Abstract
Organogenesis in plants is almost entirely a postembryonic process. This unique feature implies a strict coupling of cell proliferation and differentiation, including cell division, arrest, cell cycle reactivation, endoreplication, and differentiation. The plant retinoblastoma-related (RBR) protein modulates the activity of E2F transcription factors to restrict cell proliferation. Arabidopsis contains a single RBR gene, and its loss of function precludes gamete formation and early development. To determine the relevance of the RBR/E2F pathway during organogenesis, outside its involvement in cell division, we have used an inducible system to inactivate RBR function and release E2F activity. Here, we have focused on leaves where cell proliferation and differentiation are temporally and developmentally regulated. Our results reveal that RBR restricts cell division early during leaf development when cell proliferation predominates, while it regulates endocycle occurrence at later stages. Moreover, shortly after leaving the cell cycle, most of leaf epidermal pavement cells retain the ability to reenter the cell cycle and proliferate, but maintain epidermal cell fate. On the contrary, mesophyll cells in the inner layers do not respond in this way to RBR loss of activity. We conclude that there exists a distinct response of different cells to RBR inactivation in terms of maintaining the balance between cell division and endoreplication during Arabidopsis (Arabidopsis thaliana) leaf development.
Cell division and growth depends on a series of coordinated events strictly regulated both temporally and spatially in individual cells. In addition, multicellularity imposes extra layers of regulation, most importantly on the balance between cell proliferation, arrest, and differentiation, in coordination with the ontogenic program. However, what the relevance is of pathways regulating cell proliferation for an appropriate developmental program or whether cell cycle regulators affect the appropriate differentiation pattern in a cell type-specific manner are questions still poorly understood. For example, in animals, where organogenetic processes occur embryonically, it was anticipated that key cell cycle regulators were required for embryonic development. However, recent studies have shown that, at least for some of them, this is not the case, e.g. cyclin-dependent kinase2 (Cdk2; Ortega et al., 2003), reinforcing the need to define the role of cell proliferation in the context of a developing organism.
Contrary to the situation in animals, organogenesis in plants is almost entirely a postembryonic process. As a consequence, the production of new organs, e.g. roots, shoots, leaves, and flowers, relies on the continuous potential of particular sets of cells to proliferate and eventually undergo specific differentiation programs. Two plant-specific features make them remarkable. One is that such an organogenetic pattern extends over the entire life span of the organism, thus contributing to increase body architecture. Another is the ability of certain cells of yet unknown molecular characteristics to dedifferentiate, proliferate, and, in response to hormone signals or developmental cues, eventually generate a plethora of different cell types. Therefore, the investigation of links between cell cycle regulation and plant development can shed light on the importance of a strict balance between cell proliferation and differentiation during development.
One key cell cycle regulatory pathway depends on the plant retinoblastoma-related (RBR) protein and the E2F/DP transcription factors (Gutierrez et al., 2002; De Veylder et al., 2003; Dewitte and Murray, 2003). Arabidopsis (Arabidopsis thaliana) contains a single RBR gene and a complex family of E2F/DP proteins (Vandepoele et al., 2002). Three E2F (named a, b, and c), homologs of human E2F1-5 (Attwooll et al., 2004), contain the typical domain organization, including DNA-binding, DP heterodimerization, transactivation, and RBR-binding domains (Shen, 2002). They heterodimerize with either of the two DP proteins (named a and b) to form an active transcription factor (Kosugi and Ohashi, 2002). E2Fa/DPa heterodimers act mainly as transcriptional activators and regulate cell proliferation and endoreplication (De Veylder et al., 2002; Kosugi and Ohashi, 2003). E2Fb, likely in cooperation with DPa, regulates the entry into S phase and mitosis and seems to be a target for transducing auxin signals into the cellular decision to proliferate or arrest and enter the endocycle program (Magyar et al., 2005). E2Fc is a transcriptional repressor abundant in arrested cells and, upon cell cycle stimulation, is rapidly targeted to the proteasome by an SCFSKP2 complex (del Pozo et al., 2002). The other three E2F (named d, e, and f), whose animal homologs are E2F7/8 (Attwooll et al., 2004), are atypical since they have a duplicated DNA-binding domain and do not heterodimerize with DP (Shen, 2002) or bind RBR (Ramirez-Parra et al., 2004). E2Fe, also known as DEL1, has been implicated in regulating the endocycle program (Vlieghe et al., 2005) and E2Ff in the control of cell growth and differentiation by regulating the expression of a subset of cell wall biogenesis genes (Ramirez-Parra et al., 2004). Genome-wide analyses have revealed that genes belonging to functional categories other than cell cycle and DNA replication control (Ramirez-Parra et al., 2003, 2004; Vlieghe et al., 2003; Vandepoele et al., 2005) are also E2F targets.
RBR is required for cell proliferation arrest in tobacco (Nicotiana tabacum)-cultured cells (Gordon-Kamm et al., 2002) and during Arabidopsis gametophytic development (Ebel et al., 2004). Consistent with this, loss-of-function mutations in the RBR gene are lethal, precluding the assessment of its role in adult plants. Recently, expression of the tobacco NtRBR1 gene has been reduced using a virus-induced gene silencing approach (Park et al., 2005). These experimental conditions have allowed analyzing the effects of reducing NtRBR1 expression in organs formed above the infected leaf. Thus, the activity of the shoot apical meristem was arrested; the new leaves show growth retardation and abnormal development, and flower formation was severely retarded. Together, the data confirmed the importance of tobacco RBR in restricting cell division and also endoreplication in leaf cells (Park et al., 2005).
Our aim is to define the role of Arabidopsis RBR protein during organ formation, and here we have focused on leaves to assess RBR function in the same developing organ where RBR activity is compromised. To this end, we have developed a targeted inactivation of the RBR/E2F pathway in a temporally controlled manner using an inducible system (Aoyama and Chua, 1997). Targeting RBR protein instead of AtRBR gene expression is a useful approach since RBR function depends most significantly on posttranslational modifications, one of the most relevant being phosphorylation by CDK/cyclin complexes (Nakagami et al., 1999, 2002; Boniotti and Gutierrez, 2001). This is thought to release the activity of RBR-bound E2F transcription factors (Gutierrez et al., 2002). Thus, the experimental rationale takes advantage of the use of ectopically expressing a plant DNA virus protein, the geminivirus RepA protein. RepA interacts efficiently with RBR through an LxCxE amino acid motif (Xie et al., 1995; Grafi et al., 1996; Xie et al., 1996), regulates viral DNA replication (Gutierrez, 2000), and has been used to address the role of RBR in proliferation of cultured cells (Gordon-Kamm et al., 2002). The interaction of virus RepA with RBR bypasses the normal activity of CDK/cyclin complexes that phosphorylate RBR and release E2F activity. Consequently, the system allows relieving the endogenous set of AtE2Fa/b/c from RBR repression in an inducible manner, a situation different from constitutive overexpression of single AtE2F genes previously reported (De Veylder et al., 2002; del Pozo et al., 2002; Magyar et al., 2005). Furthermore, the use of wild-type RepA and a point mutant in which RBR interaction is abolished (Xie et al., 1996) provides a useful approach to define aspects of RBR activity strictly dependent on the release of E2F factors.
We have found that RBR restricts cell division during early leaf development when cell proliferation predominates. Shortly after the proliferative stage of leaf primordia, pavement cells retain their ability to proliferate but maintain their fate. On the contrary, other epidermal cell types, e.g. trichomes and stomata, do not change their proliferation state or fate specification, as it is also the case of mesophyll cells. At later stages, once the switch to the endocycle program has occurred, RBR largely restricts the progression through additional endocycles. Thus, we conclude that cells respond differently to RBR inactivation in terms of regulating their cell division and endoreplication potential during Arabidopsis leaf development.
RESULTS
Generation of Plants with Inducible Expression of a Geminivirus RBR-Binding Protein
Geminivirus RepA is a viral protein that participates in viral DNA replication (Gutierrez, 2000; Hanley-Bowdoin et al., 2004). It binds RBR protein through a typical LxCxE amino acid motif, and mutations in this motif abolish RBR binding and efficient viral replication in cultured cells (Xie et al., 1995, 1996). It has been proposed that interaction with RBR allows the release of RBR-bound E2F activity to facilitate viral DNA replication (Gutierrez, 2000; Hanley-Bowdoin et al., 2004), based on the similarity with the interactions of animal oncoproteins and retinoblastoma protein (Lavia et al., 2003). We reasoned that a transgenic model where expression of RepA is controlled over time could be useful to target the inactivation of RBR and modulate local increases of endogenous E2F activity. In this way, we can evaluate the relevance of the RBR/E2F pathway in differentiating cells during organ development.
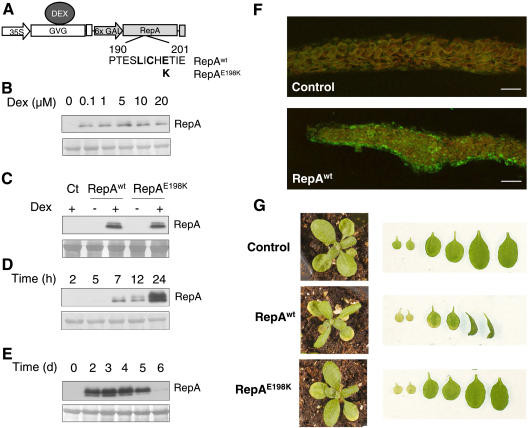
To this end, we used a dexamethasone (Dex)-inducible system (Aoyama and Chua, 1997) to generate Arabidopsis transgenic plants that can express geminivirus RepA protein (RepAwt) in an inducible manner (Fig. 1A). We also generated plants that expressed RepA containing a point mutation (E198K; RepAE198K) that almost completely abolished binding to RBR in vitro, impairing viral DNA replication (Xie et al., 1995). These constructs will allow us to determine the effects specifically associated with the RBR/E2F pathway in planta. Treatment with Dex has been shown to produce unspecific effects, especially early during development or seedling growth (Kang et al., 1999). Thus, we selected transgenic RepA plants that did not show macroscopical defects in the absence of Dex treatment. As a control we selected transgenic plants transformed with an empty vector. We evaluated the response to Dex treatment by determining the minimal Dex concentration necessary for a good induction of RepA expression. Since the protein level was not strongly dependent on the inducer concentration (Fig. 1B), we chose the lowest concentration (1 μm Dex) that repetitively gave a satisfactory induction for phenotypic studies of the transgenic plants. Figure 1C shows the results of representative lines where both wild-type and mutant RepA proteins could be readily detected in seedling extracts only after Dex treatment. This indicates that (1) any possible leakage of the induction system in the lines selected was below detectable levels, and (2) both wild-type and mutant RepA proteins accumulate after induction. Western-blotting analysis of whole-seedling extracts allowed detection of RepA as early as 7 h after induction, although the viral protein continues to accumulate at later times (Fig. 1D). We also carried out a time-course analysis of the presence of RepA after a single Dex treatment and found that high amounts of the protein were still detectable 5 d after induction (Fig. 1E). To determine the spatial distribution of RepA protein after Dex treatment, we detected the viral protein by immunofluorescence in cross sections of leaves. As shown in Figure 1F, RepA accumulates in all leaf cell layers after Dex treatment.
Figure 1.
Expression of the geminivirus RBR-binding RepA proteins in transgenic plants. A, Details of the constructs used to express the RepAwt and RepAE198K coding sequences in the Dex-inducible system (Aoyama and Chua, 1997). B, Western-blot analysis of RepA protein 7 h after induction with different concentrations of Dex (0–20 μm) in 10-d-old seedlings. Ten micrograms of protein were loaded in each lane. The bottom section is a region of the Coomassie-stained gel (molecular mass approximately 55 kD) used as loading control. C, Detection of RepAwt and RepAE198K after Dex treatment (20 μm, 20 h) in 10-d-old seedlings (20 μg protein per lane). The bottom section is the loading control as in B. D, Time course of RepAwt expression in 10-d-old seedlings (10 μg) after treatment with 20 μm Dex. The bottom section is the loading control as in B. Induction of RepAE198K protein followed a similar pattern. E, Stability of RepAwt protein after Dex treatment (1 μm). The bottom section is the loading control as in B. F, Detection of RepAwt in cross sections of leaves 3/4, 1 d after Dex treatment of control- and RepAwt-expressing plants to show that the viral protein accumulates in the epidermis as well as in the internal cell layers. Bars correspond to 50 μm. G, Phenotype at the rosette stage (18-d-old seedlings) of control (transformed with the empty vector) and transgenic plants expressing RepAwt or RepAE198K, 5 d after spraying with 1 μm Dex. The right sections show, from left to right, the two cotyledons and the first and second pairs of the leaves at the same magnification.
We then analyzed the overall phenotype of the plants after induction of RepA expression, with particular emphasis on the leaves. Thirteen-day-old seedlings, containing the two cotyledons, the first pair of leaves, and the emerging second pair of leaves, were treated with Dex and analyzed 5 d afterward. Induction of RepAwt, but not RepAE198K, produced a drastic effect since leaves 3/4 in particular showed altered growth including downward curling (Fig. 1G). It is worth noting that leaves 1/2 showed some signs of senescence several days after Dex treatment. However, since these effects were not RBR dependent, as they were observed in both RepAwt- and RepAE198K-expressing plants, they were not further studied here. These data suggest that RepAwt may affect leaf development and that this effect was dependent on an intact RBR-binding motif in the viral protein, strongly suggesting that they may be mediated by the ectopic release of endogenous RBR-bound E2F activity.
Targeting RBR with RepA Increases E2F Activity
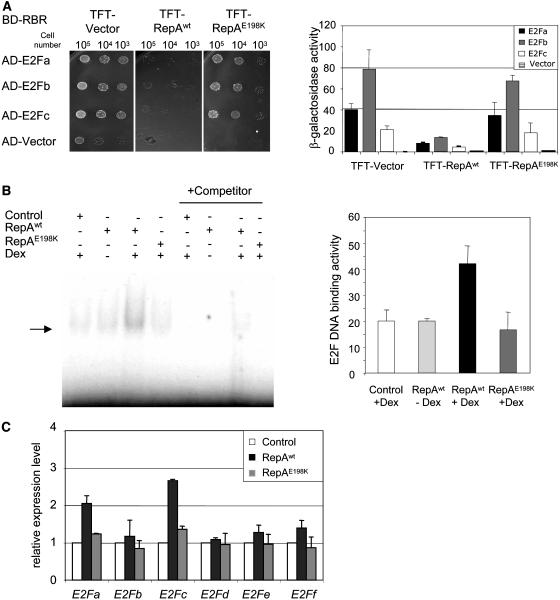
To test whether RepA was actually able to disrupt RBR/E2F interaction, we first addressed this possibility with a yeast three-hybrid protein approach (Egea-Cortines et al., 1999). We generated yeast cells expressing Arabidopsis RBR, fused to the GAL4 DNA-binding domain. Then, they were cotransformed with a plasmid that expressed each Arabidopsis E2F (a, b, and c) fused to the GAL activation domain (Fig. 2A). These combinations allowed yeast growth in selective medium, indicating a strong and specific interaction of AtRBR and each of AtE2F. To evaluate whether RepA is able to disrupt RBR/E2F interactions, we cotransformed yeast cells with a third plasmid to express either RepAwt or RepAE198K (Fig. 2A; plasmid TFT). In the absence of E2F, neither wild-type nor mutant RepA proteins alone allowed yeast cell growth (data not shown). However, transforming the third plasmid expressing RepAwt, but not the RepAE198K, into yeast cells expressing AtRBR and each of the AtE2F largely impaired the strong growth in selective medium (Fig. 2A, top section). These data were confirmed by measuring β-galactosidase activity (Fig. 2A, bottom section). Therefore, we concluded that RepAwt, but not the point mutant impaired in RBR binding, is able to disrupt the interaction of Arabidopsis RBR and E2Fa, b, and c in yeast.
Figure 2.
Disruption of RBR/E2F complexes in yeast and in planta. A, Effect of RepAwt and RepAE198K proteins on RBR/E2F interaction shown by yeast three-hybrid assay. Yeast cells expressing AtRBR-Gal4 DNA-binding domain (BD-RBR) were cotransformed, as indicated, with plasmids expressing the different AtE2F fused to the Gal4 activation domain (AD-E2F) or with the empty vector (AD), and with a vector (TFT) expressing RepAwt and RepAE198K proteins. All transformants grew normally in plates containing His (data not shown). The top sections show the growth of different transformants in selective medium (without His) at different dilutions. Galactosidase activity is shown in the bottom section and is expressed as Miller units. Data correspond to two independent experiments, which were carried out in triplicate. B, Detection of E2F DNA-binding activity in plant extracts by EMSA. Total protein extracts (15 μg) of the indicated transgenic plants, with or without Dex treatment, were incubated with a 32P-labeled double-stranded oligonucleotide containing a consensus E2F site (Ramirez-Parra and Gutierrez, 2000). Arrow points to E2F-DNA complexes. A 100-fold excess of the unlabeled probe was added as competitor (right section). Relative E2F DNA-binding activity (bottom section) was quantified using a GS-710 Calibrated Image densitometer (Bio-Rad). The analysis is based on three independent experiments. Bars show the sds. C, Expression levels of each of the six Arabidopsis E2F genes determined by real-time RT-PCR analysis in extracts of 10-d-old seedlings of controls (plants transformed with an empty vector) or transgenics expressing RepAwt and RepAE198K proteins. The analysis was carried out 7 h after treatment with Dex (20 μm). Values were first normalized to the amount of actin (AtACT2) and then made relative to the mRNA amount in the control.
To evaluate whether RepAwt could release endogenous E2F in planta, we determined E2F DNA-binding activity in extracts of control and transgenic plants with and without Dex treatment. E2F DNA-binding activity was determined by electrophoretic mobility shift assay (EMSA) using an oligonucleotide containing a consensus E2F-binding site. Induction of RepAwt, but not the RepAE198K mutant, produced a small but reproducible increase of E2F-binding activity in whole-cell extracts (Fig. 2B). Such binding activity was E2F specific, since it can be competed out with an excess of free probe (Fig. 2B) but not with a probe containing two point mutations that destroy the E2F-binding site (data not shown). Based on these data together, we conclude that RepAwt, but not the RepAE198K mutant, increases E2F DNA-binding activity in planta.
To determine whether increased availability of E2F contributes to this stimulation, we determined by real-time reverse transcription (RT)-PCR the mRNA levels of the six Arabidopsis E2F genes in plants shortly after induction of the viral protein. Expression of two E2F genes, namely E2Fa and E2Fc, an activator and a repressor of cell proliferation, respectively (De Veylder et al., 2002; del Pozo et al., 2002), was up-regulated in these plants (Fig. 2C). This transcriptional activation was totally dependent on RBR inactivation, as indicated by the lack of effect observed when the RepAE198K mutant is expressed (Fig. 2C). Consequently, we conclude that the transcriptional activation of E2Fa and E2Fc expression likely contributes to the RBR inactivation-specific increase in E2F-DNA-binding activity in planta.
Functional Inactivation of RBR Up-Regulates E2F Target Genes and Induces Cell Division in Differentiated Leaves
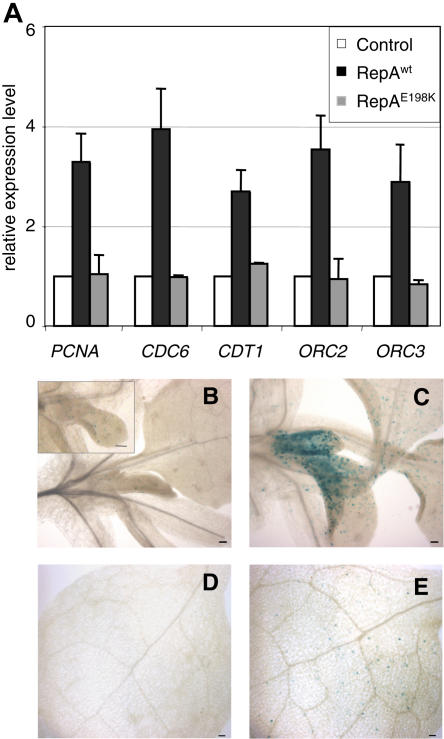
We examined if the increase of E2F activity affected the expression of selected E2F target genes. Here, we focused on S-phase genes, e.g. PCNA, CDC6, CDT1, and members of the ORC (origin recognition complex), known to be regulated by E2F (Castellano et al., 2001; Egelkrout et al., 2001; Diaz-Trivino et al., 2005). We found that expression of these genes increased in the RepAwt-expressing plants but not in the plants expressing the RepAE198K mutant (Fig. 3A). Previous studies have demonstrated the presence of E2F-binding sites in the promoter of genes belonging to various functional categories (Ramirez-Parra et al., 2003; Vlieghe et al., 2003; Vandepoele et al., 2005). We found that expression of some of these genes, in spite of containing E2F-binding sites in their promoters, was not increased by the presence of either RepAwt or RepAE198K (data not shown). Therefore, this analysis revealed that, under our experimental setting, expression of a subset of E2F target genes that play a role in cell cycle progression is up-regulated.
Figure 3.
Inactivation of RBR up-regulates the expression of a subset of E2F target genes and induces ectopic cell division. A, Expression level of the indicated E2F target genes determined by real-time RT-PCR analysis in extracts of 10-d-old seedlings of controls (plants transformed with an empty vector) or transgenics expressing RepAwt and RepAE198K proteins. The analysis was carried out 7 h after treatment with Dex (20 μm). Values were first normalized to the amount of Actin (AtACT2) and then made relative to the mRNA amount in the control. B to E, Control and transgenic plants were crossed with the cyclin B1;1-GUS marker lines (Colon-Carmona et al., 1999). Cyclin B1;1-GUS activity was detected 3 d after treating 16-d-old plants with Dex (1 μm) in the controls (B and D) and plants expressing RepAwt protein (C and E). The aerial parts around the shoot tip (B and C) and a region of leaves 1/2 (D and E) are shown. The inset in B shows a higher magnification of a growing leaf primordium showing scattered cyclin B1;1-GUS positive cells. Bars represent 50 μm.
To analyze if the increased expression of cell cycle genes affected cell division, we introduced by crossing a cell division marker into the different backgrounds. We used plants expressing a translational fusion of the β-glucuronidase (GUS) reporter gene with the destruction box of cyclin B1;1, a useful G2/M marker, since the reporter is expressed only in late G2 cells and destroyed in M phase, just as the cyclin B1;1 does (Colon-Carmona et al., 1999). GUS expression in control plants was detected in some cells of the shoot apical meristem and leaf primordia but not in mature leaves, as expected (Fig. 3, B and D). In the RepAwt background, after induction by Dex, we observed a stronger GUS expression in the leaf primordia and in the proximal zone of young leaves, indicating that more cells were entering mitosis (Fig. 3C). The increase in CYCB1;1-GUS expression was also confirmed by RT-PCR (data not shown). Moreover, scattered cells expressing GUS could also be detected in the fully differentiated first pair of leaves that had normally exited the cell proliferation program (Fig. 3E). CYCB1;1-directed GUS expression in the RepAE198K background was similar to that of the control, indicating that ectopic induction of cell division activity was dependent on RBR inactivation (data not shown).
RBR-Mediated Regulation of the Endocycle Program Is Growth Stage Dependent
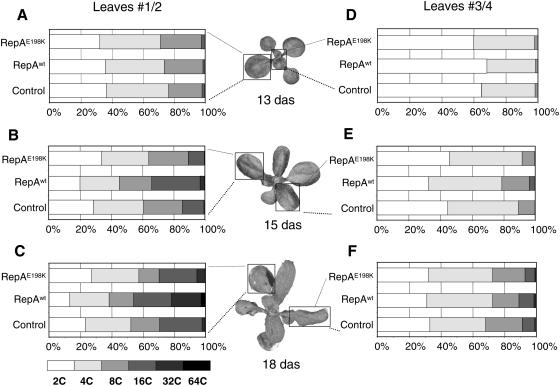
Our data show that expression of cell cycle genes increases upon RepAwt induction, and a cell division marker is detected ectopically in fully differentiated leaves. The spatial and temporal pattern of cell division during leaf development indicates that a complex balance among leaf morphogenesis, tissue-specific patterns of cell proliferation, and cell differentiation occurs (Donnelly et al., 1999). The pattern of emergence of new leaves together with their time-dependent development offers the possibility to analyze, within the same developing seedling, the effects of altering RBR/E2F-mediated gene expression at early and late growth stages. Thus, at a given stage, e.g. 13-d-old seedlings, a significant amount of nuclei in the oldest leaves (nos. 1/2) have already undergone endocycles, whereas in leaves 3/4, at an earlier stage, cells are in the process to switch to the endocycle/differentiation program, as indicated by the small peak of 8C nuclei (Fig. 4, A and D; see also Boudolf et al., 2004b; Castellano et al., 2004).
Figure 4.
Inactivation of RBR induces endoreplication in late developing leaves. Nuclear DNA ploidy distribution of control, RepAwt, and RepAE198K transgenic plants before (13 das; A and D), 2 d (15 das; B and E), and 5 d (18 das; C and F) after treatment with Dex (1 μm). Flow cytometry measurements were carried out in the first (nos. 1/2; A–C) and second (nos. 3/4; D–F) pairs of leaves. The appearance of the developmental stage of representative RepAwt-expressing plants at the time of flow cytometry analysis is also included (middle sections).
We have found previously that overriding DNA replication initiation control was dependent on CDC6 and CDT1, two E2F target genes, and this has an effect on ploidy distribution in leaf nuclei (Castellano et al., 2001, 2004). Thus, we determined ploidy levels in the leaves of transgenic plants expressing either the wild-type or the mutant RepA proteins at different developmental stages. We treated 13-d-old plants with Dex and analyzed separately nuclear ploidy levels over time in leaves 1/2 and 3/4. Nuclei of leaves 1/2, already 2 d after Dex treatment, were stimulated to develop more endocycles, as indicated by the increase in the relative amount of nuclei with 16C and the appearance of a 32C nuclear population with a concomitant decrease of the 2C population (Fig. 4B). The same effect could be observed at 5 d after Dex treatment with an increase of the 32C nuclear population and the appearance of 64C nuclei (Fig. 4C). The enhancement of endoreplication required the participation of the RBR-E2F pathway since the RepAE198K mutant was not able to induce more endocycles (Fig. 4, B and C). Nuclei of leaves 3/4 showed only a slight enhancement of the endocycle program 2 d after treatment (Fig. 4E). Five days after treatment this effect could not be observed any more and the ploidy profile of RepAwt-producing plants was comparable to that of the controls (Fig. 4F). Therefore, leaf cells respond differently to RBR inactivation depending on the developmental stage. We also measured the expression level of the set of E2F target genes, shown in Figure 3A, in leaves 1/2 and 3/4 separately after RepA induction. These genes were up-regulated, although at different levels, in an LxCxE-dependent manner, in particular in leaves 1/2 (data not shown). Differences in the E2F-mediated regulation of PCNA gene expression depending on the leaf differentiation stage have been reported (Egelkrout et al., 2002).
RBR Inactivation Alters Trichome Morphogenesis
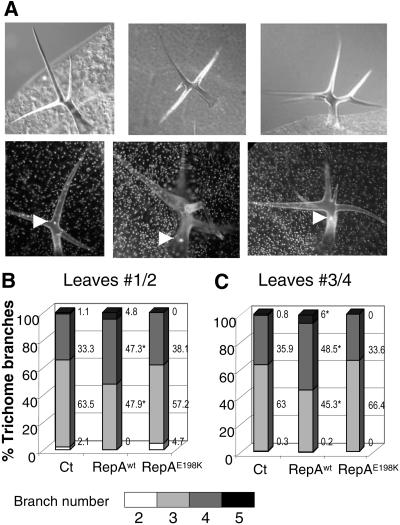
Early in leaf development, some cells in the primordia trigger a genetically defined morphogenetic pathway associated with the occurrence of endocycles. This is the case of trichomes, specialized leaf hairs located on the leaf surface with an enlarged and branched morphology and in which a rough correlation exists between nuclear ploidy and branch number (Hulskamp et al., 1999). In fact, trichome-forming cells undergo more endocycles and have more branches when CDC6 and CDT1 expression increases (Castellano et al., 2004). We asked whether expression of the viral RepA protein could alter trichome morphogenesis and, if so, whether such an effect was dependent on the inactivation of RBR. 4′,6-Diamino-phenylindole (DAPI) staining of trichome nuclei showed an increase in nuclear size of trichomes with more branches (see examples of three- to five-branched trichomes in Fig. 5A). We scored the number of trichomes with different number of branches in leaves 1/2 and 3/4, 5 d after Dex treatment. We observed an increase in the amount of four- and five-branched trichomes, and this occurred exclusively upon RepAwt expression (Fig. 5, B and C), indicating that it was an RBR/E2F-mediated effect.
Figure 5.
Inactivation of RBR alters trichome morphology. Control, RepAwt, and RepAE198K transgenic plants were sprayed with 1 μm Dex 13 d after sowing and analyzed 5 d after treatment. A, Examples of trichomes with three to five branches under light microscopy (top sections) or fluorescence microcopy after DAPI staining (bottom sections). In the latter, the arrowheads point to the nuclei. Branch number distribution of trichomes in the first pair of leaves (nos. 1/2), n > 100 in at least eight leaves (B), and in the second pair of leaves (nos. 3/4), n > 350 in at least six leaves (C). Numbers represent the percentage of each class. Asterisks indicate statistically significant differences relative to the control (at least, P < 0.05).
RBR Inactivation Produces Hyperplasia in Young Leaves
Constitutive overexpression of cell cycle activators (De Veylder et al., 2002; Dewitte et al., 2003) stimulate cell division in several Arabidopsis organs, including leaves. Conversely, ectopic expression of negative cell cycle regulators (De Veylder et al., 2001; del Pozo et al., 2002) reduces cell proliferation and cell number. Our results indicate that altering the E2F status by RBR inactivation might also have an effect on cell proliferation potential as revealed by the presence of cyclin B1-containing cells in differentiated leaves. Therefore, we analyzed leaves microscopically 5 d after the induction of either wild-type or mutant RepA using as controls plants transformed with an empty vector. As already mentioned, at this growth stage, leaves 1/2 have triggered the endocycle/differentiation program, whereas leaves 3/4 are switching to it (Boudolf et al., 2004b; Castellano et al., 2004; see also Fig. 4, A and D).
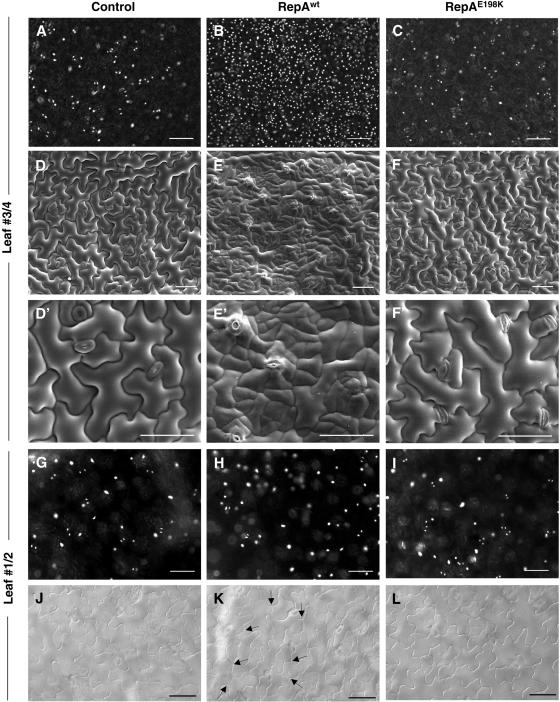
DAPI staining of leaf nuclei of leaves 3/4 revealed the typical distribution of well-separated, relatively large nuclei in control plants as well as in those induced to express the RepAE198K protein (Fig. 6, A and C). However, the leaf epidermis of plants expressing the RepAwt protein showed a large increase in the amount of small nuclei (Fig. 6B). These observations suggested that RepAwt was able to increase cell number in a RBR/E2F-dependent manner. To confirm this we analyzed, by scanning electron microscopy, leaf epidermis of plants treated in the same way. Now, it was more clearly observed that the leaf epidermis of plants expressing RepAwt contained clusters of small cells (Fig. 6, E and E′), which were not present in the controls (Fig. 6, D and D′) or in plants expressing the RepAE198K protein (Fig. 6, F and F′). We also analyzed the stomatal cells, which originate in the leaf epidermis by proliferation and further differentiation of cells of the stomatal lineage (Nadeau and Sack, 2003). Values of stomatal index are not very informative due to the large increase in nonstomatal cell number. In any case, we found that the stomatal number and morphology did not change significantly after induction of either RepAwt or RepAE198K. Altogether, these results indicate that E2F activation in leaves at the late proliferation stage has cell type-specific effects in the epidermis. Furthermore, only a subset of pavement cells can respond to ectopic activation of E2F activity leading to epidermal cell hyperplasia.
Figure 6.
Microscopic analysis of leaf epidermis of control, RepAwt, and RepAE198K transgenic plants. A to C, DAPI staining of the adaxial epidermis (leaf nos. 3/4). Note the large increase in the number of nuclei in the RepAwt plants. D to F, Cryo-scanning electron microscopy of the adaxial epidermis (nos. 3/4). Note the irregular surface of the leaf due to hyperplasia of the epidermis. D′ to F′, Close-up of D to F to show details of the clusters of small cells. G to I, DAPI staining of the adaxial epidermis (leaf nos. 1/2). Note that clusters of small cells do not appear. J to L, Light microscopy of the adaxial epidermis (leaf nos. 1/2). Note the absence of clusters of small cells, but instead the presence of ectopic cell wall dividing fully expanded pavement cells (arrows). Bars in all sections correspond to 50 μm. The study was carried out in the middle region of the leaf blade of 18-d-old plants 5 d after treatment with Dex (1 μm).
The consequences of E2F activation in more differentiated leaves (nos. 1/2) were different. Clusters of small pavement cells were not observed (Fig. 6, G–I). Instead, some fully expanded pavement cells were able to divide once after RBR inactivation, as indicated by the appearance of a new cell wall (Fig. 6, J and K), an effect that was also mediated by the RBR/E2F pathway since it was not observed in plants expressing RepAE198K (Fig. 6, J and L). The appearance of this kind of division planes in fully expanded epidermal cells was also observed after constitutive overexpression of E2Fa/DPa (De Veylder et al., 2002) or GL2-directed expression of KRP1 (Weinl et al., 2005). Moreover, this appears to be a dead-end process, as the new cells do not seem to be able to divide, explaining the lack of clusters of small cells. We also examined epidermal cells of dark-grown hypocotyls that also undergo several endocycles. Again, in this case we did not observe hyperplasia after RBR inactivation (data not shown).
Leaf Cell Layers Respond Differently to RBR Inactivation
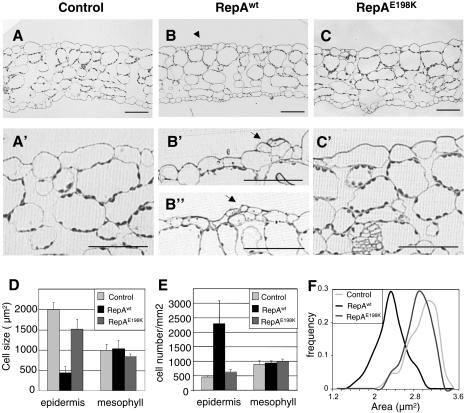
To ask whether RepA expression is able to induce cell division in other cell layers, we analyzed cross sections of leaves 3/4. This study confirmed that the leaf epidermis of plants after expression of RepAwt contained clusters of small cells (Fig. 7, B, B′, and B′′) that were not present in the controls (Fig. 7, A and A′) or in plants expressing the RepAE198K mutant (Fig. 7, C and C′). However, the morphology and number of cells in the internal cell layers did not appreciably change in any of the situations analyzed. While the average cell size of the adaxial epidermal cell layer decreased approximately 4-fold, with a concomitant increase in cell number, none of these changes was observed in the mesophyll (Fig. 7, D and E). Furthermore, the cell size distribution of leaf epidermal cells clearly indicates that a rather homogeneous population of small cells was produced in an RBR/E2F-dependent manner (Fig. 7F). We repeated these analyses, treating with Dex by watering instead of by spraying, and obtained essentially the same results (data not shown).
Figure 7.
Microscopic analysis and cell distribution in the leaves of control, RepAwt, and RepAE198K transgenic plants. A to C, Cross sections of leaf number 3. Sections below (A′ to C′) are higher magnification of A to C. Arrow in B indicates anticlinal division in the adaxial epidermis. Arrowheads in B′ and B′′; indicate periclinal and anticlinal divisions, respectively. Bars correspond to 50 μm. D, Size of adaxial epidermal and mesophyll cells (leaf nos. 3/4), n > 600 cells. E, Cell density of adaxial epidermal and mesophyll cells (leaf nos. 3/4). F, Cell size distribution of adaxial epidermal cells (leaf nos. 3/4). The study was carried out in the middle region of the leaf blade of 18-d-old plants, 5 d after treatment with Dex (1 μm).
Altogether, these data indicate that RepAwt, but not the RepAE198K mutant protein, was able to induce hyperplasia detectable even just a few days after induction and that this effect is cell layer specific. Therefore, we conclude that different cell layers respond differently to RBR inactivation. This likely reflects distinct physiological states in terms of cell division potential, as discussed below.
DISCUSSION
Plant organogenesis requires an appropriate balance between cell proliferation and differentiation (Gutierrez, 2005). Here, we have determined the relevance of the RBR/E2F pathway during Arabidopsis leaf development. Conditional inactivation of RBR function has been achieved using inducible expression of RepA, a geminivirus protein that interacts with RBR in an LxCxE-dependent manner (Xie et al., 1995, 1996). This increase in E2F activity occurred by relieving RBR-mediated repression of any of the three AtE2Fa to c factors with RBR-binding capacity (Gutierrez et al., 2002; Shen, 2002), which in turn activates a set of E2F target genes. Comparable studies in mice have demonstrated that retinoblastoma inactivation recapitulates all the phenotypes associated with HPV E7 oncoprotein-mediated activation of E2F (Balsitis et al., 2003). A large body of evidence points to RBR/E2F interactions as a major pathway affected by viral LxCxE-containing proteins. However, it should be kept in mind that the function of cellular RBR-binding proteins that use an LxCxE motif may also be affected. In any case, our results establish that inducible expression of the geminivirus RBR-binding RepA protein is a useful approach to evaluate the relevance of RBR inactivation during organogenesis in adult plants. Studies to address the role of RBR using an RNAi approach (W. Gruissem, personal communication) have yielded results consistent with ours. Therefore, we can conclude that the RBR/E2F pathway is a major player in regulating the balance between cell proliferation, endocycle program, and differentiation, but its relevance depends on the developmental stage, the tissue, and the cell type.
Distinct Impact of the RBR/E2F Pathway during Leaf Development
The increase in cell division and endoreplication are two phenotypic features of E2F activation, a consequence of transcriptional reprogramming, as suggested by an up-regulation of a set of E2F target genes. An overall role for E2Fa/DPa in controlling proliferation and endoreplication has been reported (De Veylder et al., 2002; Kosugi and Ohashi, 2003). Overexpression of upstream regulators of the pathway, e.g. cycD3;1, produces hyperplasia, reduces polyploidy, and prevents proper differentiation (Dewitte et al., 2003). Conversely, overexpression of CDK inhibitors produces hypoplasia and a reduction in endoreplication (Wang et al., 2000; De Veylder et al., 2001; Schnittger et al., 2003). However, it should be kept in mind that the effects of CDK inhibitors on endoreplication are dose dependent (Verkest et al., 2005; Weinl et al., 2005). In a different system, maize (Zea mays) endosperm, CDK inhibitors have also shown to play a role in controlling endoreplication (Coelho et al., 2005).
Previous constitutive overexpression approaches preclude the identification of temporally dependent effects. With our inducible approach, we found that the relevance of the RBR/E2F pathway depends on the developmental stage. In young leaves (nos. 3/4 at 13 d after sowing [das]), when most cell proliferation is about to finish, inactivating RBR stimulates cell division and leads to epidermal hyperplasia. This is likely a consequence of the activation of E2F targets involved in G1/S and, probably, G2/M transitions. In fact, expression of a dominant negative version of CDKB1;1, an E2F target that functions in G2, suppresses the E2Fa/DPa-mediated hyperplasia (Boudolf et al., 2004b). On the contrary, in older leaves (nos. 1/2 at 13 das), when most cells have switched to the endocycle program, more endocycles are produced instead of producing hyperplasia. It has been reported recently that the endoreplication phenotype, but not the ectopic division phenotype, observed in E2Fa/DPa overexpressing plants is counteracted by overexpression of DEL1, also known as E2Fe (Vlieghe et al., 2005). Thus, it would be important to identify E2F target genes that play specific or shared roles in cell proliferation and endoreplication.
Our results provide new insights into two aspects of cell division dynamics during leaf development: One refers to the proliferative potential of cells in different cell layers and another to the relevance of the RBR/E2F pathway in regulating the cell proliferation/differentiation balance. In particular, cells in the leaf epidermis follow different fates giving rise to pavement cells, trichomes, and stomata. While pavement cells and trichomes endoreplicate, cells of the stomatal lineage remain with a 2C DNA content. Most of the discussion in the previous section refers to pavement cells, and below we discuss aspects related to the other cell types.
Trichomes derive from cells that early in the primordia initiate their differentiation program associated with the occurrence of several endocycles (Hulskamp et al., 1999). In our experimental setting, these cells whose fate was specified early in leaf development have already switched to the endocycle program when E2F is activated. Thus, the increase observed in trichome branching suggests that more endocycles were induced. Conversely, ectopic expression of cycB1;2 in trichomes early during fate specification and before the switch to the endocycle program leads to multicellular trichomes and trichome clusters, as a result of precursor cell division (Schnittger et al., 2002).
Meristemoids, precursor cells of the stomatal lineage, retain their proliferative potential (Nadeau and Sack, 2003). However, the amount of stomata, indicative of cell division in stomatal precursor cells, does not increase in plants overexpressing constitutively E2Fa/DPa (De Veylder et al., 2002), CYCD3;1 (Dewitte et al., 2003), or under our conditions. Furthermore, the same occurs in plants overexpressing CDKB1;1, an E2F target gene required for proper stomatal development (Boudolf et al., 2004a, 2004b). We cannot exclude that the extra small cells that we observed in the epidermis are derived from stomatal precursors. Even if this were the case, we can conclude that inactivation of RBR at the developmental stage at which we have carried out the experiments prevents full differentiation of these cells into stomata. However, Park et al. (2005) have reported that abolishing tobacco NtRBR1 expression by virus-induced gene silencing leads to the appearance of stomatal clusters. In this report, RBR-specific, E2F-dependent effects cannot be separated from E2F-independent effects. Furthermore, tobacco NtRBR1 expression was abolished even before leaf primordia start to develop. The available data also suggest that restriction of cell division in stomatal precursors may be regulated by additional mechanisms. One of them may be licensing of DNA replication since constitutive AtCDC6 or AtCDT1 overexpression increases the stomatal index (Castellano et al., 2004). In any case, further restriction mechanisms, currently unknown, may operate since, under those conditions, only a doubling in the amount of stomata occurs, indicative of just one extra division to produce secondary meristemoids (Castellano et al., 2004).
Previous reports of epidermis-specific effects in cell proliferation further support our proposal. Overexpression of STRUWWELPETER (SWP) produces clusters of small cells in the leaf epidermis together with scattered fully expanded pavement cells (Autran et al., 2002), a phenotype strikingly similar to that observed by us. It is tempting to speculate that the possible function of SWP in transcription (Autran et al., 2002) may be related to that of RBR/E2F complexes. Tobacco plants silenced for the NtDEK gene, a calpain homolog, exhibit extended cell proliferation capacity and reduction of cell differentiation in the epidermis, whereas the internal cells are less affected (Ahn et al., 2004). This phenotype could be, at least in part, an indirect consequence of altering the RBR/E2F pathway since a set of cell cycle genes is transcriptionally up-regulated in these plants (Ahn et al., 2004).
The consequences of altering the levels of individual components of the RBR/E2F pathway have been described in several reports. Constitutive overexpression of AtCYCD3 (Dewitte et al., 2003) or of AtE2Fa/DPa (De Veylder et al., 2002) produces hyperplasia not only in epidermal but also in mesophyll cells. Likewise, overexpression of KRP2 leads to a reduction in cell number in different leaf cell layers (De Veylder et al., 2001). However, we have observed that RBR inactivation by overexpressing RepA in young leaves (nos. 3/4) stimulates cell proliferation in the epidermis but not in the mesophyll. These data are consistent with the observation that cell division potential lasts for longer in epidermal cells than in mesophyll cells (Donnelly et al., 1999). Similar effects have also been recently reported after reducing mRNA levels of RBR by virus-induced gene silencing (Park et al., 2005). This indicates that overexpressing single components of the pathway is substantially different from RBR inactivation. In this case, the endogenous amounts of RBR-bound E2F activity are released, and the effects are likely the consequence of the balanced action of several E2F. The possibility that other RBR-specific but E2F-independent pathways also occur cannot presently be ruled out.
Implications for Geminivirus Replication
Geminivirus DNA replication and infection require a number of cellular functions that are subverted by viral proteins (Gutierrez, 2000; Hanley-Bowdoin et al., 2004). One of them is the interference with the RBR/E2F pathway. In one geminivirus genus (Mastrevirus), this is mediated by the RepA protein through an LxCxE RBR-interaction motif (Xie et al., 1995, 1996). In the other geminivirus genera, which do not encode a homolog RepA, it is mediated by a different amino acid motif in another protein, Rep (Kong et al., 2000). These mechanisms are responsible for the up-regulation of cellular genes required for viral DNA replication, most of which are E2F targets (Hanley-Bowdoin et al., 2004). Geminivirus infection also induces nuclear DNA replication (Nagar et al., 2002), but this does not lead to activation of the cell division program. This implies that, with the exception of members of the genus Curtovirus, which encode the C4 protein able to induce local hyperplasia (Latham et al., 1997), progression through G2 and M is restricted in the infected leaf cell. The interaction of viral Rep protein with a kinesin-like motor protein (Kong and Hanley-Bowdoin, 2002) may serve to prevent progression through M and favor the occurrence of endocycles, as suggested by the increase in nuclear volume (Bass et al., 2000). Our data shed light onto this aspect since RepA induces endoreplication or cell division in an RBR-dependent manner, but depending on the developmental stage and the cell type. Future refinement of the approach used here will allow addressing some of these possibilities by the temporally controlled expression of different combinations of viral proteins.
MATERIALS AND METHODS
Plant Material and Transgenic Lines
To generate transgenic plants, the coding regions of RepAwt and RepAE198K in plasmids pBWRepA and pBWRepAE198K (Xie et al., 1995) were inserted in the pTA7002 vector (Aoyama and Chua, 1997). The constructs were introduced into Agrobacterium tumefaciens (C58C1 strain). Arabidopsis plants (Arabidopsis thaliana ecotype Landsberg erecta) were transformed by the floral dip method (Clough and Bent, 1998). Plants transformed with an empty vector were used as controls. Selection of transgenic plants was achieved by plating on Murashige and Skoog agar plates containing 25 μg/mL hygromycine. Homozygous plants were selected and the T4 was used for analysis. For phenotype analysis, seeds of control, RepAwt, and RepAE198K were grown directly in soil and maintained in a growth chamber at 22°C with a 16-h photoperiod. At day 13, plants were sprayed with a solution of 1 μm Dex (Sigma) to induce the expression of the transgene. Application of Dex was repeated 2 d later.
Antibodies and Western-Blot Assay
Antibodies against RepA protein were produced in rabbit using 50 μg of glutathione S-transferase-RepA mixed with complete Freund adjuvant. For analysis of RepA expression, homozygous plants were grown in Murashige and Skoog agar plate for 10 d and then transferred to Murashige and Skoog liquid medium containing Dex (1–20 μm during 0–20 h, as indicated). Total proteins were extracted in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.2% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride. RepA protein was detected by immunoblotting using a chemiluminescent procedure (ECLplus western-blot detection system; Amersham Bioscience).
Real-Time PCR Analysis
Total RNA was extracted using Trizol reagent (Invitrogen) and RT-PCR was carried out with the ThermoScript RT system (Invitrogen) using 500 ng of RNA as template and polydT primers. The LightCycler System with the FastStart DNA Master Green I (Roche) was used. The amount of actin (AtACT2) mRNA was determined to normalize for differences of total RNA amount. The data were generated from duplicate of three independent experiments. Primer sequences will be provided upon request.
Protein Interaction and EMSA
Plasmids pGBT-AtRBR, pGAD-AtE2Fa, b, and c were generated by cloning the full-length AtRBR (At3g12280), AtE2Fa (At2g31060), AtE2Fb (At5g22220), and AtE2Fc (At1g47870) coding sequences in-frame into the pGBT8 and pACT2 vectors (CLONTECH), respectively. Plasmids were transformed in the yeast HF7c strain, and the assays were carried out as described (Ramirez-Parra and Gutierrez, 2000). To express the third protein, the RepAwt and RepAE198K coding sequences were cloned into the pTFT1 vector (Egea-Cortines et al., 1999). Quantification of β-galactosidase assay was done in liquid culture using o-nitrophenyl-β-d-galactopyranoside (Sigma) as substrate, as described (Miller, 1972). Protein extracts for EMSA were prepared as described (Hurford et al., 1997) in a buffer containing 20 mm HEPES, pH 7.6, 0.5 m KCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm dithiothreitol, 20% glycerol, 0.2% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors. Then they were dialyzed against EMSA buffer (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm EDTA, 1 mm MgCl2, 1 mm dithiothreitol, and 12% glycerol). EMSA was carried out with 15 μg of total protein as described previously (Ramirez-Parra and Gutierrez, 2000).
Flow-Cytometry Analysis
The first (nos. 1/2) and second (nos. 3/4) pairs of leaves were harvested at 13, 15, and 18 das and chopped with a razor blade in 400 μL of nuclear isolation buffer (Galbraith et al., 1991). The suspension was filtered over 100 μm and a 30 μm nylon mesh, treated with RNaseA (200 μg/mL), and stained with propidium iodine (50 μg/mL). The nuclear DNA content was analyzed with a FACScalibur flow cytometer (BD Biosciences).
Microscopic Analysis
Tissues were placed in a solution of chloralhydrate, phenol, and lactic acid (2:1:1, w/w/w) and mounted for light microscopy observation. Samples were observed with an Axioskop2 Plus microscope (Zeiss), and the images were processed with the ImageJ software for cell size measurement. For nuclear visualization the leaves were destained overnight in ethanol, stained with DAPI (0.1 mg/mL) for 2 h, and mounted in phosphate-buffered saline (PBS)-glycerol 50% for observation. For histological observation of leaf sections, tissues were vacuum infiltrated and fixed overnight in a solution of PBS, pH 7.4, 4% paraformaldehyde, 2.5% glutaraldehyde, 0.1% Tween 20, 0.1% Triton X-100. After washing, samples were dehydrated and embedded in an Epoxy resin (TAAB 812). Semithin sections (1–2 μm) were stained with toluidine blue and observed as above. For immunofluorescence analysis, leaves were vacuum infiltrated with PBS, pH 7.4, 4% paraformaldehyde, 0.2% glutaraldehyde, 0.1% Tween 20, 0.1% Triton X-100, and then fixed overnight in a solution of PBS 4% paraformaldehyde. Tissues were then washed with PBS, soaked during 3 d in a solution of PBS-Suc 30%, and embedded in O.C.T. medium (Tissue-Tek). Cryostat sections (30 μm) were labeled with anti-RepA antibodies and anti rabbit-fluorescein isothiocyanate (Invitrogen). Observations were performed with a confocal Microradiance microscope (Zeiss). Cryo-scanning electron microscopy was carried on fresh leaves frozen in liquid nitrogen (CryoTrans Oxford CT1500).
Acknowledgments
Authors are indebted to M.B. Boniotti for developing RepA antisera, Sergio Llorens-Berzosa for technical assistance, M.T. Rejas for cross sections for microscopical analysis, and C. Ascaso and F. Pinto for help with the scanning electron microscopy. We also thank H. Sommer for the TFT plasmids, P. Doerner for the cycB1-GUS marker line, and M.B. Boniotti and E. Martinez-Salas for insightful comments on the manuscript.
This work was supported by the Spanish Ministry of Science and Technology (grant no. BMC2003–2131), by the Comunidad Autonoma de Madrid (grant no. 07B-53–2002), and by an institutional grant from Fundación Ramón Areces.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Crisanto Gutierrez (cgutierrez@cbm.uam.es).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071027.
References
- Ahn JW, Kim M, Lim JH, Kim GT, Pai HS (2004) Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J 38: 969–981 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Attwooll C, Denchi EL, Helin K (2004) The E2F family: specific functions and overlapping interests. EMBO J 23: 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inze D, Traas J (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsitis SJ, Sage J, Duensing S, Munger K, Jacks T, Lambert PF (2003) Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol Cell Biol 23: 9094–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Nagar S, Hanley-Bowdoin L, Robertson D (2000) Chromosome condensation induced by geminivirus infection of mature plant cells. J Cell Sci 113: 1149–1160 [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler Jde A, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L (2004. a) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004. b) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA (2005) Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol 138: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Joubes J, Inze D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6: 536–543 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Murray JA (2003) The plant cell cycle. Annu Rev Plant Biol 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Trivino S, Castellano MM, Sanchez MP, Ramirez-Parra E, Desvoyes B, Gutierrez C (2005) The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Res 33: 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L (2002) Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Robertson D, Hanley-Bowdoin L (2001) Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13: 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Kamm W, Dilkes BP, Lowe K, Hoerster G, Sun X, Ross M, Church L, Bunde C, Farrell J, Hill P, Maddock S, et al (2002) Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc Natl Acad Sci USA 99: 11975–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA 93: 8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C (2000) Geminiviruses and the plant cell cycle. Plant Mol Biol 43: 763–772 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (2005) Coupling cell proliferation and development in plants. Nat Cell Biol 7: 535–541 [DOI] [PubMed] [Google Scholar]
- Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC (2002) G(1) to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5: 480–486 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Robertson D (2004) Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol Plant Pathol 5: 149–156 [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Schnittger A, Folkers U (1999) Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. Int Rev Cytol 186: 147–178 [DOI] [PubMed] [Google Scholar]
- Hurford RK Jr, Cobrinik D, Lee MH, Dyson N (1997) pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev 11: 1447–1463 [DOI] [PubMed] [Google Scholar]
- Kang HG, Fang Y, Singh KB (1999) A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, et al (2000) A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol 128: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132: 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JR, Saunders K, Pinner MS, Stanley J (1997) Induction of plant cell division by beet curly top virus gene C4. Plant J 6: 1273–1283 [Google Scholar]
- Lavia P, Mileo AM, Giordano A, Paggi MG (2003) Emerging roles of DNA tumor viruses in cell proliferation: new insights into genomic instability. Oncogene 22: 6508–6516 [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bako L, Inze D, Bogre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Nadeau JA, Sack FD (2003) Stomatal development: cross talk puts mouths in place. Trends Plant Sci 8: 294–299 [DOI] [PubMed] [Google Scholar]
- Nagar S, Hanley-Bowdoin L, Robertson D (2002) Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14: 2995–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35: 25–31 [DOI] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim SJ, Kim JK, Kim WT, Pai HS (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Frundt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C (2000) Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett 486: 73–78 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Stierhof YD, Hulskamp M (2002) Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol 12: 415–420 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH (2002) The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 7: 505–511 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inze D, De Veylder L (2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inze D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inze D, De Veylder L (2003) Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJ, Nowack MK, Jakoby MJ, Hulskamp M, Schnittger A (2005) Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutierrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Suarez-Lopez P, Gutierrez C (1995) Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J 14: 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]