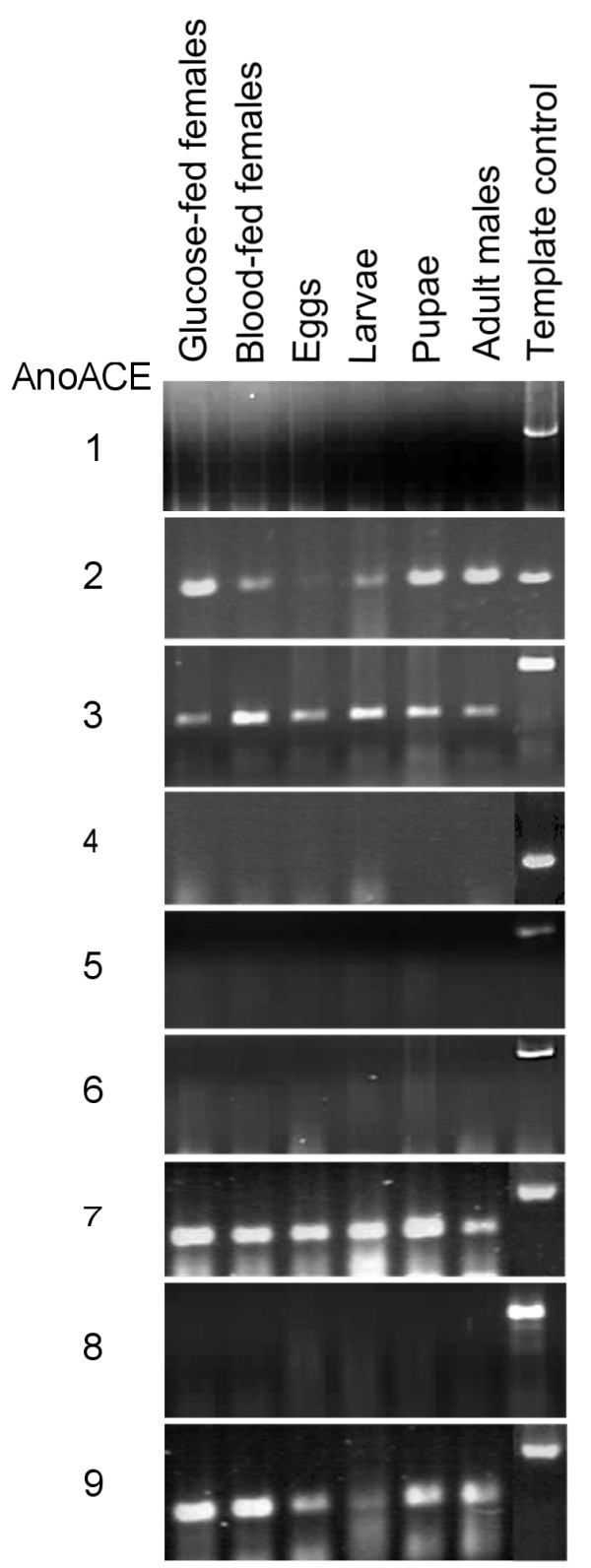
RT-PCR analysis of AnoACE gene expression. RNA was extracted from glucose fed adult females, blood fed adult females, eggs, larvae, pupae and adult males. Control templates for amplification were genomic DNA except for AnoACE2 primers when the template was DNA from cDNA clone 19600449622752 (MRA-467-39). Amplification products were observed for AnoACEs 2, 3, 7 and 9 only.
For semi-quantitative RT-PCR analysis RNA was extracted and cDNA produced from different developmental stages of mosquitoes, namely, eggs, larvae, pupae, adult males, glucose-fed females and blood-fed females. Figure 7 shows that AnoACEs 2, 3, 7 and 9 are expressed at all developmental stages tested, whereas no PCR product was observed for AnoACEs 1, 4, 5, 6 and 8. The authenticity of the products from AnoACE2, 3, 7 and 9 was confirmed by cloning into the pGEM T-Easy® vector and sequencing.
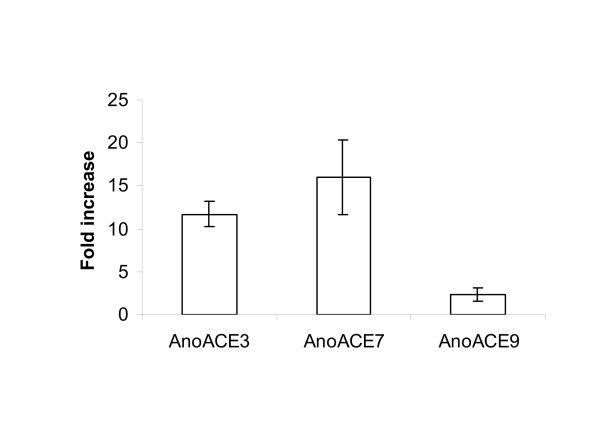
Gene expression for all the AnoACE genes was analysed in glucose and blood-fed females using quantitative real-time RT-PCR. Other than AnoACEs 2,3,7 and 9, expression levels were as for no template controls. Transcription of AnoACE3, AnoACE7 and AnoACE9 was shown to be up-regulated 48 hours following a blood meal (Fig. 8). The increase in transcription was consistent for three separate cohorts of female mosquitoes.
Figure 8.
Relative expression of AnoACE genes in glucose and blood-fed females. RNA was extracted from three different cohorts of mosquitoes, either after continuous glucose feeding or 48 hours after a blood meal, and analysed by quantitative RT-PCR. The mean increase in gene expression was 11.6 fold for AnoACE3, 16 fold for AnoACE7 and 2.4 fold for AnoACE9.
Discussion
Our analysis of the A. gambiae genome has revealed nine ACE-like genes. This is the largest number of ACE genes for any species with a complete genome sequence. Drosophila melanogaster has six ACE-like genes [15], but only two of these (Ance and Acer) are known to code for active peptidase proteins. As well as having the ACE-like genes found in D. melanogaster, D. pseudoobscura has an additional Ance-like gene which is likely to code for an active peptidase, since all the amino acids that are necessary for catalysis and coordination with the active site zinc are conserved (A.D. Shirras, unpublished work). Apis mellifera has three ACE-like genes, each with a full complement of the critical active site amino acids [24]. At present we have evidence that at least four AnoACE genes (2, 3, 7 and 9) are expressed, but have no evidence for expression of AnoACEs 1, 4, 5, 6 and 8. All of the AnoACE genes, however, have complete open reading frames and the corresponding proteins have a full complement of essential active site amino acids, so we cannot rule out the expression of the other genes at low levels, in a small number of cells or at stages that we have not examined.
Some of the AnoACE genes have clearly arisen by recent gene duplications. The cluster at 35C contains five genes that are closely related in sequence. Two of these (AnoACE5 and AnoACE6) are almost identical. Similar clusters of other duplicated genes exist within the A. gambiae genome; for example there is a cluster of M14 carboxypeptidase genes at 29D which contains adjacent genes with greater than 90% identity. In both these clusters the high levels of identity are restricted to the transcribed regions with flanking DNA showing lower levels of similarity. This suggests that selective pressures are maintaining the nucleotide sequences of these genes and that they are expressed at some stage.
Comparison of ACE protein sequences (Fig. 5) suggests that there are three functional groupings in insects. The first contains Drosophila ANCE, AnoACEs 2, 3, 4, 5 and 6 and several other insect ACEs. These are single domain, secreted enzymes. This class of ACE proteins is similar to the chordate N and C domains, both in terms of sequence and enzyme activity. The second group includes AnoACE7, Drosophila ANCE-3 and Apis 3929. These proteins are single domain, but have extended N termini. The Drosophila and mosquito proteins have a hydrophobic C terminus suggesting that they may be membrane-tethered. The active sites of these three proteins are characterised by an alanine in place of glycine at position 4 of the HExxH motif (Fig. 4). AnoACE8 and Apis 10228 form diverged members of this group.
The third group of insect ACEs, containing AnoACE1 and Drosophila ANCE-2, -4 and -5, is highly divergent. These appear to be secreted proteins and, whereas AnoACE1 may be enzymically active, the Drosophila examples lack essential active site amino acids. C. elegans ACN-1 and vertebrate ACE2 proteins form highly diverged members of this family. This result is to be expected as they have distinctive and highly evolved roles in these animals [25,11].
AnoACE9 is highly unusual. Phylogenetic analyses using several different tree construction methods (Neighbour-Joining, Maximum Parsimony, Minimum Evolution, Unweighted Pair Group Method with Arithmetic Mean) do not place it convincingly in any of the insect groups described above. In terms of overall structure it is more similar to mammalian somatic ACE than to any invertebrate ACE described so far (Fig. 6). It has two active site domains and has a hydrophobic C terminus. This similarity with vertebrate ACEs is, as far as we know, unique amongst non-chordate invertebrates. It is likely that AnoACE9 is membrane bound, either by virtue of its hydrophobic C-terminus or via a potential GPI anchor. In mammals, membrane sACE can be released from the plasma membrane by unidentified metalloproteases, known as secretases or sheddases, that cleave the Arg-Ser bond in the Ala-Arg-Ser-Glu motif of the juxtamembrane stalk [26,27]. Interestingly, a similar peptide sequence (Leu-Arg-Ser-Asp) is found in the same position in the proposed juxtamembrane stalk of AnoACE9 (Fig. 6), which might serve as a cleavage site for a mosquito ACE secretase.
In mammals, the two active sites of sACE are involved in the conversion of angiotensin I to angiotensin II and the hydrolysis of bradykinin, but only the N-domain is involved in N-acetyl SDKP metabolism [2]. Thus, duplication appears to have provided an opportunity for the N-domain to acquire distinctive substrate specificity. However, this does not explain why mammalian sACE is a single protein with two catalytic domains joined by a peptide linker, rather than single genes encoding single domain proteins as occurs in D. melanogaster. Our phylogenetic analysis indicates that the gene duplication event leading to the two-domain vertebrate somatic ACE, occurred around 450 million years ago, before the divergence of amphibians and fishes, and that two other very similar events occurred independently to give rise to a two-domain enzyme: one in the lineage leading to A. gambiae and one in the urochordate lineage leading to Ciona intestinalis (Fig. 5). That is, two-domain ACE proteins have been selected for on three separate occasions, suggesting that there is a distinct functional advantage to having two active sites organised in this manner over two single-domain proteins. Support for this hypothesis comes from several studies on mammalian sACE that suggest that the two active sites can have cooperative effects on ACE activity [5-7]. Recent studies indicate that the N-domain of human somatic ACE also has a negative effect on the shedding of the protein from the cell surface by cleavage within the stalk region that separates the C-domain from the membrane anchor [28]. More detailed biochemical studies on AnoACE9 are now required to determine whether similar cooperative phenomena occur in mosquitoes.
The expression of AnoACEs 3, 7 and 9 is induced by a blood meal. Two of these genes (AnoACE 3 and 7) are induced more than ten-fold. Neuropeptides are known to be important for mediating several behavioural and physiological responses to a blood meal in mosquitoes [29-34]. Our previous work showing that ACE inhibitors block egg-laying, but not egg development, in blood fed A. stephensi [19] led us to propose that the elevated peptidase activity might be important for regulating a peptide signal involved in egg-laying. The up-regulation of AnoACEs 3, 7 and 9 following a blood meal in A. gambiae, raises the possibility that ACE may also be involved in regulating post feeding oviposition in this species.
A role for at least some AnoACEs in the immune response is suggested by recent microarray data [35]. AnoACE7 (ENSANGT00000022325) was shown to be 2.4-fold upregulated by challenge with Salmonella typhimurium. AnoACE1 (ENSANGT0000010993) was 3.5-fold upregulated by Staphylococcus aureus infection, whereas the expression of AnoACE9 (ENSANGT00000028617) was reduced 2.4-fold by Beauveria bassiana. We were unable to detect AnoACE1 expression in a variety of developmental stages. However, the expression of this gene suggested by the infection microarray may mean that this gene plays a specific role as a component of the immune response.
Conclusion
The Anopheles gambiae genome contains 9 genes coding for proteins with similarity to mammalian ACE. Six of these genes are found in a tandem cluster situated at 35C on chromosome 3R. Five of the members of this cluster code for proteins which are similar to previously described insect ACEs: they contain a single active site domain and lack a C-terminal hydrophobic region. AnoACE7, situated at the 3' end of the 35C cluster, is an orthologue of Drosophila Ance-3 and of Apis gene ENSAPMG00000014390. In all three species, there is a similar gene structure with the bulk of the coding exons separated from the C-terminal exons by a large intron which contains other genes. The AnoACE7 protein, like Drosophila ANCE-3, has a hydrophobic C-terminus and is likely to be membrane bound. AnoACEs 8 and 10 are also predicted to be single domain proteins but AnoACE9 is unique amongst insect ACEs described so far in that it contains two active site domains and has a hydrophobic C-terminus, a structure similar to that of mammalian somatic ACE. Expression of only 4 of the Anopheles ACE genes (2, 3, 7 and 9) could be detected by RT-PCR in the stages tested. Expression of AnoACEs 3, 7 and 9 increased following a blood meal.
Methods
Insects
Male and female Anopheles gambiae were maintained at a temperature of 26°C and at a relative humidity of 80 %. Newly hatched adult mosquitoes were fed for seven days on sterile 5% (w/v) glucose/0.5% (w/v) p-aminobenzoic acid solution [20]. On the seventh day female mosquitoes were harvested and divided into two cohorts. Mosquitoes in cohort A were maintained on the glucose/p-aminobenzoic acid solution, whilst mosquitoes in cohort B were fed mouse blood. After the blood meal any unfed female mosquitoes (from cohort B) were discarded. Mosquitoes from cohort B were collected 48 hours after the blood meal. Insects from cohort A were collected 48 hours after cohort B was given the blood meal.
Extraction of nucleic acids
Nucleic acids were extracted using either the Qiagen DNeasy® or RNeasy® kits as per manufacturer's directions. During RNA extraction the optional RNase-free DNase I from Qiagen® was used to digest DNA. After quantification, extracted DNA was stored at 4°C and RNA at -80°C until required.
PCR
Primers were designed for each possible mosquito ACE gene, with each pair crossing exon boundaries, where this was possible (see Table 1 for primer sequences).
Table 1.
Primers for PCR of AnoACE genes
| Gene | Forward Primer sequence (5'-3') | Reverse Primer Sequence (5'-3') | Predicted Product Size From Genomic DNA(bp) |
| AnoACE1 | GGAATGCATCGTGTGTGTTC | TACTCGAAGGGAATGGTTGG | 943 |
| AnoACE2 | GGTACCGATCAACCAGTGCT | AAATCGCATCGACAAGCTCT | 440 |
| AnoACE3 | CCCTGTGCAAGTGCTGATAA | ATACGGAACGCCATTGGATA | 214 |
| AnoACE4 | CAGCCGAGATGCTGAAGAGT | GAACTCGCGCAACCTATACC | 328 |
| AnoACE5 | TTGCTGCAGCGAATGCTAT | CACACCAGCCCAATCTTGT | 754 |
| AnoACE6 | CAGCGAATGCTGTGTTCCT | CAATGTTGCCCCACTTCTGT | 748 |
| AnoACE7 | CGCAGTGGAACTTTGAGACA | GTCTATTCGGCGCATTTGAT | 782 |
| AnoACE8 | GGTCTGGGCGATACCAAGTA | CGCGTCAAATCGGAGTTAAT | 220 |
| AnoACE9 | GCACACGTATCGGAAAATGA | GCGCTTCTCGATGAACTACC | 803 |
The primers were also designed so that the product size from cDNA would be approximately 100 bp as appropriate for use with QPCR, except for AnoACE1 where the product was 943 bp. Amplification reactions were prepared in 20 μl volumes and contained; 100 ng of template, 2 μl of PCR buffer (670 mM Tris-HCl, pH 8.8; 166 mM [NH4]2SO4; 67 mM MgCl2), 200 μM of dNTPs, 330 ng of BSA, 0.14 μl of 5% (v/v) β-mercaptoethanol, 1 unit of Promega® Taq polymerase, 20 pmol of forward primer, 20 pmol of reverse primer. The following cycling conditions were used: one incubation at 94°C for 5 min; 35 cycles of 94°C for 30 s; Tm for 30 s; 74°C for 1 min: followed by one incubation at 74°C for 5 min (where the Tm is appropriate for each set of primers).
RT-PCR
To amplify the 5' end of AnoACE7, cDNA was prepared using an Invitrogen Cloned AMV kit as per the manufacturer's instructions. Using 2 μg total RNA from blood fed A. gambiae and 10 mM gene specific primer (ACE7 REV: 5'-GTCTATTCGGCGCATTTGAT-3'), first strand synthesis was performed at 50°C for 1 hour. PCR was then carried out using 1 μl of the first strand reaction, 2 mM dNTPs, 0.5 mM each primer (ACE7 REV and ACE7 FOR3: 5'-AGTGAAAACGTAACCATTGCAGAA-3') and 2.5 units Proofstart enzyme (Qiagen). PCR conditions were as follows: initial denaturation at 95°C, 5 min. then 35 cycles of 95°C for 30s, 54°C for 30s, 72°C for 3 min., and a final extension at 72°C for 5 min. A 1.1 kb product was purified using the Qiagen® gel purification kit as per the manufacturer's instructions, cloned into pGEM®T-Easy and sequenced using M13 forward and reverse, and internal primers. The accession number of this sequence is [EMBL:AM085517]. For semi-quantitative RT-PCR, first strand synthesis of cDNA took place in a 20 μl reaction volume using 1 μg of RNA and Promega ImPromII RT® kit as per the manufacturer's instructions and either random or gene-specific primers. PCR was carried out using the method described above with 2 μl of cDNA, primers as in Table 1, and the following cycling conditions: 94°C for 5 min. then 35 cycles of 94°C for 15 s, Tm for 15 s, 74°C for 30 s followed by incubation at 74°C for 5 min. PCR products were separated by agarose gel electrophoresis. To confirm identity of the PCR products, bands were purified using the Qiagen® gel purification kit, cloned into pGEM® T-Easy and sequenced using M13 forward and reverse primers.
Quantitative RT-PCR
RNA (3 μg) was reverse transcribed using the Promega ImPromII® kit and the resulting cDNA quantified using a Biorad iCycler instrument. cDNA (5 μl) was diluted in water to a total of 200 μl. Three-fold serial dilutions were made from this mixture as templates for the calibration curve. The remaining 15 μl of each experimental cDNA was made up to 400 μl with water. Reactions were performed in duplicate using 10 μl of template, 12.5 μl of ABsolute™ QPCR SYBR® green and 2.5 μl of primers at a concentration determined to be optimum for each primer pair. Reactions were subjected to the following amplification cycles; 95°C for 15 min, once; 95°C for 15 s; 60°C for 30 s for 40 cycles. Melt curves were also performed to confirm that a single PCR product and no primer dimers were produced. The melt curves were performed using the following conditions; one incubation at 95°C for 30 s, 55°C for 30 s, once; followed by incubation at 55°C for 10 s 82 times, with a temperature increase of 0.5°C every cycle after cycle 2. Transcript levels of the AnoACE genes were normalised against the levels of mRNA for the ribosomal protein RPS17, amplified using primers 5'-TTGACCATGGATTTCGACAC-3' and 5'-TGATGGAAATACCACGCACT-3', forward and reverse, respectively.
List of abbreviations
ACE: angiotensin I-converting enzyme, AnoACE: Anopheles ACE
Authors' contributions
SB and AJL identified AnoACE genes and carried out expression studies. SB produced the initial draft of the manuscript. JAS cloned and sequenced the 5' end of AnoACE7. REI helped draft the manuscript. ADS refined the AnoACE annotation, carried out sequence comparisons and phylogenetic analysis, and re-drafted the manuscript. REI and ADS jointly planned the study and supervised experiments.
Supplementary Material
AnoACE protein sequences in FASTA format.
Acknowledgments
Acknowledgements
This work was supported by the Wellcome Trust (Grant GR071921). We thank Debbie Evans and Martin Looker for providing mosquitoes. The Malaria Research and Reference Reagents Resource Center (MR4) supplied cDNA clones deposited by R. A. Holt. NAP1 cDNA clones were supplied by George Christophedes, European Molecular Biology Laboratory. We also acknowledge valuable discussion with Ed Parkin of the School of Biochemistry and Molecular Biology, University of Leeds, about ACE secretase.
Contributor Information
Susan Burnham, Email: bgysbu@leeds.ac.uk.
Judith A Smith, Email: j.a.smith@lancaster.ac.uk.
Alison J Lee, Email: a.j.lee@leeds.ac.uk.
R Elwyn Isaac, Email: r.e.isaac@leeds.ac.uk.
Alan D Shirras, Email: a.shirras@lancaster.ac.uk.
References
- Erdos EG. Angiotensin I-converting enzyme and the changes in our concepts through the years. Hypertension. 1990;16:363–370. doi: 10.1161/01.hyp.16.4.363. [DOI] [PubMed] [Google Scholar]
- Corvol P, Williams TA, Soubrier F. Peptidyl dipeptidase A: Angiotensin I-converting enzyme. Methods In Enzymology. 1995;248:283–305. doi: 10.1016/0076-6879(95)48020-x. [DOI] [PubMed] [Google Scholar]
- Corvol P, Eyries M, Soubrier F. Peptidyl-dipeptidase A/angiotensin I-converting enzyme. In: Barrett AJ, Rawlings ND, Woessner JF, editor. Handbook of Proteolytic Enzymes. 2. Vol. 1. Amsterdam: Elsevier Academic Press; 2004. pp. 332–346. [Google Scholar]
- Hubert C, Houot AM, Corvol P, Soubrier F. Structure of the angiotensin I-converting enzyme gene – 2 alternate promoters correspond to evolutionary steps of a duplicated gene. J Biol Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- Binevski PV, Sizova EA, Pozdnev VF, Kost OA. Evidence for the negative cooperativity of the two active sites within bovine somatic angiotensin-converting enzyme. FEBS Lett. 2003;550:84–88. doi: 10.1016/S0014-5793(03)00825-1. [DOI] [PubMed] [Google Scholar]
- Georgiadis D, Beau F, Czarny B, Cotton J, Yiotakis A, Dive V. Roles of the two active sites of somatic angiotensin-converting enzyme in the cleavage of angiotensin I and bradykinin: insights from selective inhibitors. Circ Res. 2003;93:148–154. doi: 10.1161/01.RES.0000081593.33848.FC. [DOI] [PubMed] [Google Scholar]
- Andújar-Sánchez M, Cámara-Artigas A, Jara-Pérez V. A calorimetric study of the binding of lisinopril, enalaprilat and captopril to angiotensin-converting enzyme. Biophys Chem. 2004;111:183–189. doi: 10.1016/j.bpc.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Langford KG, Shai SY, Howard TE, Kovac MJ, Overbeek PA, Bernstein KE. Transgenic mice demonstrate a testis-specific promoter for angiotensin-converting enzyme. J Biol Chem. 1991;266:15559–15562. [PubMed] [Google Scholar]
- Hagaman JR, Moyer JS, Bachman ES, Sibony M, Magyar PL, Welch JE, Smithies O, Krege JH, O'Brien DA. Angiotensin-converting enzyme and male fertility. Proc Natl Acad Sci USA. 1998;95:2552–2557. doi: 10.1073/pnas.95.5.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac RE, Ekbote U, Coates D, Shirras AD. Insect angiotensin-converting enzyme. A processing enzyme with broad substrate specificity and a role in reproduction. Ann NY Acad Sci. 1999;897:342–347. [Google Scholar]
- Macours N, Hens K. Zinc-metalloproteases in insects: ACE and ECE. Insect Biochem Mol Biol. 2004;34:501–510. doi: 10.1016/j.ibmb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Houard X, Williams TA, Michaud A, Dani P, Isaac RE, Shirras AD, Coates D, Corvol P. The Drosophila melanogaster-related angiotensin-I-converting enzymes Acer and Ance: Distinct enzymic characteristics and alternative expression during pupal development. Eur J Biochem. 1998;257:599–606. doi: 10.1046/j.1432-1327.1998.2570599.x. [DOI] [PubMed] [Google Scholar]
- Siviter RJ, Taylor CAM, Cottam DM, Denton A, Dani MP, Milner MJ, Shirras AD, Isaac RE. Ance, a Drosophila angiotensin-converting enzyme homologue, is expressed in imaginal cellsduring metamorphosis and is regulated by the steroid, 20-hydroxyecdysone. Biochem J. 2002;367:187–193. doi: 10.1042/BJ20020567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D, Rylett CM, Isaac RE, Shirras AD. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev Biol. 2003;254:238–247. doi: 10.1016/S0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- Coates D, Isaac RE, Cotton J, Siviter R, Williams TA, Shirras A, Corvol P, Dive V. Functional conservation of the active sites of human and Drosophila angiotensin I-converting enzyme. Biochemistry. 2000;39:8963–8969. doi: 10.1021/bi000593q. [DOI] [PubMed] [Google Scholar]
- Siviter RJ, Nachman RJ, Dani MP, Keen JN, Shirras AD, Isaac RE. Peptidyl dipeptidases (Ance and Acer) of Drosophila melanogaster : major differences in the substrate specificity of two homologs of human angiotensin I-converting enzyme. Peptides. 2002;23:2025–2034. doi: 10.1016/S0196-9781(02)00190-0. [DOI] [PubMed] [Google Scholar]
- Taylor CAM, Coates D, Shirras AD. The Acer gene of Drosophila codes for an angiotensin-converting enzyme homologue. Gene. 1996;181:191–197. doi: 10.1016/S0378-1119(96)00503-3. [DOI] [PubMed] [Google Scholar]
- Ekbote U, Coates D, Isaac RE. A mosquito (Anopheles stephensi) angiotensin I-converting enzyme (ACE) is induced by a blood meal and accumulates in the developing ovary. FEBS Lett. 1999;455:219–222. doi: 10.1016/S0014-5793(99)00870-4. [DOI] [PubMed] [Google Scholar]
- Ekbote U, Looker M, Isaac RE. ACE inhibitors reduce fecundity in the mosquito, Anopheles stephensi. Comp Biochem Physiol B Biochem Mol Biol. 2003;134:593–598. doi: 10.1016/S1096-4959(03)00019-8. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- EnsemblMosquito http://www.ensembl.org/Anopheles_gambiae/
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- GPI Lipid Anchor Project http://mendel.imp.univie.ac.at/sat/gpi/gpi_server.html
- Ensembl Honeybee http://www.ensembl.org/Apis_mellifera/
- Brooks DR, Appleford PJ, Murray L, Isaac RE. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol Chem. 2003;278:52340–52346. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- Ramchandran R, Sen I. Cleavage processing of angiotensin-converting enzyme by a membrane-associated metalloprotease. Biochemistry. 1995;34:12645–12652. doi: 10.1021/bi00039a021. [DOI] [PubMed] [Google Scholar]
- Woodman ZL, Oppong SY, Cook S, Hooper NM, Schwager SL, Brandt WF, Ehlers MR, Sturrock ED. Shedding of somatic angiotensin-converting enzyme (ACE) is inefficient compared with testis ACE despite cleavage at identical stalk sites. Biochem J. 2000;347:711–718. doi: 10.1042/0264-6021:3470711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman ZL, Schwager SL, Redelinghuys P, Carmona AK, Ehlers MR, Sturrock ED. The N domain of somatic angiotensin-converting enzyme negatively regulates ectodomain shedding and catalytic activity. Biochem J. 2005;389:739–744. doi: 10.1042/BJ20050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements AN. The biology of mosquitoes. London: Chapman & Hall; 1992. [Google Scholar]
- Stracker TH, Thompson S, Grossman GL, Riehle MA, Brown MR. Characterization of the AeaHP gene and its expression in the mosquito Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2002;39:331–342. doi: 10.1603/0022-2585-39.2.331. [DOI] [PubMed] [Google Scholar]
- Brown MR, Graf R, Swiderek KM, Fendley D, Stracker TH, Champagne DE, Lea AO. Identification of a steroidogenic neurohormone in female mosquitoes. J Biol Chem. 1998;273:3967–3971. doi: 10.1074/jbc.273.7.3967. [DOI] [PubMed] [Google Scholar]
- Hayes TK, Holman GM, Pannabecker TL, Wright MS, Strey AA, Nachman RJ, Hoel DF, Olson JK, Beyenbach KW. Culekinin depolarizing peptide: A mosquito leucokinin-like peptide that influences insect malpighian tubule ion transport. Regul Pept. 1994;52:235–248. doi: 10.1016/0167-0115(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Isolation and identification of three leucokinins from the mosquito Aedes aegypti. Biochem Biophys Res Commun. 1994;202:715–719. doi: 10.1006/bbrc.1994.1989. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Costes L. Isolation and identification of a peptide and its cDNA from the mosquito Aedes aegypti related to Manduca sexta allatotropin. Peptides. 1999;20:1145–1151. doi: 10.1016/S0196-9781(99)00117-5. [DOI] [PubMed] [Google Scholar]
- Aguilar R, Jedlicka AE, Mintz M, Mahairaki V, Scott AL, Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem Mol Biol. 2005;35:709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Mosquito AnoACE annotation http://www.ensembl.org/mosquito/contigview?data_URL=http://biol.lancs.ac.uk/ads/AnoACEs.bed
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AnoACE protein sequences in FASTA format.