Abstract
Chlamydia spp. are obligate intracellular bacterial pathogens that alternate between two metabolically and morphologically distinct developmental forms, and differentiation depends on transcriptional regulation. Genome sequencing of Chlamydia trachomatis revealed an ORF, CT630 (chxR), whose amino acid sequence contains a winged helix–turn–helix motif similar to the DNA-binding domain of response regulators in the OmpR subfamily. ChxR differs from many response regulators in that essential residues in the receiver or phosphorylation domain are lacking. ChxR functions as a transcriptional regulator because it activated transcription of ompF and ompC when expressed in Escherichia coli. In vitro transcription combined with microarray analysis also demonstrated that ChxR activates its own expression by binding directly to sites upstream of chxR; it also activates infA, tufA, oppA, and CT084. DNase I protection studies showed that ChxR bound to sites in the ompF and ompC promoter proximal regions that overlap but were distinct from OmpR binding sites. Both proteins could bind simultaneously to their nonoverlapping binding sites. This report identifies a stage-specific transcriptional regulator and some of its target genes in Chlamydia.
Keywords: OmpR, response regulator, transcription, winged helix–turn–helix protein
Many bacterial species undergo developmental transitions that are essential for growth and survival. Bacterial development is largely regulated at the level of transcription, mediated by both activators and repressors. A large group of transcription factors are the response regulators of bacterial two-component systems, which allow bacteria to sense and adapt to changing environmental conditions (1). Upon perceiving an environmental signal, a sensor histidine kinase is autophosphorylated and transfers the phosphoryl group to the response regulator. The activated response regulator elicits the appropriate response (activation or repression) based on the signal input.
Chlamydiae are obligate intracellular bacterial pathogens that undergo metabolic and morphological developmental changes in response to unknown intracellular conditions as a requirement for completing their developmental cycle (2). Developmental changes for growth and sensing the successful entry into a host cell to initiate differentiation likely require the presence of regulatory mechanisms involving environmental sensing and adaptation by the organism. These processes in Chlamydia are poorly understood.
Several putative transcription factors were identified and annotated in the chlamydial genome (3), including a member from the NtrC subfamily of response regulators (4). Transcriptional activators have not previously been identified in Chlamydia; thus, another gene of particular interest was CT630, whose predicted amino acid sequence contains a winged helix–turn–helix motif characteristic of the DNA binding domains of response regulators in the OmpR subfamily. By using Escherichia coli as a heterologous system, it was determined that CT630 is a transcriptional activator. The gene was named according to its function as a chlamydial expression regulator (chxR).
Results
Sequence Analysis of CT630. The complete genome sequences of Chlamydia trachomatis (3) and Chlamydia pneumoniae (5) revealed ORFs CT630 and CPn0750, respectively, initially designated “cpxR” based on its closest E. coli homolog by blast analysis and annotation convention. The chxR gene encodes a protein of 227 aa with similarity (27% identity) to E. coli CpxR (6) and other OmpR subfamily members. ChxR orthologs were present and conserved for all chlamydial species described to date, including C. trachomatis (3), C. pneumoniae (5), murine C. trachomatis (MoPn) (7), Chlamydia psittaci (8), and a very distantly related chlamydiae isolated from amoebae (9). The N-terminal domain of ChxR, which corresponds to the phosphorylation or receiver domain of response regulators (amino acids 1–108), lacked essential residues in the active site of phosphorylation (10), especially the conserved aspartate residues, suggesting that activation by phosphorylation was unlikely (see supporting information, which is published on the PNAS web site). The predicted phosphorylated aspartate residue was replaced with glutamate for each of the chlamydial strains except the strain isolated from amoebae, which retained the conserved aspartate; however, the amoebal strain separated phylogenetically from each of the other chlamydial strains over a billion years ago (11). The C-terminal half of the protein (amino acids 133–227) has a predicted winged helix–turn–helix motif, characteristic of the DNA-binding domain of the OmpR subfamily of response regulators (12). The homology suggests that ChxR functions as a transcription factor (13), although it is likely not differentially regulated by phosphorylation.
Developmental Expression of ChxR. Chlamydia has a unique developmental cycle dependent on stage-specific transcriptional events (14). Determining the time at which a particular gene is first transcribed can provide clues to its functional role in chlamydial development. Genome DNA array data suggested that this gene is not significantly regulated; however, the level of transcription is very low (14). Total RNA from Chlamydia-infected cells was isolated from different time points after infection, and RT-PCR was used to detect chxR-specific transcripts. A chxR transcript was detected at 10 h after infection (Fig. 1), corresponding to the time at which chlamydiae undergo active metabolism and multiplication, suggesting a likely role for ChxR as a regulator of genes important for shifts in mid-cycle developmental stage-specific processes.
Fig. 1.
RT-PCR detects chxR transcripts by 10 h after infection of L929 cells. PCR controls included reactions containing no template (–) and chlamydial DNA (+). Each RT-PCR time point was assessed by using equal amounts of RNA and was accompanied by a reaction containing no reverse transcriptase, as indicated above the gel; the times in hours postinfection (hpi) are also indicated.
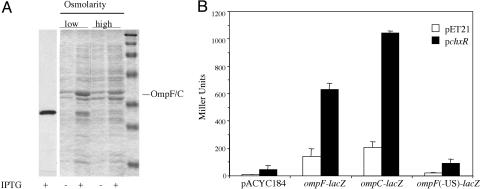
ChxR Expression and Its Interaction with the ompF and ompC Promoters. To test its function, chxR was expressed in E. coli for ChxR purification and functional assays. Interestingly, we observed that outer-membrane fractions of E. coli clones expressing ChxR had increased expression of one or more E. coli proteins of ≈40,000 Da (Fig. 2A). Their relative abundance, size, and location in the outer-membrane fraction were consistent with the likelihood that these were outer-membrane porin proteins. Because ChxR was homologous to OmpR, we reasoned that it might affect expression of the OmpR-regulated porin genes ompF and ompC in E. coli. A lacZ reporter system was used in which lacZ was placed downstream of E. coli ompF and ompC promoters and their respective upstream regulatory regions. Even in the presence of wild-type OmpR, the induction of ChxR stimulated both ompF–lacZ and ompC–lacZ expression ≈4-fold (Fig. 2B). Activation of ompF and ompC by ChxR was a surprising result; most other response regulators in the OmpR subfamily, such as PhoB, do not activate expression of the porin genes (V. Tran, D.W., and L.J.K., unpublished data). However, recent studies have shown that the CpxR/A and EnvZ/OmpR regulons overlap and that both response regulators affect porin gene expression by binding to the regulatory regions (15).
Fig. 2.
ChxR activates transcription of ompF and ompC in E. coli.(A) Coomassie brilliant blue-stained SDS/polyacrylamide gels of outer-membrane preparations of E. coli before and after induction of ChxR expression by IPTG. The outer membranes were prepared as described in Materials and Methods. E. coli harboring pChxR were grown in LB (low osmolarity) or LB containing 20% sucrose (high osmolarity) at 37°C until a OD600 of ≈0.6. No IPTG or 1 mM IPTG was added, and cells were grown for an additional 2 h before centrifugation. The first lane is an immunoblot showing induction of ChxR. The antibody used for detection is anti-hexaHIS. The molecular mass markers are 202, 116, 94, 53, 37, 29, and 20 kDa. (B) β-Galactosidase assays measured transcriptional activity from ompF–lacZ and ompC–lacZ fusions after induction of strains containing the vector alone (pET21) or chxR (pChxR). Little activity was detected in an ompF–lacZ fusion in which the upstream regulator sequences had been removed [ompF(-US)-lacZ].
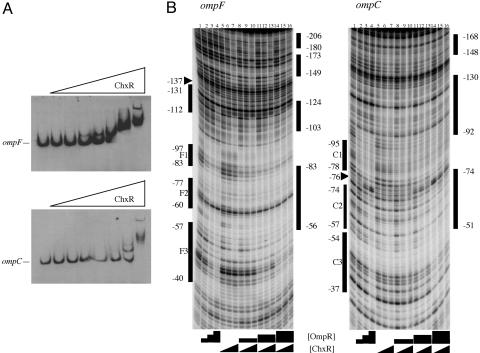
DNA Binding by ChxR. Because ChxR activated expression of the ompF–lacZ and ompC–lacZ fusions, we determined whether activation was mediated by ChxR binding to the porin regulatory regions. Electrophoretic mobility-shift assays using DNA fragments containing the ompF and ompC promoter regions and increasing concentrations of ChxR resulted in electrophoretic shifts of both ompF and ompC DNA (Fig. 3A). Multiple band-shifting patterns were observed with increasing concentrations of ChxR, suggesting the likelihood of multiple binding sites, as is typical for OmpR-like transcription factors (16–19). Furthermore, ChxR has higher affinity for ompF compared with ompC DNA (Fig. 3A).
Fig. 3.
ChxR binds ompF and ompC DNA. (A) ChxR binds DNA upstream of ompF and ompC in gel mobility-shift assays containing increasing concentrations of ChxR. (B) DNase I protection footprinting of ChxR at the ompF promoter (Left) and ompC promoter (Right). The lanes are identical with respect to proteins in each panel. Lane 1 contains a DNase I-only ladder. Lanes 2–4, 2-fold increases of phosphorylated OmpR from 31 to 125 nM; lanes 5–7, 2-fold increases of ChxR from 12.5 to 50 nM. For the remaining lanes containing various concentrations of both OmpR and ChxR, the black triangles and boxes under the panel are representative of the protein concentrations described for lanes 2–7. The numbered bars on the right of each panel represent regions of DNA protected from DNase I cleavage in the presence of ChxR. The bars on the left indicate the OmpR binding sites at each promoter. Arrows indicate hypersensitive sites observed in the presence of high OmpR concentrations.
The expression and electrophoretic mobility-shift assay results demonstrated that ChxR interacted directly with DNA sequences upstream of ompF and ompC. Thus, DNase I protection assays were performed to compare OmpR binding to ChxR binding at ompF and ompC (Fig. 3B). In lanes 2–4, the protection in the presence of increasing concentrations of OmpR is shown, whereas ChxR alone is shown in lanes 5–7. Differences in the protection pattern are clearly evident when comparing the two response regulators. At ompF, ChxR binds farther upstream (–206 to –149) than does OmpR, and ChxR does not bind as close to the promoter (only down to –56) as does OmpR (to –40).
At ompC, a pattern of ChxR protection similar to that observed at ompF was evident. Sites upstream of the high-affinity OmpR binding site C1 were protected (–168 to –148 and –130 to –92), whereas C1 (–95 to –78) is not protected. The furthest downstream site that was protected by ChxR is C2 (–74 to –51). The precise boundary of the promoter proximal site was difficult to discern because of the paucity of DNase I sites in this region (see Discussion). The footprinting results indicate that ChxR can recognize similar (but not identical) sequences compared with those bound by OmpR and are consistent with ChxR transcriptional activation of both ompF and ompC (Fig. 2B). However, the results from DNase I protection demonstrated that ChxR binds to regions of ompF and ompC that are farther upstream than the promoter-proximal OmpR site. In a previous study using OmpR mutants, it was shown that sites closest to the –35 region (ompC3 and ompF3) needed to be occupied for OmpR-dependent transcription to occur (20, 21). Either ChxR is capable of activating transcription from this upstream position on the DNA or it interacts with OmpR to activate transcription.
To determine whether OmpR and ChxR could bind simultaneously at ompF or ompC, cofootprinting experiments were performed in which the same concentrations of OmpR were used (as shown in Fig. 3B, lanes 2–4) and the ChxR concentration was varied (Fig. 3B, lanes 8–16). At every OmpR concentration, ChxR protection was evident, especially at the upstream sites and those overlapping F2 and C2. At the highest OmpR concentration, the OmpR protection pattern at F3 and C3 was observed. When both proteins were present, the protection pattern demonstrated features of each protein alone, consistent with the conclusion that both proteins were bound. Therefore, both proteins can bind simultaneously to their nonoverlapping binding sites, but they appear to compete for binding to the overlapping sites. At the ompC promoter, similar results were observed.
Genome-Wide Screen for Targets of ChxR. Having demonstrated that ChxR can function as a transcriptional activator of E. coli genes, gene targets of ChxR were sought in Chlamydia. The inability to make mutants in Chlamydia precludes such strategies as the isolation of chxR-deficient or overexpression mutants to aid in identification of its target genes. Although only a small percentage of chlamydial genes are capable of being transcribed by E. coli σ70 (unpublished observations), we used a microarray approach using mRNA isolated from in vitro transcription of chlamydial genomic DNA by E. coli σ70. The rationale was that, because ChxR was a transcriptional activator for E. coli genes when expressed in E. coli, it should activate transcription of target genes in the in vitro σ70-based system.
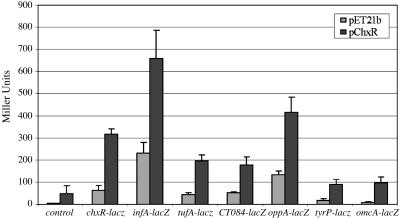
With a few exceptions, differences between transcript levels from reactions with and without ChxR were all determined to be low (≈2-fold or less), and the level of transcription did not show a dose-response with higher concentration of ChxR (see supporting information). Of the genes that appeared to be significantly up-regulated (P < 0.05), five were selected for further study (chxR, oppA, infA, tufA, and CT084), and lacZ reporter fusions were constructed. Two additional genes, tyrP and omcA, which showed almost no change in transcription in response to ChxR, were included as additional controls. The chxR-dependent activity of the tyrP–lacZ and omcA–lacZ fusions varied (28–42 Miller units), and the stimulation above that level varied from 2-fold (tufA) to 9-fold (infA). Although the tyrP–lacZ and omcA–lacZ fusions exhibited an increase in activity in the presence of chxR, this low level was not considered to be meaningful, nor was it substantially different from the control (first and second columns in Fig. 4).
Fig. 4.
ChxR activates transcription of chlamydial genes chxR, infA, tufA, CT084, and oppA. β-Galactosidase assays measured transcriptional activity from chxR–lacZ, infA–lacZ, tufA–lacZ, oppA–lacZ, and CT084–lacZ. The lacZ reporter gene was cloned downstream of each gene and expressed in an E. coli strain containing IPTG-inducible chxR on a compatible plasmid. The background activity in the presence of the vector alone is shown in the lightly shaded bars, and the activity in the presence of chxR is shown in the darker bars. The standard error of the mean is indicated by the error bars; measurements were performed in triplicate. The “control” shown was the pACYC184 vector lacking a promoter region in front of the lacZ gene.
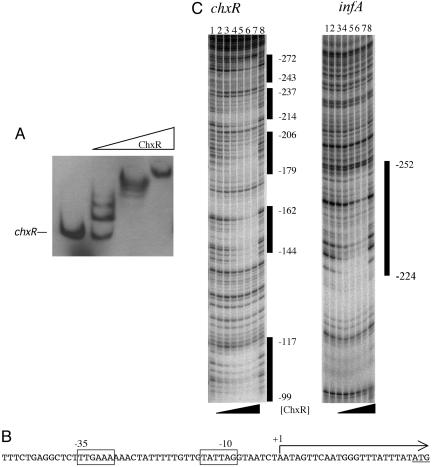
ChxR Activates Transcription of chxR. One of the most highly regulated genes was chxR itself. It displayed an 11-fold change over transcripts produced in the absence of ChxR, suggesting that chxR is autoregulated. The in vitro microarray results were confirmed by RT-PCR (data not shown) and by testing promoter–lacZ reporter fusions in E. coli (Fig. 4). To determine whether ChxR was capable of binding to the regulatory regions of chxR, electrophoretic mobility-shift assays were performed. Increasing concentrations of ChxR added to DNA sequences upstream of chxR resulted in a mobility shift (Fig. 5A), indicating that ChxR acts directly to activate its own transcription by binding to DNA. Like the interaction with ompC and ompF, ChxR showed multiple shifted species, suggesting the existence of multiple binding sites at the regulatory region.
Fig. 5.
ChxR binds and activates transcription of chlamydial chxR. (A) ChxR binds DNA upstream of chlamydial genes in gel mobility-shift assays containing increasing concentrations of ChxR. Arrows indicate shifted complexes. (B) Transcriptional start site (+1) for chxR extended 23 bp upstream of the translation initiation codon (underlined). –10 and –35 promoter consensus sequences are boxed. (C) DNase I protection footprinting of ChxR at the chxR (Left) and infA (Right) promoters. (Left) Lanes 1 and 8 contain DNase I-only ladders. Lanes 2–6 show 2-fold increases of ChxR from 450 nM to 7.2 μM. Lane 7 contains 10.2 μM. (Right) Lanes 1 and 8 show DNase I-only ladders. Lanes 2–7 show 2-fold increases of ChxR from 225 nM to 7.2 μM. The numbers to the right of each panel represent the coordinates of ChxR protection from the transcription and translation start sites of chxR and infA, respectively.
The transcriptional start site for chxR in Chlamydia was determined and thus permitted the prediction of the promoter region that contains a σ70-like –10 and –35 promoter sequence (Fig. 5B). Interestingly, despite similarity to σ70 promoters, there was little transcription without ChxR in E. coli or in vitro. To identify the precise regions of ChxR binding, a DNase I protection assay was performed by using chxR and infA, because infA was also highly up-regulated (Fig. 4). ChxR protected five distinct regions spanning a total of 173 nucleotides upstream of the chxR coding sequence. At infA, ChxR protected one region between –252 and –224 upstream from the translational start site. When the regions protected by ChxR at the chxR and infA regulatory regions were aligned, there was some limited homology to OmpR binding sites.
Discussion
Two-component signal transduction systems enable microorganisms to sense changes in the external environment and translate this input into changes in transcriptional response. Chlamydiae are deeply separated from other bacteria (22) and have been genetically isolated from other bacteria for hundreds of millions of years (11). Nevertheless, chlamydiae contain a conserved ortholog of the OmpR subfamily of response regulators, called ChxR. The function of OmpR in gene regulation is complex and is not completely understood (12). Recent evidence suggests that other subfamily members, such as CpxR, may additionally modulate OmpR regulation of target genes by competitive or cooperative interaction at DNA-regulatory binding sites (15, 23).
The N terminus of ChxR has limited similarity to receiver domains of response regulators. The essential residues in the catalytic site are not conserved, suggesting that ChxR is not a classical response regulator. Moreover, no cognate histidine kinase was identified for ChxR in the chlamydial genome. Only one histidine kinase-response regulator pair, CtcB–CtcC, has thus far been identified (4). The presence of a glutamate residue in place of the conserved phosphorylated aspartate suggests that ChxR inherently mimics the structure of an activated response regulator, enabling it to function constitutively (24). Thus, ChxR is a transcriptional activator whose OmpR-like DNA-binding domain is located in the C-terminal half of the protein, but whose N-terminal domain is unique in comparison to most other previously characterized activators.
OmpR binding sites are not well conserved; aligning the areas of DNase I protection by OmpR and ChxR identifies few common features, apart from A-T-rich regions. No consensus sequence for ChxR binding was identified from the DNase I protection assays. At ompF, ChxR protects three regions upstream from where OmpR binds (–206 to –180, –173 to –149, and –124 to –103). The high-affinity OmpR binding site F1 (–97 to –83) is not protected by ChxR, but the F2 site (–83 to –56) is protected by ChxR. Unlike OmpR, ChxR does not protect closer to the RNA polymerase binding sites (e.g., C3 and F3); these are sites where OmpR is required to bind to maximally activate transcription (20, 25).
If the ChxR2 binding site is compared with the 18-bp OmpR binding sites at ompF and ompC, the best fit is to C3, a very-low-affinity site, in which 12 of 18 bases are identical (see supporting information). However, ChxR did not protect at this site (Fig. 5), suggesting that the variable nucleotides might be important for making base contacts with ChxR.
A commonly held view now being challenged is that homologous response regulators activate a unique repertoire of genes (ref. 15 and this work). Of special interest was that ChxR activated transcription of E. coli genes ompF and ompC but protected regions of ompF and ompC that were shifted with respect to the OmpR binding sites. Activation by ChxR was observed in E. coli in the presence of OmpR and in vitro at chlamydial promoters without OmpR, suggesting that ChxR binds promoter-proximal activating sites at ompF and ompC and activates transcription. Genetic evidence indicates that OmpR activates transcription by interaction with RpoA, the α-subunit of RNA polymerase (26–28), and it is tempting to speculate that ChxR might employ a similar mechanism of activation. However, members of the same subfamily of response regulators can interact with different subunits to activate transcription (26, 29, 30). The fact that ChxR activated transcription in the presence of OmpR and that their DNA interaction sites overlapped suggests that ChxR and OmpR may interact or that they compete with one another for binding. Similar findings have also been reported for CpxR regulation of the porin genes (15).
The replacement of active-site aspartate residues with glutamate suggests that ChxR activity is not regulated by posttranslational activation and, when present, is capable of binding DNA and activating transcription. Although autoactivation of chxR provides an explanation for its regulation of expression during the midcycle phase of chlamydial growth, how chxR transcription is initially activated is unknown. Because a large percentage of the chlamydial genome is activated at the same time as chxR (14), there may exist a common mechanism of transcriptional control for this stage that is presently undefined. Alternatively, small amounts of ChxR may be produced by low basal transcription until a critical level is reached, allowing for optimal transcriptional activation by ChxR. Transcripts of chxR were first detected in Chlamydia by 10 h after infection, corresponding to the midcycle metabolic burst (14). ChxR has been conserved throughout the chlamydial phylogenetic tree. Thus, it is an essential protein and, by temporal association, is likely responsible for activating a subset of genes necessary for midcycle stage-specific processes essential for chlamydial development. The regulatory role for ChxR in chlamydiae is enigmatic; however, if ChxR regulates gene expression required for developmental stage progression, it likely represents a key regulator for modulating chlamydial persistence in vivo.
Materials and Methods
Bacterial Strains. C. trachomatis serovar D was propagated in monolayers of HeLa 229 cells, and serovar L2 was propagated in L929 cells in suspension cultures. RPMI medium 1640 (Invitrogen) was supplemented with 5–10% heat-inactivated FBS plus 50 μg/ml vancomycin. Chlamydial organisms were isolated by sonic treatments of cell suspensions and purified by ultracentrifugation as described in ref. 31.
Construction of C. trachomatis chxR Expression Vector. chxR from C. trachomatis serovar D was PCR-amplified by using primers 5′-GAATTCATGCAGGGCCTAAACATGTG and 5′-CTCGAGAGAAAGCTTTGTATCTTGTTGAGAG (Operon, Valencia, CA). The product was cloned into the TOPO-TA vector (Invitrogen) and digested with EcoRI and XhoI, and excised chlamydial DNA was cloned into pET21 (Novagen). pChxR expressed from the predicted first methionine was very insoluble; thus, several ChxR clones were selected, and ChxR initiating translation at the first methionine residue in the vector (containing the T7 tag sequence) was significantly more soluble and was used for most experiments. The translational start (MASMTGG) was confirmed by N-terminal sequencing of purified ChxR from E. coli (Protein Sequencing Facility, Columbia University, New York). Outer-membrane fractions of E. coli were prepared as described in ref. 32.
Purification of ChxR. pChxR was transformed into E. coli BL21(DE3) grown at 37°C in 100 ml of Luria broth with 100 μg/ml ampicillin to an OD600 of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was added, and cells were harvested after 2 h. ChxR was purified on a nickel resin column (Novagen) or a HiTrap HP column (Amersham Pharmacia). E. coli harboring pChxR were grown in LB (low osmolarity) or LB containing 20% sucrose (high osmolarity) at 37°C to an OD600 of 0.6. No IPTG or 1 mM IPTG was added, and cells were grown for 2 h. Pellets were frozen and sonicated, and proteins were separated on a SDS polyacrylamide gel before Coomassie blue staining.
Construction of lacZ Reporter Fusions. Transcriptional fusions of promoter regions to lacZ were constructed by placing 100–200 bp of DNA sequence upstream of the E. coli ompF and ompC transcriptional start sites adjacent to a promoterless lacZ gene in pACYC184 (New England Biolabs, Beverly, MA). E. coli lacZ gene was amplified with Amplitaq-long (Stratagene) by using primers 5′-GCATGCATGACCATGATTACGGATTC and 5′-GTCGACTTATTTTGACACCAGACC. Primer pairs used for amplifying were as follows: ompF, 5′-GGATCCGACGGTGTTCACAAAGTTCC and 5′-GTCGACTATTTATTACCCTCATGG; ompF (US), 5′-GGATCCCGGTAGCGAAACGTTAGTTT and 5′-GTCGACTATTTATTACCCTCATGG; ompC,5′-AAGCTTGCTTATTTCGCCATTCCGA and 5′-GTCGACGTTATTAACCCTCTGTTATA. Each lacZ reporter fusion was cotransformed with pChxR into BL21(DE3). Overnight cultures were subcultured in Luria broth, and chxR expression was induced for 2 h with 1 mM IPTG at an OD600 of 0.6. β-Galactosidase activity was measured as described in ref. 33.
For chlamydial genes, transcriptional fusions were constructed by placing the upstream regions of chlamydial chxR, infA, tufA, CT084, oppA, tyrP, and omcA adjacent to the lacZ gene in pACYC184. The following primers were used for amplification: chxR, 5′-AAGCTTTCAACGGCTATAGAAGCTATAG and 5′-GCATGCAACCCATTGAACTATTAGATTAC; CT084, 5′-AAGCTTAAATCTTGTTTCTTCTCGCTG and 5′-CATGCTTGATTGTGTTTAQGCTCCTTG; oppA, 5′-AAGCTTTATGCCCAGACTGAGCAGAA and 5′-GCATGCTCAGCAAACAATCAAATAATGTTG; infA, 5′-AAGCTTCATGGCCTATCTACTCTAC and 5′-GCATGCAACATTCTATCTCTTGATCCC; tufA, 5′-AAGCTTTACAGACATCGTTAAGGTTG and 5′-GCATGCAAATTAGTTTGCTACCAATAATC; tyrP, 5′-AAGCTTTTGCCCAGAAGTTTCTAGG and 5′-GCATGCATTCCGCHXRCACATCCTC; omcA, 5′-AAGCTTCTTCCAGACTCCTTTCTAG and 5′-GCATGCTGAGACAATTCTTCAAGACTTG.
chxR Transcription in Chlamydia. C. trachomatis serovar L2 was used to infect 8 × 105 L929 cells per ml at a multiplicity of infection of 1 in a 1-liter flask. Total RNA was extracted from infected cells at 0, 4, 10, 24, and 48 h after infection by using TRIzol reagent (Invitrogen), followed by DNase I (Roche Molecular Biochemicals, Indianapolis) treatment. Primer pairs were designed according to predicted ORFs from the C. trachomatis serovar D genome and used to amplify regions of ≈400 bp within coding sequences. The euo and omcB genes were used as early and late expression controls, respectively. cDNA synthesis was performed by using reverse transcriptase (Invitrogen) and 1 μg of RNA for each time point at 37°C for 1 h. PCRs used Amplitaq polymerase (PerkinElmer) and consisted of 30 cycles of 95°C, 52–55°C, and 72°C. Positive chlamydial DNA controls were used for each primer pair. The chxR transcriptional start site in Chlamydia was determined by using the 5′ RACE system (Invitrogen). cDNA was generated from RNA isolated from purified C. trachomatis organisms 36 h after infection (14) by using SuperScript III (Invitrogen), annealing at 55°C, and the chxR 3′ RACE primer 5′-GAAAGAAACAAGCTGATTGCGG. Terminal deoxynucleotidyltransferase reactions were incubated at 37°C, 30 min before heat inactivation (65°C, 10 min). Primary PCR used 30 cycles of 94°C, 50°C, and 72°C, each for 30 seconds, using the chxR-nested RACE primer 5′-GAATTAAATACCAAAAACCACGATCC. Secondary PCRs (30 μl) used 2 μl of a 1:500 dilution of the primary PCR (annealing at 60°C). Amplicons were separated on a 1.8% agarose gel, and DNA was isolated from excised bands (Qiagen, Valencia, CA). Direct sequencing of amplicons was performed by using RACE nested primer. The 5′ end of the RNA was confirmed by using amplicons that were cloned into pTOPO-TA 2.1 and by sequencing individual clones.
In Vitro Transcription and Microarray Analysis. In vitro transcription reactions were performed by adding transcription buffer (40 mM Tris-Cl, pH 8.0/10 mM MgCl2/5mMDTT/50 mM KCl/50 μg/ml BSA), 0.5 mM ribonucleoside NTPs, and 20 units of RNasin (Promega) to 2 μg of EcoRI-digested C. trachomatis L2 genomic DNA. Eσ70 (178 nM; Epicentre, Madison, WI) was added to 0, 160 nM, or 1.6 μM ChxR and incubated for 60 min at 37°C. DNA was removed by the addition of 20 units of RQ1 DNase (Promega) and RNA-purified using RNeasy columns (Qiagen). Eluted RNA was fluorescently labeled and hybridized to chlamydial DNA microarrays as described in ref. 11.
Electrophoretic Mobility-Shift Assays. TOPO-TA vectors containing ompF and ompC upstream regulatory regions were digested with HindIII and SphI (New England Biolabs), and each fragment was gel-purified. Fragments were digoxigenin-labeled by using 3′-end labeling (Roche, Indianapolis). Reactions containing 10 ng of DNA and increasing concentrations of ChxR (16 nM, 72 nM, 160 nM, 720 nM, 1.6 μM, 7.2 μM, and 16 μM) in reaction buffer {40 mM Tris-Cl, pH 8.0/0.1 mM EDTA/100 mM NaCl/250 mM KCl/1mM DTT/1 μg of poly[d(I-C)]/10% glycerol} were incubated at 30°C for 15 min. Reactions were mixed with 2 μl of 50% sucrose and separated by electrophoresis in 5% TBE gel, 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 50 V for 2 h. Gels were transferred to nylon membranes in an electrotransblotter in 0.5× TBE at 400 mA for 40 min. After UV-crosslinking DNA to membranes, digoxigenin-labeled DNA was detected by using digoxigenin-specific Fab antibody conjugated to alkaline phosphatase (1:10,000), and immune reactions were detected by chemiluminescence (Roche).
DNase I Protection Assays. Genomic DNA from C. trachomatis serovar L2 was used to amplify the chxR and infA promoters. Plasmids pDW99 (chxR) and pDW110 (infA) served as PCR templates to generate the DNase I footprinting templates. Before PCR, the oligonucleotide was phosphorylated with [γ-32P]ATP (3000 Ci/mmol, PerkinElmer) by using T4 polynucleotide kinase (New England Biolabs). PCR products were purified with QIA-quick columns and a 5% TAE polyacrylamide gel. DNA was extracted from polyacrylamide fragments by using QIAex II gel purification (Qiagen). Template DNA was quantified, and 30,000 cpm was used per reaction. Binding reactions were performed in 12% glycerol, 4 mM Tris (pH 7.6), 20 mM KCl, 2 mM EDTA (pH 8.0), 1 mM DTT, and 1 μg of poly[d(I-C)] (Roche) for 10 min at room temperature. DNase I (Roche) was diluted in binding buffer containing 10 mM MgCl2 and added to each reaction for 30 seconds (0.1 units of DNase I/30,000 cpm labeled DNA). Cleavage was stopped by the addition of 0.375 M sodium acetate (pH 5.2), 20 mM EDTA, and 0.5 μg of glycogen, and ethanol precipitation followed. DNase I reactions were resolved on an 8% urea-TBE acrylamide gel with sequencing ladders generated by Thermo Sequenase Cycle Sequencing Kit (USB, Swampscott, MA). DNase I footprinting templates of the ompF and ompC promoters were generated as described in ref. 21 and performed as above, except that 3 μg of poly[d(I-C)] was used in each binding reaction.
Supplementary Material
Acknowledgments
We thank T. Nicholson for microarray analysis and C. Shields for technical support of this research. This research was supported by National Institutes of Health Fellowship F32-GM68364 (to D.W.), National Institutes of Health Grants AI042156 (to R.S.S.) and GM 58746 (to L.J.K.), and National Science Foundation Grant MCB0243085 (to L.J.K.).
Conflict of interest statement: No conflicts declared.
Abbreviation: IPTG, isopropyl-β-d-thiogalactopyranoside.
References
- 1.Parkinson, J. S. & Kofoid, E. C. (1992) Annu. Rev. Genet. 26, 71–112. [DOI] [PubMed] [Google Scholar]
- 2.Hatch, T. P. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity, ed. Stephens, R. S. (Am. Soc. for Microbiology, Washington, DC), pp. 29–67.
- 3.Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L., Zhao, Q., et al. (1998) Science 282, 754–759. [DOI] [PubMed] [Google Scholar]
- 4.Koo, I. C. & Stephens, R. S. (2003) J. Biol. Chem. 278, 17314–17319. [DOI] [PubMed] [Google Scholar]
- 5.Kalman, S., Mitchell, W., Marathe, R., Lammel, C., Fan, J., Hyman, R. W., Olinger, L., Grimwood, J., Davis, R. W. & Stephens, R. S. (1999) Nat. Genet. 21, 385–389. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., Plunkett, G. R., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 7.Read, T. D., Brunham, R. C., Shen, C., Gill, S. R., Heidelberg, J. F., White, O., Hickey, E. K., Peterson, J., Utterback, T., Berry, K., et al. (2000) Nucleic Acids Res. 28, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Read, T. D., Myers, G. S., Brunham, R. C., Nelson, W. C., Paulsen, I. T., Heidelberg, J., Holtzapple, E., Khouri, H., Federova, N. B., Carty, H. A., et al. (2003) Nucleic Acids Res. 31, 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn, M., Collingro, A., Schmitz-Esser, S., Beier, C. L., Purkhold, U., Fartmann, B., Brandt, P., Nyakatura, G. J., Droege, M., Frishman, D., et al. (2004) Science 304, 728–730. [DOI] [PubMed] [Google Scholar]
- 10.Stock, J. B., Ninfa, A. J. & Stock, A. M. (1989) Microbiol. Rev. 53, 450–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens, R. S. (2002) in Chlamydial Infections: Proceedings of the Tenth International Symposium on Human Chlamydial Infections, eds. Schachter, J., Christiansen, G., Clarke, I. N., Hammerschlag, M. R., Kaltenboeck, B., Kuo, C. C., Rank, R. G., Ridgway, G. L., Saikku, P., Stamm, W. E., et al. (Int. Chlamydial Symp., San Francisco), pp. 3–12.
- 12.Kenney, L. J. (2002) Curr. Opin. Microbiol. 5, 135–141. [DOI] [PubMed] [Google Scholar]
- 13.Bajaj, V., Hwang, C. & Lee, C. A. (1995) Mol. Microbiol. 18, 715–727. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson, T. L., Olinger, L., Chong, K., Schoolnik, G. & Stephens, R. S. (2003) J. Bacteriol. 185, 3179–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batchelor, E., Walthers, D., Kenney, L. J. & Goulian, M. (2005) J. Bacteriol. 187, 5723–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, K. J. & Igo, M. M. (1996) J. Mol. Biol. 262, 615–628. [DOI] [PubMed] [Google Scholar]
- 17.Head, C. G., Tardy, A. & Kenney, L. J. (1998) J. Mol. Biol. 281, 857–870. [DOI] [PubMed] [Google Scholar]
- 18.Harlocker, S. L., Bergstrom, L. & Inouye, M. (1995) J. Biol. Chem. 270, 26849–26856. [DOI] [PubMed] [Google Scholar]
- 19.Rampersaud, A., Harlocker, S. L. & Inouye, M. (1994) J. Biol. Chem. 269, 12559–12566. [PubMed] [Google Scholar]
- 20.Mattison, K., Oropeza, R. & Kenney, L. J. (2002) J. Biol. Chem. 277, 32714–32721. [DOI] [PubMed] [Google Scholar]
- 21.Mattison, K., Oropeza, R., Byers, N. & Kenney, L. J. (2002) J. Mol. Biol. 315, 497–511. [DOI] [PubMed] [Google Scholar]
- 22.Pace, N. R. (1997) Science 276, 734–740. [DOI] [PubMed] [Google Scholar]
- 23.Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., Lejeune, P. & Dorel, C. (2005) J. Bacteriol. 187, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klose, K. E., Weiss, D. S. & Kustu, S. (1993) J. Mol. Biol. 232, 67–78. [DOI] [PubMed] [Google Scholar]
- 25.Maeda, S., Takayanagi, K., Nishimura, Y., Maruyama, T., Sato, K. & Mizuno, T. (1991) J. Biochem. 110, 324–327. [DOI] [PubMed] [Google Scholar]
- 26.Slauch, J. M., Russo, F. D. & Silhavy, T. J. (1991) J. Bacteriol. 173, 7501–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo, F. D., Slauch, J. M. & Silhavy, T. J. (1993) J. Mol. Biol. 231, 261–273. [DOI] [PubMed] [Google Scholar]
- 28.Bowrin, V., Brissette, R., Tsung, K. & Inouye, M. (1994) FEMS Microbiol. Lett. 115, 1–6. [DOI] [PubMed] [Google Scholar]
- 29.Igarashi, K. & Ishihama, A. (1991) Cell 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- 30.Makino, K., Amemura, M., Kim, S. K., Nakata, A. & Shinagawa, H. (1993) Genes Dev. 7, 149–160. [DOI] [PubMed] [Google Scholar]
- 31.Koehler, J. E., Burgess, R. R., Thompson, N. E. & Stephens, R. S. (1990) J. Biol. Chem. 265, 13206–13214. [PubMed] [Google Scholar]
- 32.Nikaido, H. (1994) Methods Enzymol. 235, 225–234. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Woodbury, NY).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.