Abstract
Adult hippocampal neurogenesis is highly variable and heritable among laboratory strains of mice. Adult neurogenesis is also remarkably plastic and can be modulated by environment and activity. Here, we provide a systematic quantitative analysis of adult hippocampal neurogenesis in two large genetic reference panels of recombinant inbred strains (BXD and AXB/BXA, n = 52 strains). We combined data on variation in neurogenesis with a new transcriptome database to extract a set of 190 genes with expression patterns that are also highly variable and that covary with rates of (i) cell proliferation, (ii) cell survival, or the numbers of surviving (iii) new neurons, and (iv) astrocytes. Expression of a subset of these neurogenesis-associated transcripts was controlled in cis across the BXD set. These self-modulating genes are particularly interesting candidates to control neurogenesis. Among these were musashi (Msi1h) and prominin1/CD133 (Prom1), both of which are linked to stem-cell maintenance and division. Twelve neurogenesis-associated transcripts had significant cis-acting quantitative trait loci, and, of these, six had plausible biological association with adult neurogenesis (Prom1, Ssbp2, Kcnq2, Ndufs2, Camk4, and Kcnj9). Only one cis-acting candidate was linked to both neurogenesis and gliogenesis, Rapgef6, a downstream target of ras signaling. The use of genetic reference panels coupled with phenotyping and global transcriptome profiling thus allowed insight into the complexity of the genetic control of adult neurogenesis.
Keywords: gene array, hippocampus, precursor, quantitative trait loci, stem cell
Neurogenesis in adult mammals is modulated by complex interactions among genetic and environmental factors. Our goal is to use a systems genetics approach to determine how the development of new neurons and glia is modulated by gene polymorphisms, activity, and environmental stimuli. A first step toward this goal is to analyze natural variation and genetic covariance among key neurogenesis parameters such as proliferation, survival, and differentiation of new cells. We have shown that natural variation is substantial among different strains of mice (1–3). The normal range of variation often exceeds the effects of single gene mutations in engineered lines of mice.
In this study, we extended this analysis of normal variation and applied a systems genetics approach to study the covariance structure of four key parameters of adult hippocampal neurogenesis across two large genetic reference populations consisting of a total of 52 recombinant inbred strains. This approach allowed us to exploit covariance of diverse traits to demonstrate biological linkage and pleiotropy (4, 5). The analysis, however, extends beyond a correlative approach. Both of the genetic reference populations used, the BXD and AXB/BXA sets, are standard mapping panels, making it possible to search for gene loci, so-called quantitative trait loci (QTL), that produce common variation and pleiotropy among traits linked to adult hippocampal neurogenesis.
Adult hippocampal neurogenesis originates from proliferating neural-precursor cells in the adult dentate gyrus and proceeds over a number of identifiable stages to the long-term survival of a new granule cell (6–8). In the course of this development, two key phases can be distinguished: an expansion phase, in which the number of immature postmitotic neurons is increased, followed by a phase of selective, long-lasting survival (9, 10).
Strain differences in adult hippocampal neurogenesis in mice are observed in cell proliferation and survival (3), the distribution of phenotypes among the newly generated cells (1, 2), and how different stages of neuronal development are activated by stimuli that induce adult neurogenesis (11). Strains DBA/2J and A/J show particularly large differences in adult neurogenesis from the canonical mouse strain, C57BL/6J (1). In this study, we explored natural variation in adult hippocampal neurogenesis in two complementary sets of recombinant inbred (RI) strains of mice (BXD and AXB/BXD), which represent inbred progeny of F2 crosses from C57BL/6J and DBA/2J (BXD), and C57BL/6J and A/J (AXB/BXA) (12–14). On average, and in large panels, each RI strain shows an ≈50:50 pattern of genome inherited from the parental strains and is homologous at every locus. The GeneNetwork (www.genenetwork.org) is an open-access resource that contains genomic and phenotypic information on RI strains (15). Complex traits like neuronal development are likely to show associations with many gene loci. The high number of candidate genes and the lack of sufficiently large breeding panels to increase mapping precision still limit the use of RI strains for the direct identification of quantitative trait genes (16, 17). The combination of expression genetics with classical linkage analysis, however, allows the in silico identification of candidate genes controlling polygenic phenotypes as complex as adult neurogenesis and, at the same time, reveals insights into regulatory transcriptional networks underlying such phenotypes (18).
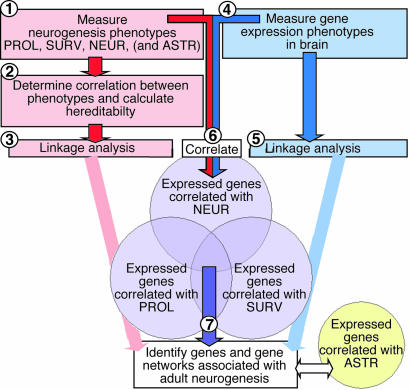
Genetic polymorphisms influence systems-level phenotypes through a network of genes. The small molecular variation is a naturally occurring perturbation of this network that can reveal the genes that comprise it. Discovering this network and the consequences of this variation are facilitated by the use of genetic reference populations. This systems-genetics approach allows one to relate gene-expression data to phenotypes across biological scale. Adult neurogenesis is a phenotype that is particularly amenable to this type of dissection. Fig. 1 outlines the experimental design and the rationale for our study.
Fig. 1.
Flow-chart diagram of the experimental design and the underlying rationale. The study combines a classical linkage study with expression genetics, that is, the genetics of how genes control genes. At the end of the present experiment stands a number of genes whose variation in expression across the BXD breeding panel is correlated with adult hippocampal neurogenesis.
Results
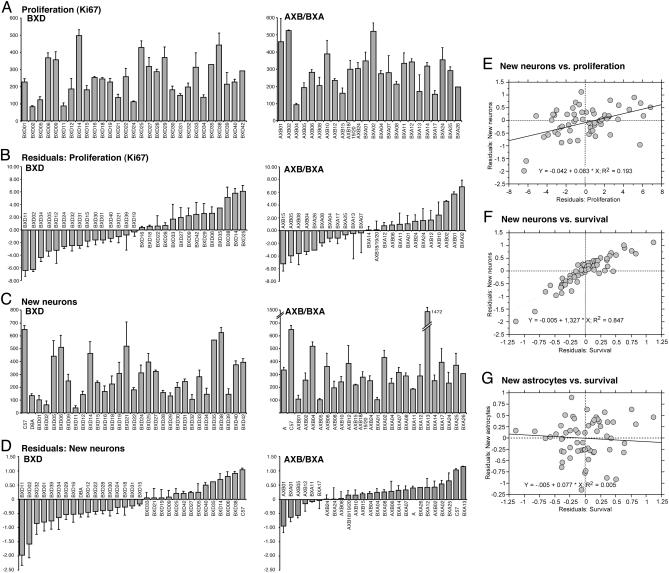
Strain Differences in Adult Neurogenesis. We quantified adult hippocampal neurogenesis in 29 BXD and 23 AXB/BXA strains and in the three parental strains, C57BL/6J, DBA/2J, and A/J (see Table 2, which is published as supporting information on the PNAS web site). Proliferation (PROL) was assessed by the absolute number of Ki67-positive cells in the subgranular zone of the dentate gyrus. Neurogenesis was measured by determining the fraction of cells among the number of surviving cells that expressed neuronal marker NeuN 4 weeks after labeling with the proliferation marker BrdUrd. We found large strain differences in both PROL (Fig. 2A) and the number of new neurons (NEUR) (Fig. 2C) as well as in surviving (SURV) and the number of new astrocytes (ASTR) (see Fig. 5, which is published as supporting information on the PNAS web site). Because age is a strong negative regulator of adult hippocampal neurogenesis (19, 20), all data were corrected for age effects (Figs. 2 B and D and 5). A general linear model fitting age was run for each phenotype, and residuals were obtained. Each residual distribution was assessed for normality (Kolmogorov–Smirnov's D, n = 174, P < 0.01). Data were transformed, and modeling was rerun where necessary to satisfy assumptions of normality. Cell counts of SURV, NEUR, and ASTR were log transformed. PROL numbers were square-root transformed. Residual diagnostics revealed homoscedastic residuals to be normally distributed about 0 and uncorrelated with age (Fig. 2 B and D).
Fig. 2.
PROL and number of new NEUR in BXD and AXB/BXA strains of mice. (A) The raw data as assessed by Ki67 immunohistochemistry in the subgranular zone of the dentate gyrus. (B) The residuals after correction for age effects (square transformation). (C) The raw data as assessed by BrdUrd and NeuN immunohistochemistry in the subgranular zone of the dentate gyrus, 4 weeks after the BrdUrd injections. (D) The residuals after correction for age effects (log transformation). The analogous information for SURV and ASTR is found in Fig. 5. (E–G), Predictive power of PROL and SURV for NEUR and ASTR. Regression analysis of the residuals was performed. (E) Cell proliferation was significantly correlated with the number of new NEUR (P = 0.0006), but r2 = 0.193. (F) SURV, as assessed by the total number of BrdUrd-labeled cells, irrespective of their phenotype, was also significantly correlated with the number of new NEUR(P <0.001), but, here, r2 = 0.847, indicating a high predictive power of SURV for NEUR. (G) This link was not apparent for ASTR, where SURV was not correlated with NEUR (P = 0.5628; r2 = 0.005).
Assessing the Predictive Power of BrdUrd-Based Numbers. We had proposed that proliferation in the subgranular zone is a poor predictor of net neurogenesis (2). There was a significant correlation between PROL and NEUR (P = 0.0019) with a coefficient of determination (R2) of 0.19 (Fig. 1E). Thus, PROL explained only 20% of the variance in NEUR. The correlation between SURV and NEUR was stronger (R2 = 0.85). At 4 weeks after BrdUrd, 85% of the variance in NEUR was explained by different rates of survival, and only 15% was explained by other factors. This same link did not exist for ASTR (P = 0.58; R2 = 0.005; Figs. 2G and 5).
Heritability of Variation in Adult Hippocampal Neurogenesis. We hypothesized that regulation of adult hippocampal neurogenesis has a strong heritable component (3). If heritability is high, the likelihood that a genetic source of variation can be identified will be increased. Heritability was calculated as the ratio of between-strain variance to total variance (between-strain variance plus withinstrain variance) for each of the four phenotypes. Heritability for PROL was 0.53 ± 0.06 (T ± standard error), for SURV 0.68 ± 0.05, for NEUR 0.70 ± 0.05, and for ASTR 0.23 ± 0.04. These data indicate that ≈70% of the variation in neurogenesis in the hippocampus is accounted for by heritable traits, compared with only 23% of the variation in gliogenesis.
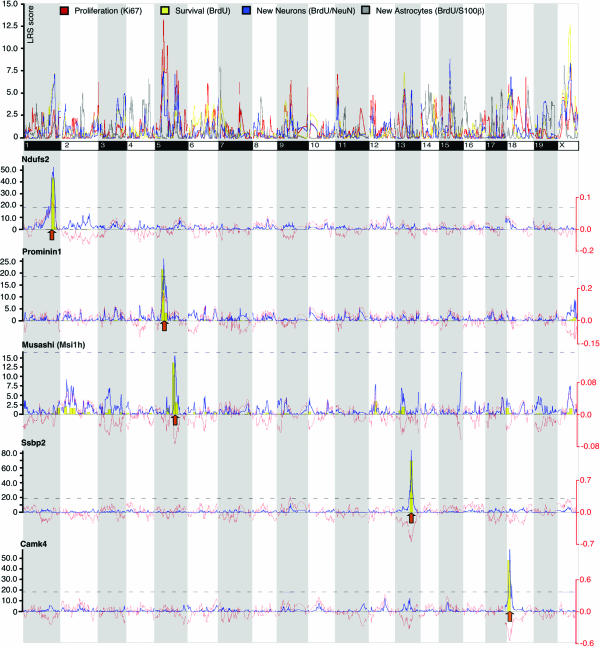
Genome-Wide Interval Mapping for Neurogenesis Phenotypes. We generated genome-wide linkage maps for residuals of PROL, SURV, NEUR, and ASTR with WebQTL (Fig. 3). In this study, we did not pursue the identification of QTL associated with adult neurogenesis but focused on phenotypic covariance. The similarities and dissimilarities of the four curves reflect the genetic covariance associated with the phenotypic covariance. As expected, the curve for ASTR differed the most from the other three, whereas SURV and NEUR showed a large overlap. The overall pattern with many overlapping peaks implies that there will not be strong QTL with independent effects specific to, for example, only PROL or only SURV. Rather, the interaction between many shared loci and many shared genes will determine the phenotypes. The distinct histological phenotypes are closely linked genetically. By using a “forward genetics” approach that allows us to relate geneexpression data to measured phenotypes, we next tried to elucidate the molecular bases underlying phenotypic covariance.
Fig. 3.
Interval mapping for PROL, SURV, NEUR, and ASTR. Age-corrected residuals for the three phenotypes were analyzed in WebQTL. The entire genome is depicted from the first base of chromosome 1 on the left to the last base on chromosome X on the right. The colored lines indicate the LRS score at each genome segment. Below are interval mappings for five of the cis-acting candidates with significant QTL colocalizing with peaks in the linkage curves for the neurogenesis phenotypes. The yellow bars are results of a bootstrap analysis; higher bars indicate higher stability of the LRS score against random permutations of the data. LRS levels of significance (P < 0.05) were determined for the individual data sets and marked by the dashed lines. The thin red line shows additive effects: Negative values indicate an influence of the C57 allele, positive values of DBA.
Covariance of mRNA Expression in the Brain. The GeneNetwork database contains data from a microarray analysis of whole brain in which mRNA levels were treated as quantitative traits (4). We correlated our four phenotypes with mRNA-expression data in a new expression database referred to as “INIA Brain mRNA M430 (Apr05) RMA.” Above a correlation threshold of 0.5 (Pearson), we found the expression of 190 genes correlated with two or more of our phenotypes (see Table 3, which is published as supporting information on the PNAS web site).
Cis-acting genes are self-regulating genes and tend to have a large, often monogenic, effect on trait variation. We performed interval mapping for all 190 transcripts and found a total of 21 to be cis-acting (Table 1). The criterion for cis action was whether, in genome-wide linkage analysis, a peak with a likelihood ratio statistic (LRS) score above 8.0 was found within 10 Mb of the physical location of the probe set.
Table 1. Cis-acting transcripts covarying with adult neurogenesis.
| Name | Symbol | Description | Ch. | Position | PROL | SURV | NEUR |
|---|---|---|---|---|---|---|---|
| PROL, SURV, NEUR | |||||||
| 1 | 1444667_at_B Brdt | bromodomain, testis-specific | 5 | 106.44 | -0.6146 | -0.7413 | -0.8008 |
| 11 | 1444306_at_B Msi1h | Musashi homologue 1(Drosophila) | 5 | 114.56 | 0.5342 | 0.6618 | 0.6387 |
| 39 | 1449435_at_A B4galt3 | UDP-Gal:betaGlcNAc β 1,4-galactosyltransferase, polypeptide 3 | 1 | 171.21 | 0.5123 | 0.5405 | 0.5667 |
| SURV, NEUR | |||||||
| 2 | 1434933_at_B 5730557L09Rik | RIKEN cDNA 5730557L09 gene | 1 | 160.88 | 0.7043 | 0.6278 | |
| 16 | 1417024_at_A Hars | histidyl-tRNA synthetase | 18 | 36.99 | -0.5944 | -0.5821 | |
| 26 | 1429951_at_B Ssbp2 | single-stranded DNA binding protein 2 | 13 | 87.70 | 0.5576 | 0.5871 | |
| 32 | 1453170_at_B 2610206C24Rik | RIKEN cDNA 2610206C24 gene | 18 | 32.15 | -0.5677 | -0.5614 | |
| 37 | 1420800_a_at_A Kcnq2 | potassium voltage-gated channel, subfamily Q, 2 | 2 | 180.83 | 0.5795 | 0.5346 | |
| 52 | 1451096_at_A Ndufs2 | NADH dehydrogen, (ubiquinone) Fe-S protei 2 | 1 | 171.16 | -0.5561 | -0.5364 | |
| 58 | 1439843_at_B Camk4 | calcium/calmodulin-dependent protein kinase IV | 18 | 33.42 | 0.5136 | 0.5708 | |
| 70 | 1459001_at_B Vps33a | vacuolar protein sorting 33A (yeast) | 5 | 122.73 | 0.5426 | 0.5328 | |
| 73 | 1417697_at_A Soat1 | sterol O-acyltransferase 1 | 1 | 156.34 | 0.5382 | 0.5341 | |
| 78 | 1450712_at_A Kcnj9 | potassium inwardly-rectifying channel, J, 9 | 1 | 172.39 | -0.5280 | -0.5375 | |
| 84 | 1433690_at_A 2210016L21Rik | RIKEN cDNA 2210016L21 gene | 5 | 114.06 | -0.5465 | -0.5132 | |
| 90 | 1431493_at_B 9530046B11Rik | RIKEN cDNA 9530046B11 gene | 5 | 116.25 | -0.5176 | -0.5350 | |
| 94 | 1424748_at_A Galnt11 | UDP-N-acetyl-α-d-galactosamine | 5 | 23.73 | -0.5369 | -0.5112 | |
| 97 | 1418839_at_A Glmn | glomulin, FKBP associated protein | 5 | 106.62 | -0.5268 | -0.5185 | |
| PROL, NEUR | |||||||
| 13 | 1433646_at_B Mrps27 | mitochondrial ribosomal protein S27 | 13 | 95.60 | 0.5569 | 0.5198 | |
| 25 | 1460175_at_A Rps23 | ribosomal protein S23 | 13 | 87.06 | -0.5041 | -0.5029 | |
| PROL, SURV | |||||||
| 2 | 1419700_a_at_A Prom1 | prominin 1 | 5 | 42.75 | -0.6526 | -0.5097 | |
| NEUR, ASTR | |||||||
| 1 | 1427412_s_at_A Rapgef6 | Rap guanine nucleotide exchange factor (GEF) 6 | 11 | 54.45 | -0.6026 | 0.6251 |
Cis-acting transcripts whose expression is correlated with two or more of the four neurogenesis phenotypes. Transcripts with significant QTL at the site of their physical location are in boldface type.
The influence of trans-acting genes on a trait is indirect and more difficult to assess. The list of trans-acting genes whose expression correlated with adult neurogenesis parameters contained a number of interesting candidates, such as CD36, Spry1, Dock1, Snap25, Chrna2, NT3 (Ntf3), netrin G1 (Netng1), Nedd8, Unc5c, Akt1, and others. There were only four transcripts that linked gliogenesis with neurogenesis (with R > 0.5), one of which (Rapgef6) was cis-acting. All four transcripts showed an inverse link with neuro- and gliogenesis: if positively correlated with one, they were negatively correlated with the other.
We focused on the cis-acting genes and searched for QTL associated with the expression of these genes. The peak associations of 12 of these transcripts were significant at the site of the physical location of the gene in the genome (boldface type in Table 1).
Two of the cis-acting genes, Msi1h (LRS not significant, see Fig. 2) and prominin1/CD133 (Prom1, significant), have well established functions in neural stem-cell activity. The LRS peaks of both genes fall into regions with a high density of SNP (data not shown). We used the information about SNP density in the physical mapping tool of WebQTL and found that all 21 cis-acting genes were located in SNP-rich regions of the genome.
Discussion
In this study, we demonstrate how information can be obtained from cumulative knowledge in a public database about genetics of adult neurogenesis in relation to the patterns of gene expression in the adult brain. Our study linked the classical analysis of a physiological quantitative trait with the concept of “expression genetics” (21), that is, the genetics of how genes control other genes. The linkage analysis for our four phenotypes (Fig. 3) revealed that a very large number of gene loci are associated with adult neurogenesis, making it unlikely that one QTL with dominating influence on the phenotype will be identifiable. By investigating F1 generations, so-called recombinant intercrosses (22), or congenic lines (23), the search for master regulatory QTL for adult neurogenesis could, in principal, be refined. The need for very large breeding panels to achieve the precision required to confirm single loci makes this approach problematic. This method has been successfully exploited in only relatively few cases (24). Despite the general relevance of direct QTL mapping, the strategy to study associations across physiological and expression phenotypes adds an entirely different dimension to the genetic analysis of complex traits. The goal is not so much to describe the function of isolated genes (as would be the case in classical QTL mapping) but rather to understand the contribution of regulatory networks to a given phenotype. Irrespective of this, the sensitivity of QTL mapping depends on the precision with which the quantitative trait can be defined and measured. With the identification of further specified subphenotypes in adult neurogenesis (e.g., the dynamics of particular stages of development), the identification of narrow QTLs will become possible.
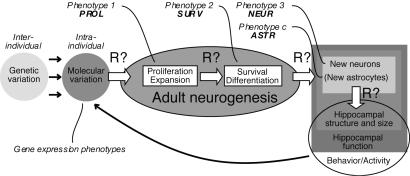
We show that PROL has a low predictive power (≈19%) for NEUR, despite the fact that both traits are correlated and causally related. Similarly, we found that ASTR is quantitatively independent of NEUR. The finding that the genetic determinants of SURV explained 85% of the variance in NEUR supports our hypothesis that quantitative regulation of neurogenesis occurs on the level of a postmitotic selection process (9). The aligned linkage curves (Fig. 3) reveal how the phenotypic covariance and correlation relate to covariance on a genetic level. We currently lack the tools to directly interpret differences in the LRS score between traits for many loci simultaneously and to extract which regulatory principles link phenotypes (Fig. 4).
Fig. 4.
Conceptual scheme highlighting the relationship between genomic factors, gene expression, and measured phenotypes of adult neurogenesis.
The correlation of the traits PROL, SURV, NEUR, and ASTR with gene-expression data yielded a total of 756 genes correlated with one of the traits above a threshold of R = 0.5 (data not shown). Focusing on intersecting associations between related phenotypes reduced the number of candidates to 190, 169 of which were trans-acting. The identification of transcript QTL associated with the potential QTL for adult neurogenesis (Fig. 3) allows the identification of plausible candidates within a given interval. The association is not evidence of causality, but transcript QTL with high correlations are stronger candidates (18). This feature is particularly true for cis-acting genes.
Among the cis-acting genes whose expression correlated with measures of adult hippocampal neurogenesis were two candidates, musashi (Msi1h) and prominin1/CD133 (Prom1), which have been related to stem-cell activity in the developing and adult brain. Both appear to play a major role in the control of stem-cell maintenance (self renewal) and asymmetric stem-cell divisions (25, 26). Msi1h showed the second highest of all correlations and was correlated to PROL, SURV, and NEUR.
Another stem-cell gene not previously related to neural stem cells is Camk4, which is a maintenance factor for hematopoietic stem cells (27). Camk4 is expressed in the adult brain; deletion of Camk4 causes a cerebellar defect (28). A role of Camk4 is assumed in the consolidation of long-term memory, a centrally hippocampal function (29). In ganglionic neurons, Camk4 promoted survival and axonal growth (30). We found an association with SURV and NEUR, which might hint at a similar role in the context of hippocampal neurogenesis.
Ndufs2 mutations are part of complex I deficiency disorders such as Leigh disease and mitochondrial diseases that include enceph-alopathies (31). A particular hippocampal function has not yet been reported, but in a rat model of depression, NADH dehydrogenase was up-regulated in the hippocampus (32), and a role for NADH dehydrogenases has been suggested in the context of neurodegenerative disorders (33). Ndufs2 was inversely related to the adult-neurogenesis phenotypes, consistent with the idea of neurogenesis being down-regulated in depression and neurodegeneration.
Ssbp2 is a tumor-suppressor gene whose overexpression in hematopoietic stem cells is accompanied by C-myc down-regulation (34). Ssbp2 was positively correlated with SURV and NEUR, suggesting that, as in myeloid cells, Ssbp2 might be linked to growth arrest and cellular differentiation (35).
Mutation of Kcnq2 is associated with benign familial neonatal convulsions, a self-remitting developmental disorder. A role in neuronal maturation has been hypothesized, because Kcnq2 might mediate inhibitory action during that phase of development of glutamatergic neurons in which GABAergic interneurons provide a transient excitatory input to the new cells (36). A transient GABAergic innervation has also been found in hippocampal progenitor cells in vivo (37). A short splice variant of Kcnq2 is detectable in undifferentiated neuronal cells (38).
Kcnj9 has been identified as the likely quantitative trait gene controlling basal locomotor behavior (39). Kcnj9 is highly expressed in strains with low basal locomotor activity. Physical activity induces and maintains adult neurogenesis (40). We found a negative correlation between Kcnj9 expression and adult neurogenesis.
Five of the cis-acting transcripts did not suggest any known link to neuronal development. Of all these transcripts, Brdt showed the strongest, albeit negative, association with adult neurogenesis (R = –0.8). Brdt was considered to be specifically expressed in mid-to late spermatocytes (41), but is also expressed during brain development (42). Polymorphisms in Soat1 (ACAT1) have been identified as risk factors in Alzheimer's disease (43). The function of Hars lies in protein biosynthesis; a negative correlation with adult neurogenesis is not very plausible based on the currently available information. Two transcripts were related to the Golgi system. Galnt11 is a calcium-binding protein of the Golgi membrane. Vps33a is involved in the formation of cellular vacuoles and the trafficking between Golgi and lysosomes (44).
The only cis-acting gene whose expression levels linked neurogenesis and astrogenesis was Rapgef6, a downstream target of m-Ras and, as such, involved in cytoskeleton reorganization. Increased Ras in neural stem cells was found to lead to an increase in gliogenesis (45). Rapgef6 expression was positively linked with astrogenesis and negatively linked to neurogenesis.
Our study shows that several genes with correlating expression patterns rather than single master switches control complex phenotypes like adult neurogenesis. The structure of the genetic network built by these genes remains unknown at present. These data suggest that scientific approaches studying one gene at a time will not reveal much about the main principles governing the genetic control of complex phenotypes like adult neurogenesis.
Methods
Animals. Twenty-nine BXD/Ty and 23 AXB/BXA RI strains, and the three parental strains C57BL/6J, DBA/2J, and A/J were obtained from The Jackson Laboratory. The BXD strains were generated by crossing C57BL/6J and DBA/2J mice. BXD strains are inbred lines derived from brother–sister matings starting from an F2 cross (12, 13). AXB/BXA are based on reciprocal intercrosses between C57BL/6J and A/J (14). Animals were kept in conventional laboratory cages with standard food and access to food and water ad libitum. We studied five animals from each of the parental strains, three each of the great majority of recombinant inbred strains, two each of AXB1, AXB2, AXB4, BXA12, BXD2, BXD18, and BXD35, and one BXA26 animal. The mice were between 36 and 106 days old, but most were close to 70 days (Table 2). All were females.
Immunohistochemistry. All mice received one daily i.p. injection of BrdUrd for five consecutive days. Four weeks after the final BrdUrd injection, the mice were killed and their brains processed as described in ref. 10. Immunohistochemistry for proliferation marker Ki67 [polyclonal rabbit antibody, Novocastra (Newcastle Upon Tyne, U.K.) 1:500], BrdUrd [monoclonal rat, Harlan Sera-lab (Loughborough, Leicestershire, U.K.) 1:400], neuronal marker NeuN (monoclonal mouse, Chemicon, 1:100), and astrocytic marker S100β (polyclonal rabbit, Swant, 1:2,500) was performed and analyzed quantitatively as described in ref. 10.
Correction for Age Effects. Because age is a strong negative regulator of adult neurogenesis (19), data were corrected for age effects. Modeling was performed by using sas proc glm, v 9.0 (SAS Institute, Cary, NC). Residual diagnostics were performed by using proc plot, proc univariate, and proc corr.
Heritability-Estimation Procedures. For each trait, heritability was estimated by use of sas proc varcomp, v 9.0 (SAS Institute). Variance components were estimated by using the restricted-maximum-likelihood method. The heritability was calculated as the ratio of between-strain variance to total (between-strain plus within-strain) variance.
Correlation Comparisons and QTL Analysis. Assessment of phenotypic covariance and QTL analysis were performed with the GeneNetwork (www.genenetwork.org). Phenotypes measured in our study were deposited in the public GeneNetwork database (record IDs 10795–10798). Pearson product-moment correlations for the corrected numerical values were calculated as implemented in GeneNetwork. Details of the transcript expression data are described in ref. 4. Linkage analysis in WebQTL (the mapping module of the GeneNetwork) is based on a set of >7,482 informative SNPs (Wellcome-CTC SNP data set) plus 1,500 microsatellite markers distributed across all autosomes and the X chromosome. Genomewide significance levels for assessing confidence in linkage statistics were estimated by comparing the peak LRS of correctly ordered data sets with LRS computed for 1,000 or 2,000 permutations.
Supplementary Material
Acknowledgments
We thank Irene Thun and Daniela Gast for technical assistance, Sebastian Jessberger for important discussions, and Mary Lynn Gage for editing the manuscript. This work was funded by Deutsche Forschungs-gemeinschaft (G.K.) and the Max Planck Award (Alexander von Hum-boldt Foundation) (to F.H.G.). The M430 forebrain array data were generated by the Integrative Neuroscience Initiative on Alcoholism (www.iniastress.org; U01AA13499). GeneNetwork is funded by P20-DA21131, U01CA105417, and R24RR021760.
Conflict of interest statement: No conflicts declared.
Abbreviations: ASTR, astrocytes; LRS, likelihood ratio statistic; NEUR, neurons; PROL, proliferation; RI, recombinant inbred; SURV, surviving; QTL, quantitative trait loci.
Data deposition: Phenotypes measured in this study were deposited in the GeneNetwork database, www.genenetwork.org (IDs 10795–10798).
References
- 1.Kempermann, G. & Gage, F. H. (2002) Brain Res. Dev. Brain Res. 134, 1–12. [DOI] [PubMed] [Google Scholar]
- 2.Kempermann, G. & Gage, F. H. (2002) Eur. J. Neurosci. 16, 129–136. [DOI] [PubMed] [Google Scholar]
- 3.Kempermann, G., Kuhn, H. G. & Gage, F. H. (1997) Proc. Natl. Acad. Sci. USA 94, 10409–10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesler, E. J., Lu, L., Shou, S., Qu, Y., Gu, J., Wang, J., Hsu, H. C., Mountz, J. D., Baldwin, N. E., Langston, M. A., et al. (2005) Nat. Genet. 37, 233–242. [DOI] [PubMed] [Google Scholar]
- 5.Chesler, E. J., Wang, J., Lu, L., Qu, Y., Manly, K. F. & Williams, R. W. (2003) Neuroinformatics 1, 343–357. [DOI] [PubMed] [Google Scholar]
- 6.Altman, J. & Das, G. D. (1965) J. Comp. Neurol. 124, 319–335. [DOI] [PubMed] [Google Scholar]
- 7.Seki, T. & Arai, Y. (1999) J. Comp. Neurol. 410, 503–513. [DOI] [PubMed] [Google Scholar]
- 8.Seri, B., Garcia-Verdugo, J. M., McEwen, B. S. & Alvarez-Buylla, A. (2001) J. Neurosci. 21, 7153–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempermann, G., Jessberger, S., Steiner, B. & Kronenberg, G. (2004) Trends Neurosci. 27, 447–452. [DOI] [PubMed] [Google Scholar]
- 10.Kempermann, G., Gast, D., Kronenberg, G., Yamaguchi, M. & Gage, F. H. (2003) Development (Cambridge, U.K.) 130, 391–399. [DOI] [PubMed] [Google Scholar]
- 11.Kempermann, G., Brandon, E. P. & Gage, F. H. (1998) Curr. Biol. 8, 939–942. [DOI] [PubMed] [Google Scholar]
- 12.Taylor, B. A., Wnek, C., Kotlus, B. S., Roemer, N., MacTaggart, T. & Phillips, S. J. (1999) Mamm. Genome 10, 335–348. [DOI] [PubMed] [Google Scholar]
- 13.Taylor, B. A. (1989) in Genetic Variants and Strains of the Laboratory Mouse, eds. Lyon, M. L. & Searle, A. G. (Oxford Univ. Press, Oxford), pp. 773–796.
- 14.Marshall, J. D., Mu, J. L., Cheah, Y. C., Nesbitt, M. N., Frankel, W. N. & Paigen, B. (1992) Mamm. Genome 3, 669–680. [DOI] [PubMed] [Google Scholar]
- 15.Wang, J., Williams, R. W. & Manly, K. F. (2003) Neuroinformatics 1, 299–308. [DOI] [PubMed] [Google Scholar]
- 16.Consortium, C. T., Abiola, O., Angel, J. M., Avner, P., Bachmanov, A. A., Belknap, J. K., Bennett, B., Blankenhorn, E. P., Blizard, D. A., Bolivar, V., et al. (2003) Nat. Rev. Genet. 4, 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchill, G. A., Airey, D. C., Allayee, H., Angel, J. M., Attie, A. D., Beatty, J., Beavis, W. D., Belknap, J. K., Bennett, B., Berrettini, W., et al. (2004) Nat. Genet. 36, 1133–1137. [DOI] [PubMed] [Google Scholar]
- 18.Broman, K. W. (2005) Nat. Genet. 37, 209–210. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn, H. G., Dickinson-Anson, H. & Gage, F. H. (1996) J. Neurosci. 16, 2027–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron, H. A. & McKay, R. D. (1999) Nat. Neurosci. 2, 894–897. [DOI] [PubMed] [Google Scholar]
- 21.Jansen, R. C. & Nap, J. P. (2001) Trends Genet. 17, 388–391. [DOI] [PubMed] [Google Scholar]
- 22.Zou, F., Gelfond, J. A., Airey, D. C., Lu, L., Manly, K. F., Williams, R. W. & Threadgill, D. W. (2005) Genetics 170, 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadeau, J. H., Singer, J. B., Matin, A. & Lander, E. S. (2000) Nat. Genet. 24, 221–225. [DOI] [PubMed] [Google Scholar]
- 24.Korstanje, R. & Paigen, B. (2002) Nat. Genet. 31, 235–236. [DOI] [PubMed] [Google Scholar]
- 25.Kosodo, Y., Roper, K., Haubensak, W., Marzesco, A. M., Corbeil, D. & Huttner, W. B. (2004) EMBO J. 23, 2314–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okano, H., Kawahara, H., Toriya, M., Nakao, K., Shibata, S. & Imai, T. (2005) Exp. Cell Res. 306, 349–356. [DOI] [PubMed] [Google Scholar]
- 27.Kitsos, C. M., Sankar, U., Illario, M., Colomer-Font, J. M., Duncan, A. W., Ribar, T. J., Reya, T. & Means, A. R. (2005) J. Biol. Chem. 280, 33101–33108. [DOI] [PubMed] [Google Scholar]
- 28.Ribar, T. J., Rodriguiz, R. M., Khiroug, L., Wetsel, W. C., Augustine, G. J. & Means, A. R. (2000) J. Neurosci. 20, RC107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang, H., Sun, L. D., Atkins, C. M., Soderling, T. R., Wilson, M. A. & Tonegawa, S. (2001) Cell 106, 771–783. [DOI] [PubMed] [Google Scholar]
- 30.Hansen, M. R., Bok, J., Devaiah, A. K., Zha, X. M. & Green, S. H. (2003) J. Neurosci. Res. 72, 169–184. [DOI] [PubMed] [Google Scholar]
- 31.Martin, M. A., Blazquez, A., Gutierrez-Solana, L. G., Fernandez-Moreira, D., Briones, P., Andreu, A. L., Garesse, R., Campos, Y. & Arenas, J. (2005) Arch. Neurol. 62, 659–661. [DOI] [PubMed] [Google Scholar]
- 32.Nakatani, N., Aburatani, H., Nishimura, K., Semba, J. & Yoshikawa, T. (2004) Pharmacogenomics J. 4, 114–126. [DOI] [PubMed] [Google Scholar]
- 33.Aksenov, M. Y., Tucker, H. M., Nair, P., Aksenova, M. V., Butterfield, D. A., Estus, S. & Markesbery, W. R. (1999) Neurochem. Res. 24, 767–774. [DOI] [PubMed] [Google Scholar]
- 34.Castro, P., Liang, H., Liang, J. C. & Nagarajan, L. (2002) Genomics 80, 78–85. [DOI] [PubMed] [Google Scholar]
- 35.Liang, H., Samanta, S. & Nagarajan, L. (2005) Oncogene 24, 2625–2634. [DOI] [PubMed] [Google Scholar]
- 36.Okada, M., Zhu, G., Hirose, S., Ito, K. I., Murakami, T., Wakui, M. & Kaneko, S. (2003) Epilepsy Res. 53, 81–94. [DOI] [PubMed] [Google Scholar]
- 37.Wang, L. P., Kempermann, G. & Kettenmann, H. (2005) Mol. Cell. Neurosci. 29, 181–189. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. S., Iannotti, C. A., Dargis, P., Christian, E. P. & Aiyar, J. (2001) J. Neurosci. 21, 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hitzemann, R., Malmanger, B., Reed, C., Lawler, M., Hitzemann, B., Coulombe, S., Buck, K., Rademacher, B., Walter, N., Polyakov, Y., et al. (2003) Mamm. Genome 14, 733–747. [DOI] [PubMed] [Google Scholar]
- 40.Van Praag, H., Christie, B. R., Sejnowski, T. J. & Gage, F. H. (1999) Proc. Natl. Acad. Sci. USA 96, 13427–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang, E., Salazar, G., Crowley, T. E., Wang, X., Lopez, R. A., Wang, X. & Wolgemuth, D. J. (2004) Gene Expr. Patterns 4, 513–519. [DOI] [PubMed] [Google Scholar]
- 42.Gray, P. A., Fu, H., Luo, P., Zhao, Q., Yu, J., Ferrari, A., Tenzen, T., Yuk, D. I., Tsung, E. F., Cai, Z., et al. (2004) Science 306, 2255–2257. [DOI] [PubMed] [Google Scholar]
- 43.Papassotiropoulos, A., Wollmer, M. A., Tsolaki, M., Brunner, F., Molyva, D., Lutjohann, D., Nitsch, R. M. & Hock, C. (2005) J. Clin. Psychiatry 66, 940–947. [PubMed] [Google Scholar]
- 44.Gissen, P., Johnson, C. A., Gentle, D., Hurst, L. D., Doherty, A. J., O'Kane, C. J., Kelly, D. A. & Maher, E. R. (2005) Hum. Mol. Genet. 14, 1261–1270. [DOI] [PubMed] [Google Scholar]
- 45.Dasgupta, B. & Gutmann, D. H. (2005) J. Neurosci. 25, 5584–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.