Abstract
Interactions between plant cell walls and plasma membranes are essential for cells to function properly, but the molecules that mediate the structural continuity between wall and membrane are unknown. Some of these interactions, which are visualized upon tissue plasmolysis in Arabidopsis (Arabidopsis thaliana), are disrupted by the RGD (arginine-glycine-aspartic acid) tripeptide sequence, a characteristic cell adhesion motif in mammals. In planta induced-O (IPI-O) is an RGD-containing protein from the plant pathogen Phytophthora infestans that can disrupt cell wall-plasma membrane adhesions through its RGD motif. To identify peptide sequences that specifically bind the RGD motif of the IPI-O protein and potentially play a role in receptor recognition, we screened a heptamer peptide library displayed in a filamentous phage and selected two peptides acting as inhibitors of the plasma membrane RGD-binding activity of Arabidopsis. Moreover, the two peptides also disrupted cell wall-plasma membrane adhesions. Sequence comparison of the RGD-binding peptides with the Arabidopsis proteome revealed 12 proteins containing amino acid sequences in their extracellular domains common with the two RGD-binding peptides. Eight belong to the receptor-like kinase family, four of which have a lectin-like extracellular domain. The lectin domain of one of these, At5g60300, recognized the RGD motif both in peptides and proteins. These results imply that lectin receptor kinases are involved in protein-protein interactions with RGD-containing proteins as potential ligands, and play a structural and signaling role at the plant cell surfaces.
In plants, proteins connecting the cell wall, the plasma membrane, and the cytoskeleton are believed to participate in a monitoring system that is required for the perception and transduction of environmental and developmental signals (Wyatt and Carpita, 1993). The continuum between wall, membrane, and cytoplasm is important not only for cell growth (Schindler et al., 1989) and cell differentiation (Roberts and Haigler, 1989) but also during abiotic and biotic stress (Levitt, 1983; Zhu et al., 1993), when the continuum has to function properly. During pathogen attack, for example, disruption of cell wall-plasma membrane adhesions may lead to a reduction of cell wall-associated defense responses, thereby making the plant more susceptible to disease (Lee-Stadelmann et al., 1984; Mellersh and Heath, 2001). The cell wall also may transmit chemical signals that direct specific developmental pathways, and some of these signals are thought to arise from the wall itself (Berger et al., 1994; McCabe et al., 1997). Other cues that are mechanical in origin, such as relaxation of the cell wall, can influence cell behavior (Lintilhac and Vesecky, 1984; Fleming et al., 1997). In all events, the cell wall and the plasma membrane act as the functional interface for chemical and mechanical signal exchange.
A variety of proteins have the potential to mediate wall-membrane interactions. Examples are arabinogalactan proteins, cellulose synthases, and endo-1-4-β-d-glucanases (Kohorn, 2000). Also, the wall-associated kinases (WAKs) have been described to physically link the plasma membrane to the plant cell wall. The WAK extracellular domain is variable between the five isoforms found in Arabidopsis (Arabidopsis thaliana), but all contain at least two epidermal growth factor repeats and a cytoplasmic kinase. This suggests that the plasma membrane-cell wall interacting molecules not only have a structural role through their extracellular domains but also a signaling role through their cytoplasmic domains, and can thus act in the communication between the apoplasm and the cytoplasm (Kohorn, 2001). However, evidence for a role of these proteins in the physical continuum between the cell wall and the plasma membrane is still lacking.
Adhesions between the cell wall and the plasma membrane during plasmolysis of Arabidopsis suspension cells were described previously (Canut et al., 1998). The cytoplasm and plasma membrane remained attached to the cell wall at some points, resulting in concave pockets. Such adhesions could be disrupted by the application of synthetic peptides containing the RGD (Arg-Gly-Asp) tripeptide motif, a cell adhesion motif present in several mammalian extracellular matrix proteins. Recently, we showed that plants have proteins with RGD-binding activity (Senchou et al., 2004). We found an 80-kD Arabidopsis plasma membrane protein that specifically binds to RGD-containing peptides and, in addition, shows high affinity binding to in planta induced-O (IPI-O), an RGD-containing protein secreted by the oomycete plant pathogen Phytophthora infestans. The 80-kD Arabidopsis plasma membrane protein recognizes IPI-O via its RGD motif, and, similar to RGD-peptides, IPI-O is able to disrupt plasma membrane-cell wall adhesions.
The aim of this study was to identify RGD-binding proteins in plants. As a first step, we used phage display as a tool to select peptides capable of interacting with the RGD tripeptide motif of the IPI-O protein. Combinatorial phage display peptide libraries provide powerful molecular tools to study protein-protein interactions, and they have been used extensively to discover bioactive peptides and ligands for receptors and to characterize enzymes (Bernal and Willats, 2004). The phage library we used here displayed a large repertoire of randomized heptapeptides expressed at the surface of M13 bacteriophages in the context of a coat protein. Two RGD-binding peptides were identified, and their sequence led to the identification of 12 proteins that were considered as candidates for natural RGD-binding proteins. Eight of these belong to the large family of receptor-like kinases (RLKs) in Arabidopsis. In this study, we show that the lectin receptor kinase (LecRK) encoded by At5g60300 is a potential interacting molecule involved in plasma membrane-cell wall adhesions. The possibility that RGD-containing proteins act as ligands for LecRK in plants is discussed.
RESULTS
Screening a Phage Library for Peptides Binding to IPI-O
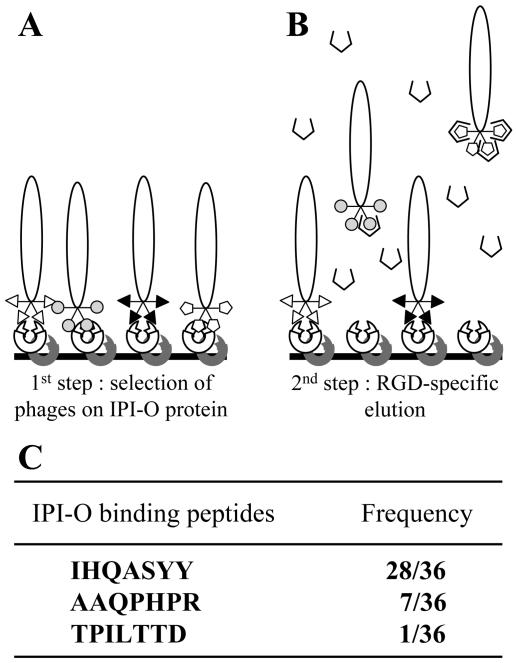
To find peptides capable of interacting with the RGD tripeptide motif in IPI-O protein, we screened a combinatorial phage display peptide library. Recombinant maltose-binding protein (MBP)-IPI-O protein was immobilized by adsorption onto microtiter plates saturated with bovine serum albumin (BSA). A suspension of the phage library was preincubated with BSA to eliminate phages expressing BSA-binding peptides and subsequently incubated in the coated microtiter plates (Fig. 1A). Given the relatively large size of the MBP-IPI-O fusion protein and the expected high number of potential peptide-binding sites in the protein for random peptides, we performed an elution with the RGDS peptide to specifically select phages that bind to the RGD tripeptide motif in IPI-O (Fig. 1B). Only a single round of selection was performed. Of the 36 phage plaques that were selected for sequence analysis, the majority, i.e. 28, expressed the peptide IHQASYY, seven the peptide AAQPHPR, and only one TPILTTD (Fig. 1C). These three peptides were used for further analysis.
Figure 1.
Selection of phages displaying random heptamer peptides that interact with the RGD sequence of the IPI-O protein. A, The library of phage-displayed peptides preincubated with BSA was incubated into microtiter plates coated with the IPI-O protein (lobster claw; first step). B, After extensive washing to remove unbound phages, competition with an RGD-containing peptide (broken square) was carried out to release a specific subset of the bound phages (second step). C, After a single round of bio-panning selection, 36 phages were randomly picked and the inserts were sequenced. The three different heptapeptides obtained are shown with their frequency.
Two Peptides Act as Inhibitors of RGD-Binding Activity in Arabidopsis
The three peptides, selected from the phage display library, were assayed for their ability to interfere with RGD binding to Arabidopsis plasma membrane proteins. In a previous study, we used photoaffinity cross-linking of RGD-containing peptides to demonstrate that an 80-kD plasma membrane protein in Arabidopsis has RGD-binding activity (Senchou et al., 2004). If we assume that the sequences of the peptides expressed by the selected phages occur in plasma membrane proteins and as such participate in the recognition of an RGD motif, then these peptides should inhibit the binding of an RGD-containing photoaffinity probe to its target, i.e. the 80-kD plasma membrane protein. Therefore, we synthesized the IHQASYY, AAQPHPR, and TPILTTD peptides and tested them using photoaffinity assays (Fig. 2A). The IHQASYY and AAQPHPR peptides clearly blocked cross-linking of an iodinated azido-RGD heptapeptide to the 80-kD polypeptide: Less than one-quarter of the label (24% and 22%, respectively) was linked to the 80-kD plasma membrane protein, and this inhibitory activity was in the same range as that of the recombinant MBP-IPI-O protein (29%). In contrast, the mutated MBP-IPI-O-D56A and -D56E proteins, as well as the TPILTTD peptide, did not inhibit cross-linking of the RGD heptapeptide. It should be noted that the two inhibitory peptides were those found with the highest frequency in the phage display selection and it is therefore likely that these two peptides, IHQASYY and AAQPHPR, have RGD-binding properties.
Figure 2.
Photoaffinity labeling of plasma membrane proteins from Arabidopsis with an azido RGD heptapeptide derivative, and effect of the RGD-binding heptapeptides selected by phage display. Plasma membrane proteins were separated in SDS-11% polyacrylamide gels after photolysis of the radioiodinated RGD-photoaffinity probe as described in “Materials and Methods.” Autoradiography revealed label associated with a protein of 80 kD. To quantify cross-linking, gel slices corresponding to the 80-kD polypeptide were excised and γ counted. The values of remaining label are indicated below each band as percentages of the control that was set at 100% in each independent experiment. Fifty micrograms of protein were deposited in each lane. A, Competition by isolated RGD-binding heptapeptides (IHQASYY, TPILTTD, AAQPHPR), a c-myc peptide with unrelated sequence (EQKLISEEDL; each peptide was applied at the concentration of 100 μm), and wild-type and mutated (-D56A, -D56E) MBP-IPI-O proteins (each protein was applied at the concentration of 0.3 μm). B, Competition by the two active RGD-binding heptapeptides (AAQPHPR and IHQASYY) and derived tetrapeptides. Each peptide was applied at the concentration of 100 μm.
Comparison of the RGD-Binding Peptides with Arabidopsis Protein Sequences
The sequences of the two RGD-binding peptides, IHQASYY and AAQPHPR, were compared to protein sequences present in the Arabidopsis genome database. First, we selected Arabidopsis proteins containing at least one transmembrane domain and a signal peptide, i.e. plasma membrane proteins, and looked for the presence of the RGD-binding sequences in their extracellular domains. When searching with the full-length heptapeptide sequences, or partial hexa- and pentapeptide sequences, no hit was obtained. Only with tetrapeptides was a number of proteins with matching sequences found (in total 14 hits corresponding to 12 proteins, of which eight belong to the large family of RLKs in Arabidopsis; Shiu and Bleecker, 2001). The results are summarized in Table I. The 12 proteins are all suitable candidates able to recognize and bind RGD tripeptide motifs in proteins, especially the RGD motif in IPI-O. To further investigate the significance of these proteins, we selected one candidate for further studies. We choose At5g60300 for several reasons: first, because it is a member of the RLK family that comprises the majority the candidates (eight out of 12); and, second, because At5g60300 contains not only the ASYY motif, the most represented sequence that was selected in the phage display, but also the second most represented sequence, PHPR. Each of these tetrapeptides could bind the RGD motif of IPI-O independently in the phage display experiment, and their inhibitory activity of these tetrapeptides was confirmed by photoaffinity assays (Fig. 2B). The ASYY and PHPR tetrapeptides blocked cross-linking of an RGD-containing photoaffinity probe to the 80-kD plasma membrane protein: The inhibition was 67% and 27%, respectively. In contrast, the IHQA and AAQP tetrapeptides had no effect. The third reason is that the UNIGENE database at National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) reported several full-length mRNAs and expressed sequence tags isolated from different plant tissues for At5g60300. In contrast, no expressed sequence tag was found for At5g60310, the RLK family member that also contains both motifs. Finally, the extracellular domain of At5g60300 appeared to be a legume-type lectin. Because of already known structures of legume lectins, a structural characterization of At5g60300 by molecular modeling was feasible.
Table I.
Arabidopsis plasma membrane proteins sharing amino acid sequences found in the RGD-binding peptides
The sequence source was the Munich Information Center for Protein Sequences Arabidopsis thaliana Genome Database. PSORT and TMHMM programs were used for the prediction of signal peptides and transmembrane domains. Identification of protein families and domains was performed using the Plant Receptor Kinase Resource (Shiu and Bleecker, 2001), and the Pfam and InterPro databases (Mulder et al., 2003; Bateman et al., 2004). The amino acids identical to those found in the phage sequence are shown in bold.
| Proteins | Sequences |
|---|---|
| Proteins selected from the IHQASYY sequence | |
| At1g07550 RLK (LRR) | AVKNIQASYGLNRI |
| At5g39000 RLK (CrRLK1L) | ASFTAQASYQESGV |
| At1g16380 putative cation-proton exchanger, CPA2 subfamily (AtCHX1) | LQKDSASYYIFFSF |
| At3g21630 RLK (LysM) | CPLALASYYLENGT |
| At3g45330 RLK (L-Lectin) | VESASASYYSDKEG |
| At3g45390 RLK (L-Lectin) | VVSASASYYSDREG |
| At5g60300 RLK (L-Lectin) | VAIASASYYSDMKG |
| At5g60310 RLK (L-Lectin) | VGTASASYYSDIKG |
| Proteins selected from the AAQPHPR sequence | |
| At5g06050 unknown function | GYFVWAAQPVYKHE |
| At1g10540 putative permease (AtNAT8) | KQEDLQPHPVKDQL |
| At5g10290 RLK (LRR) | NCGGRQPHPCVSAV |
| At1g73810 unknown function | WSRRGPHPRKYTTR |
| At5g60300 RLK (L-Lectin) | KLPEVPHPRAPHKK |
| At5g60310 RLK (L-Lectin) | RLPEVPHPRAEHKN |
At5g60300 Is a RLK with a Legume-Type Lectin Extracellular Domain
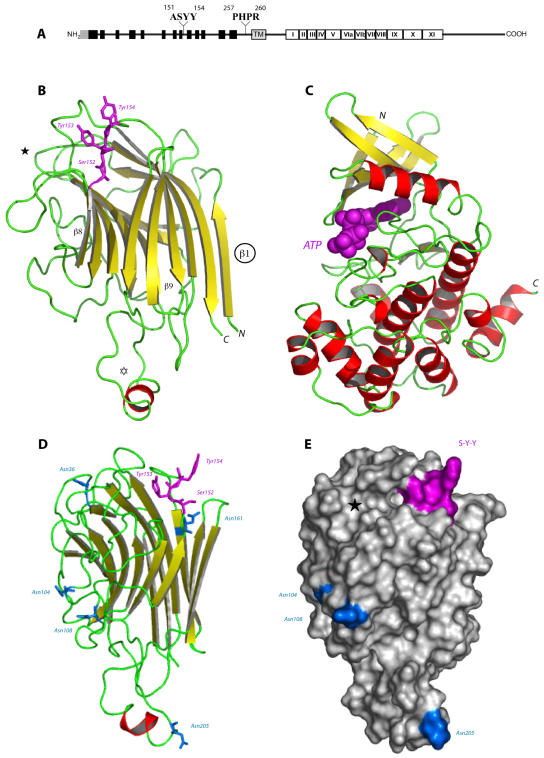
As deduced from its amino acid sequence, At5g60300 obviously belongs to the class of Ser/Thr-kinase receptors with a legume-type lectin extracellular domain. The extracellular lectin domain and the intracellular kinase domain are connected to a central hydrophobic transmembrane helix by two linker regions (Fig. 3A).
Figure 3.
Schematic representations of the lectin-like receptor kinase encoded by At5g60300. A, The NH2-terminal signal peptide is displayed as a gray box: It is presumably absent in the mature protein. The legume lectin domain is composed of 13 β-strands (black boxes) that form antiparallel β-sheets. The transmembrane domain is shown as a gray box (TM). The 12 subdomains of the kinase region are numbered in Roman numerals (I–XI). The amino acid sequences found in the isolated phage-displayed peptides are given above. B, Ribbon diagram of the modeled lectin-like domain of At5g60300 showing the overall β-sandwich fold organized in a flattened back face of a six-stranded β-sheet connected by turns and loops to a curved front face of a seven-stranded β-sheet. β-Strands are represented by yellow arrows, and coil structures are colored green. The additional extended loop (open star) contains a short α-helical stretch (colored red). Residues Ser-152, Tyr-153, and Tyr-154 (magenta ball-and-sticks) involved in the recognition of the RGD motif of IPI-O are located in an exposed loop connecting strand β8 to β9. The location of the putative carbohydrate-binding site is indicated (★). Strand β1 could participate in the dimerization of the lectin-like domain of At5g60300. C, Ribbon diagram of the modeled Ser/Thr-kinase domain of At5g30600 built up from a small β-rich N-terminal lobe (top part) connected to a large α-helical C-terminal lobe (bottom part). ATP (magenta CPK, Corey-Pauling-Koltun) is docked into the catalytic cavity located at the interface of the two lobes. D, Sagital view of the modeled lectin-like domain showing the location of the Asn residues belonging to the possibly glycosylated putative N-glycosylation sites. E, Molecular surface of the modeled lectin-like domain showing the location of the RGD-binding site S-Y-Y (colored magenta), the putative N-glycosylation sites (colored blue), and the apparently inactive carbohydrate-binding cavity (★). The lectin-like domain is similarly oriented in D and E.
The modeled lectin domain of At5g60300 exhibits the typical β-sandwich fold of the legume lectins, which consists of a flattened six-stranded β-sheet (back face) connected by turns and loops to an incurved seven-stranded β-sheet (front face; Fig. 3B). The lectin domain differs from the canonical legume lectin three-dimensional scaffold by the occurrence of an additional extended loop of 17 amino acid residues and a higher degree of N-glycosylation. Seven putative N-glycosylation sites, Asn-36-Ala-Ser, Asn-104-Ala-Ser, Asn-108-Gly-Ser, Asn-161-Glu-Ser, Asn-184-Val-Ser, Asn-205-Leu-Thr, and Asn-212-Arg-Ser, occur along the amino acid sequence of the lectin-like domain (Fig. 3D). With the exception of the buried Asn-184-Val-Ser and Asn-212-Arg-Ser, all other putative N-glycosylation sites are nicely exposed to the solvent and are expected to be glycosylated. Even though Asn-161-Glu-Ser does not occur in a loop region but is located at the beginning of strand β9, it is sufficiently exposed to be glycosylated (Fig. 3D). Accordingly, the exposed N-glycan chains should significantly restrict the accessibility of a large portion of the molecular surface of the lectin domain.
The Ala-151-Ser-Tyr-Tyr (ASYY) sequence stretch involved in the interaction with the RGD motif of IPI-O is located in the exposed loop connecting strand β8 to β9 (Fig. 3B) and occupies a shallow depression on the surface of the lectin-like domain (Fig. 3E). Ser-152 and the two Tyr-153 and Tyr-154 residues are readily exposed to interact with the RGD motif, most probably via hydrogen bonds. The N-glycan chains supposed to cover the lectin domain are too far from the RGD-binding residues to hamper the RGD recognition process. Also, interference with the putative carbohydrate-binding site of the lectin-like domain located in the vicinity of the RGD-binding residues seems unlikely (Fig. 3E). The putative carbohydrate-binding site is apparently devoid of any activity as predicted from docking experiments performed with simple sugars (data not shown). The invariant Asp-81 key residue responsible for the sugar recognition process of the canonical legume lectins is replaced by a His-79 residue in At5g60300. This prevents the formation of two hydrogen bonds with O3 and O6 of the pyranose ring upon anchoring a hexose into the carbohydrate-binding cavity. The lectin-like domain of At5g60300 is thereby suspected to be devoid of any significant monosaccharide-binding activity.
The three-dimensional model built for the Ser/Thr-kinase domain of At5g60300 exhibits the 12 subdomains characteristic of the catalytic kinase domain arranged in a small β-rich N-terminal lobe connected to a large α-helical C-terminal lobe (Fig. 3C). The catalytic loop connecting strand β6 to β7 contains the conserved Asp-448, Lys-450, and Asn-453 triad involved in both the ATP-binding and catalytic mechanism. The Gly-rich loop connecting strand β1 to β2, which participates in the binding of ATP, and the activation T-loop lying between strand β9 and α-helix F are also conserved. Accordingly, the kinase domain readily accommodates ATP in docking experiments (Fig. 3C) and is therefore suspected to be fully active.
The Lectin Domain of At5g60300 Binds RGD/RGE-Containing Peptides
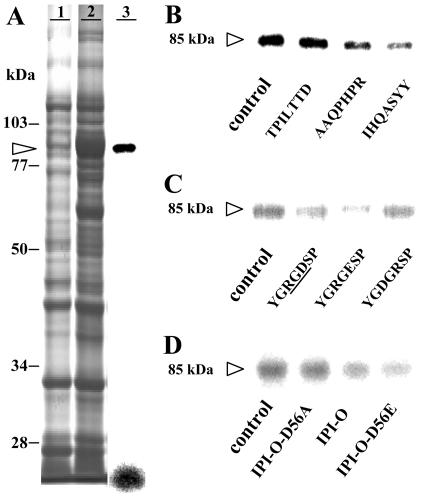
To verify that the lectin domain of At5g60300 can potentially recognize and bind RGD, we produced a recombinant protein in Escherichia coli consisting of intein and the lectin domain and tested this fusion protein with the photoactivatable RGD peptide (Fig. 4). The lectin domain indeed bound the RGD sequence (Fig. 4A, lane 3). Most of the label was associated with the recombinant protein of 85 kD. The label migrating at the front of the gel represents a free photoaffinity probe, which is released upon electrophoresis. The three heptapeptides selected from the phage library were then assayed as competitors (Fig. 4B): The IHQASYY and AAQPHPR peptides were strong inhibitors, whereas the TPILTTD peptide had no effect. The specificity of inhibition among the peptides is similar to that shown in Figure 2A for the 80-kD plasma membrane protein.
Figure 4.
Specificity of the photoaffinity labeling of crude extracts obtained from an E. coli strain expressing the lectin domain of At5g60300. A, Lanes 1 and 2 are electrophoregrams obtained from total proteins of E. coli containing either an empty pTYB12 vector (lane 1) or a pTYB12 vector with a partial open reading frame of At5g60300 encoding the extracellular lectin domain as insert (lane 2): The recombinant protein appeared as a major band at a molecular mass of 85 kD (arrowhead). Lane 3 is the labeling pattern observed after autoradiography when the radioiodinated RGD-photoaffinity probe was photolysed as described in “Materials and Methods.” Ten micrograms were deposited in lanes 1 and 2, 0.8 μg of protein in lane 3. B, Labeling patterns and competition by RGD-binding peptides: Each peptide was applied at the concentration of 200 μm. A total of 0.8 μg of recombinant protein was deposited in each lane. C, Labeling patterns and competition by peptides containing a full or modified RGD motif: Each peptide was applied at the concentration of 200 μm. A total of 0.8 μg of protein was deposited in each lane. D, Labeling patterns and competition by the wild-type and mutant MBP-IPI-O proteins: Each protein was applied at the concentration of 0.3 μm. A total of 0.8 μg of protein was deposited in each lane.
When testing the specificity toward the RGD motif, however, a marked difference between the 80-kD plasma membrane protein and the lectin domain of At5g60300 was observed. Both the YGRGDSP and YGRGESP peptides blocked cross-linking to the lectin domain (Fig. 4C), whereas on the plasma membrane protein only RGD-containing peptides were effective (Fig. 2). This difference in specificity between the 80-kD plasma membrane protein and the lectin domain of At5g60300 was confirmed when using MBP-IPI-O and the mutated MBP-IPI-O proteins as competitors for binding to the lectin domain of At5g60300 (Fig. 4D). Both the MBP-IPI-O and the mutated MBP-IPI-O-D56E inhibited cross-linking to the recombinant lectin domain by 61% and 83%, respectively, while the mutated MBP-IPI-O-D56A had no effect. These results indicate that the recombinant lectin domain recognizes not only the RGD tripeptide motif but also the RGE motif.
Effect of the RGD-Binding Peptides on Plasma Membrane-Cell Wall Adhesions
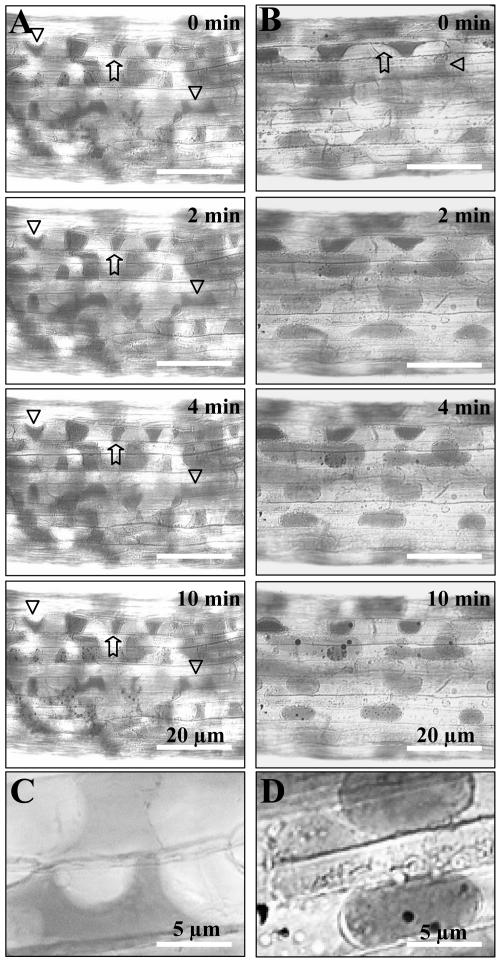
In earlier studies we demonstrated that RGD-containing peptides and the MBP-IPI-O protein promote the disruption of plasma membrane-cell wall adhesions (Senchou et al., 2004). Since the RGD-binding peptides that were selected from the phage library compete with the 80-kD plasma membrane protein for binding to RGD-containing peptides, we were urged to assay the capacity of the RGD-binding peptides to interfere in vivo with the plasma membrane-cell wall adhesions. Upon plasmolysis of etiolated hypocotyls of Arabidopsis, the plasma membrane readily separated from the cell wall, but at some points adhesions between cell wall and plasma membrane were maintained, resulting in pockets that are concave with respect to cells (Fig. 5, A and C). This pattern of plasmolysis occurred in all the hypocotyl cells and remained stable for at least 10 min. An identical pattern of plasmolysis was observed in the presence of the TPILTTD peptide (1 mm). However, in the presence of the IHQASYY and AAQPHPR peptides, the hypocotyl cells showed convex forms of plasmolysis: The plasma membrane quickly separated from the wall to make spherical protoplasts (Fig. 5, B and D). This pattern of plasmolysis is similar to what we observed previously when adding RGD-containing peptides or IPI-O protein to cell suspensions (Canut et al., 1998; Senchou et al., 2004), suggesting that RGD-containing peptides and RGD-binding peptides disrupt similar types of plasma membrane-cell wall adhesions in vivo.
Figure 5.
Plasma membrane-cell wall adhesions in Arabidopsis hypocotyls. Hypocotyls from 8-d-old etiolated seedlings were prepared and stained with neutral red as described in “Materials and Methods.” Upon addition of 0.4 m CaCl2, time courses of plasmolysis were observed for 10 min. Plasmolysis was induced in the absence of additives. A and B show a portion of entire hypocotyls. A, Control series without additives: The plasmolysed cells revealed plasma membrane-cell wall adhesions (arrows) and concave forms of plasmolysis (triangles). B, RGD-binding peptides (1 mm IHQASYY and AAQPHPR) were added to the plasmolysed hypocotyls: The plasma membrane quickly separates from the wall to make spherical protoplasts and convex forms of plasmolysis. C and D are enlarged images to show wall-to-membrane adhesions and concave forms of plasmolysis (C), as well as convex forms of plasmolysis in the presence of IHQASYY and AAQPHPR peptides (D).
DISCUSSION
The aim of this study was to obtain a molecular characterization of the plasma membrane-cell wall adhesions in Arabidopsis and, in particular, to identify proteins having an RGD-binding activity at the plasma membrane. A phage display approach allowed us to select 12 proteins as candidates to be RGD-binding proteins, and among them was the LecRK At5g60300. The latter contains in its extracellular lectin domain both the ASYY and PHPR sequences, which were able to interact with an RGD sequence. While ASYY strongly inhibited the RGD-binding activity located at the plasma membrane of Arabidopsis, inhibition by PHPR was less strong. It is very well possible that the ASYY sequence is the major determinant of the RGD-binding properties of the extracellular domain of At5g60300, with PHPR playing a less significant role. The PHPR sequence is predicted to be located at the emerging part of the molecule, close to the membrane lipid bilayer. On the other hand, three-dimensional modeling of the lectin domain of At5g60300 revealed that the ASYY sequence is located in a loop at the surface of the molecule, in a position to recognize ligands. Consistently, the lectin domain of At5g60300, obtained as a recombinant protein, showed an RGD-binding activity in vitro. LecRKs are members of the large family of the RLKs and, based on the identity of the extracellular domains, belong to a gene subfamily with at least 46 members (Shiu and Bleecker, 2001; Barre et al., 2002). From the compiled Arabidopsis Genome Initiative (2000), homologs to At5g60300 define an even smaller family of 11 members clustered in chromosomes 3 and 5. The ASYY or PHPR sequences are also found in At5g60310, At3g45330, and At3g45390 (Table I). Others exhibit an ASYF, AAYF, or PSYF sequence (At3g45410, At3g45420, At3g45430, At3g45440, At5g60270, At5g60280, At5g60320), while the PHPR sequence is less conserved. Identity or strong similarity to these sequences was also found in LecRKs from Medicago truncatula and Oryza sativa.
The biological function of LecRKs remains a pending question (Van Damme et al., 1998). LecRKs are most likely plasma membrane proteins (Hervé et al., 1999; Navarro-Gochicoa et al., 2003). The structural homology they share with genuine sugar-binding legume lectins suggests they are involved in the recognition of oligosaccharide or lipochitooligosaccharide signals (Barre et al., 2002; Navarro-Gochicoa et al., 2003). Docking experiments performed with simple sugars indicated that the carbohydrate-binding cavity of the modeled lectin-like domain of At5g60300 is apparently nonfunctional. However, since other soluble legume lectin-like proteins occur in Arabidopsis, one cannot exclude that the lectin-like domain of LecRKs might recruit another lectin domain via hydrogen bonds and Van der Waals interactions between strands β1 to form the homodimeric 12-stranded β-sandwich structure commonly found in typical legume lectins, e.g. in pea (Pisum sativum) or lentil (Lens culinaris) lectins (Van Damme et al., 1998). This association could restore a functional homodimer with an active carbohydrate-binding site susceptible to specifically interact with simple or complex sugars. The occurrence of a highly conserved hydrophobic cavity in both the typical legume lectins and the LecRKs similarly suggests a possible recognition of hormone ligands, such as auxins or cytokinins, by the lectin-like domain of LecRKs (Barre et al., 2002). The results shown in this study on the specific interaction of the lectin domain with the RGD/RGE motif of IPI-O show that, in addition to possible involvement in the specific recognition of small ligand molecules, the lectin-like domain of At5g60300 functions as a protein-protein interaction domain. Until now, such a function of protein recognition has rather been devoted to proteins of the RLK family containing Leu-rich repeat (LRR) domains (Torii, 2000).
We have previously shown that the RGD motif in synthetic peptides or in the IPI-O protein from the plant pathogen P. infestans disrupted the plasma membrane-cell wall adhesions of Arabidopsis (Canut et al., 1998; Senchou et al., 2004). Similar observations were reported for the brown alga Pelvetia (Henry et al., 1996), onion (Allium cepa; Canut et al., 1998), and tobacco (Nicotiana tabacum; Zhu et al., 1993). Other observations suggest the existence of multiple plasma membrane-cell wall interacting molecules involved in different functions in plants (Mellersh and Heath, 2001). Indeed, when the rust fungus Uromyces vignae attacked cowpea (Vigna unguiculata), its natural host, RGD-containing peptides disrupted the plasma membrane-cell wall adhesions. However, in nonhost pea cells, the plasma membrane remained attached to the cell wall during the pathogen attack. Also, when powdery mildew (Erysiphe polygoni) fungi attacked host or nonhost cowpea plants, the plasma membrane-cell wall adhesions appeared to be stronger than in noninfected plants. An increase in adhesions has also been reported for powdery mildew-barley (Hordeum vulgare) interactions (Lee-Stadelmann et al., 1984). Consistent with these observations, a variety of RGD-binding proteins may exist in plants, and they may differ in their specificity profiles. In this study, we selected 12 candidates from Arabidopsis, including the At5g60300 LecRK, that contain sequences covered by IHQASYY and/or AAQPHPR (Table I), and we expect more to show up when other plant genome sequencing projects are completed. Also, other LecRKs homologous to At5g60300 may be able to bind the RGD motif. We showed previously that the photolabeled 80-kD plasma membrane protein exhibited a strict specificity toward the RGD motif and not toward RGE. In contrast, the lectin domain of At5g60300 recognized both the RGD and RGE motifs. Since previous studies showed that peptides containing the RGE motif were unable to compete for the plasma membrane RGD-binding activity (Canut et al., 1998), the RGE binding of the lectin domain of At5g60300 may not be representative for the in vivo situation. The lack of specificity for RGD may be due to the fact that the lectin domain is a recombinant protein produced in a heterologous expression system (E. coli) that differs in structure with the corresponding native protein.
In this study, we showed that the peptides selected from the phage library are active in disrupting plasma membrane-cell wall adhesions in Arabidopsis. Since At5g60300 is expressed in hypocotyl tissues as determined by RT-PCR experiments (data not shown) and since its lectin domain contains the RGD-binding sequences, our microscopic observations indicate that the At5g60300 lectin domain may be an RGD-interacting partner in the plasma membrane-cell wall adhesions. To find further evidence that At5g60300 acts as a RGD-binding protein in planta, we also tested the recombinant lectin domain for its ability to disrupt plasma membrane-cell wall adhesions. Unfortunately, we did not observe any changes as compared to the control (data not shown). The recombinant lectin domain is 85 kD in size, and it is questionable whether such a large molecule can reach the membrane-wall interface to find its targets. Future experiments involving, for example, At5g60300-overexpressing lines, knockout mutants, and tagged versions of At5g60300 should clarify the in vivo role of this LecRK in RGD binding.
Interestingly, both LecRKs and WAKs, which are also putative linkers between the plasma membrane and the cell wall, have a kinase domain. It suggests that adhesions are part of signal transduction cascades in plant cells. The nature of the linkage between the cell wall and the plasma membrane is not known. The ligands of WAKs have been described, namely, pectins and secreted Gly-rich proteins (Kohorn, 2001). The ligands of LecRKs are not known. Our results indicate protein-protein interactions through a lectin domain, and based on that we hypothesize that RGD-containing proteins present either in the cell wall or in the plasma membrane are potential ligands for the At5g60300 LecRK. RGD-containing proteins in the cell wall can serve as a direct link between the cell wall and the plasma membrane, whereas RGD-containing proteins present in the plasma membrane can participate in membrane complexes with At5g60300 LecRK, which through such interactions gain the potential to recognize cell wall components or to activate downstream signaling pathways. Such an RGD-mediated interaction between membrane proteins was already described for RGD-binding proteins in mammals, i.e. the integrins (Papadopoulos et al., 1998; Erb et al., 2001). In plants, the application of RGD synthetic peptides led to numerous effects in very different physiological processes: The multiplicity of RGD-binding proteins and their potential interactions may explain that. Determining the partners of the LecRK lectin domains will help to unravel the structural and signaling role of such proteins at the plant cell surfaces.
MATERIALS AND METHODS
Phage Display Screening
The ipiO1 open reading frame from Phytophthora infestans (GenBank accession no. L23939), encoding the RGD-containing IPI-O protein without the putative signal sequence, was cloned into the pMALc vector in fusion with MBP (Senchou et al., 2004). Within the ipiO1 open reading frame, two individual mutations in the RGD tripeptide motif were obtained using PCR-mediated mutagenesis. The recombinant MBP-IPI-O fusion protein as well as the recombinant mutated proteins MBP-IPI-O-D56A and MBP-IPI-O-D56-E were expressed in Escherichia coli, extracted, and purified as described previously (Senchou et al., 2004).
Phage display screening was carried out using the Ph.D.-7 phage display library kit (New England Biolabs). The library consists of 2.8 × 109 electroporated sequences amplified once to yield an average of 70 copies of each sequence in 10 μL of the supplied phage suspension. The random 7-mer peptides were expressed in the context of the minor coat protein of M13 phage. The affinity selection of phages on the recombinant MBP-IPI-O protein was done as described by Koivunen et al. (1993) with the following modifications. A 10-μL portion of the library (2 × 1011 transducing units) was first incubated for 2 h at 4°C in a 1.5-cm-diameter well coated with BSA in 500 μL of Tris-buffered saline (TBS). The phages unbound to BSA were transferred to a similar well that had been coated with 100 μg/mL recombinant MBP-IPI-O protein and saturated with BSA. After incubation for 1 h at 4°C, the unbound phages were removed by washing 10 times with TBS buffer containing 0.1% Tween 20. The bound phages were specifically eluted with 0.5 mm RGDS peptide (Bachem) in TBS buffer. A single round of bio-panning selection was performed, and the affinity-purified phages were isolated on Luria-Bertani agar plate supplemented with Xgal/IPTG using the E. coli strain ER2738. From blue plaques, randomly picked phages were individually amplified and prepared for DNA sequencing.
Production of a Recombinant Lectin Domain
The open reading frame of At5g60300, encoding the extracellular lectin domain without the putative signal sequence (amino acids 14–275), was cloned from the F15L12 bacterial artificial chromosome provided by the Arabidopsis Biological Resource Center (Ohio State University). It was cloned into the XhoI-SmaI sites of the pTYB12 vector (New England Biolabs), which produced a fusion protein with intein containing a chitin-binding domain. The ligation products were introduced into E. coli strain ER2566. Plasmid DNA was isolated and checked by sequencing. Expression of the pTYB12-based plasmid was induced by activating the lac promoter with IPTG (0.5 mm) for 3 h 30 min at 37°C. Cells were harvested and resuspended in lysis buffer (20 mm Tris, pH 7.4 [HCl], 500 mm NaCl, 1 mm EDTA, 0.1% Triton X-100). Level of protein expression, isolation of the protein, and amino acid sequence were confirmed by SDS-PAGE, western-blot analysis, and mass spectrometry, respectively.
Plant Material and Purification of Plasma Membrane
Etiolated seedlings of Arabidopsis (Arabidopsis thaliana) ecotype Columbia were grown in 1-L Erlenmeyer flask containing 150 mL of Murashige and Skoog basal medium with the addition of Suc to 10 g L−1 (Bardy et al., 1998). They were harvested after 2 weeks of culture, leading to 20 to 30 g (fresh weight) of plant material. The purified plasma membrane vesicles were isolated from microsomes of Arabidopsis seedlings by preparative free-flow electrophoresis. The purity was assessed both by the determination of marker enzyme activities and by the reactivity of immunological probes. Based on the measurement of ATPase latency, the plasma membrane fraction appeared to consist essentially of cytoplasmic side-in vesicles (Bardy et al., 1998).
Peptides and Photoaffinity Assays
The synthesis of four peptides, AGRGDSP, YGRGDSP, YGRGESP, and YGDGRSP, was done automatically by stepwise Fmoc-t-butyl solid phase synthesis in a Synergy Applied Biosystems peptide synthesizer (Senchou et al., 2004). They were purified by reverse-phase HPLC. The RGDS peptide was purchased from Bachem. Other peptides were synthesized by the MilleGen Company.
The synthesis of the photoaffinity probe, N-(4-azido-salicylyl) AGRGDSP heptapeptide, started with the addition of the photoreactive heterobifunctional reagent N-hydroxysuccinimidyl-4-azido-salicylic acid (Pierce) to the NH2 group of the Ala residue of the AGRGDSP peptide. The photoaffinity probe was radioiodinated as described, to yield the mono-iodinated N-(4-azido-salicylyl) AGRGDSP peptide with a specific radioactivity of 78.6 TBq mmol−1. The iodinated peptide was purified by reverse-phase HPLC. Its integrity was confirmed by mass spectrometry and amino acid analysis (Senchou et al., 2004).
The photolabeling of the recombinant lectin domain with the mono-iodinated N-(4-azido-salicylyl) AGRGDSP heptapeptide was carried out in 50 mm MES, pH 5.5 (HCl), as follows. Incubation mixtures contained crude bacterial extracts (0.8 μg of protein) and 125I-labeled N-(4-azidosalicylyl) peptide (28 kBq, 0.35 pmol) in a total volume of 0.1 mL. The samples were incubated for 5 min on ice and then photoilluminated for 30 s on ice with high UV intensity light (312 nm) from a 176 W Spectroline lamp (Spectronics), situated 15 cm away from the sample. When additives were present, as indicated in the legends of the figures, the samples were preincubated for 10 min on ice. Laemmli sample buffer was added to the samples, and after SDS-PAGE the 11% acrylamide gels were dried under vacuum at 70°C for 2 h. Radioactivity in the protein bands was detected with a PhosphorImager (Molecular Dynamics). A similar procedure was used for the photolabeling of plasma membrane vesicles (Senchou et al., 2004).
Microscopy
Hypocotyls were separated from cotyledons and roots from 8-d-old seedlings, and were allowed to float on 50 mm Tris, pH 8.0 (HCl), for at least 1 h at room temperature. After a rinse in distilled water, hypocotyls were transferred to a solution of 0.05% neutral red for 5 min. After a new rinse in distilled water, hypocotyls were placed on a microscope slide into 100 μL of CaCl2 0.4 m to plasmolyze the cells. The observation was without coverslip. Directly under the microscope, stock solutions of peptides (50 mm) were applied to the hypocotyls as a drop with a pipette to a final concentration of 1 mm. Hypocotyls were observed with a Leitz DB-IRBE inverted microscope in bright field. Images were acquired with a Color Coolview CCD camera (Photonic Science) and analyzed with the Image-Pro Plus image analysis software (Media Cybernetics).
Molecular Modeling and Docking Experiments
Homology modeling of both the lectin and kinase domains of At5g60300 was performed on a Silicon Graphics O2 10000 workstation, using InsightII, Homology, and Discover3 programs (Accelrys). The atomic coordinates of the legume lectin LoLI of Lathyrus ochrus (PDB code 1LOB; Bourne et al., 1990a) and the kinase from Zea mays (PDB code 1LR4) were taken from the RCSB Protein Data Bank (Berman et al., 2000) and used to build the three-dimensional models. The amino acid sequence alignment was performed with ClustalX (Thompson et al., 1997), and the structurally conserved regions were inferred from the comparison of the HCA plots (Gaboriaud et al., 1987) generated with the program drawhca (http://smi.snv.jussieu.fr/hca/hca-form.html). Steric conflicts resulting from the replacement or the deletion of some residues in the modeled proteins were corrected during the model building using the rotamer library (Ponder and Richards, 1987) and the search algorithm (Mas et al., 1992) of Homology to maintain proper side chain orientation. Energy minimization and relaxation of the loop regions were carried out by several cycles of steepest descent. After correction of the geometry of the loops using the minimize option of TurboFrodo (Roussel and Cambillau, 1989), a final energy minimization step was performed by 50 cycles of conjugate gradient using Discover3. PROCHECK (Laskowski et al., 1993) was used to check the stereochemical quality of the three-dimensional models. Cartoons were drawn with PyMOL (“The PyMOL Molecular Graphics System”; DeLano Scientific LLC, http://www.pymol.org).
Docking of simple sugars into the carbohydrate-binding site of the lectin-like domain of At5g60300 was performed with InsightII. The lowest apparent binding energy (Ebind expressed in kcal mol−1) compatible with the Van der Waals interactions and hydrogen bonds found in the L. ochrus lectin/sugar complex (PDB code 1LOE; Bourne et al., 1990b) was calculated using the forcefield of Discover3. The position of the sugar observed in the lectin/sugar complex was used as a starting position to anchor the sugars into the binding site of the modeled At5g60300 lectin domain. Anchoring of ATP in the kinase domain of At5g60300 was similarly checked using the Z. mays kinase/ATP complex (PDB code 1LR4; Van Damme et al., 1998) as a template.
Sequence data from this article can be found in the GenBank/EMBL data libraries under IPI-O accession L23939 and the RLK accession numbers in Table I.
This work was supported by the Université Paul Sabatier, Toulouse; by the Centre National de la Recherche Scientifique; by GABI/Génoplante (contract no. AF–2001091); and by the Netherlands-French bilateral exchange program Van Gogh (NWO-VGP 85–343).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rafael Pont-Lezica (lezica@scsv.ups-tlse.fr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066464.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bardy N, Carrasco A, Galaud JP, Pont-Lezica R, Canut H (1998) Free-flow electrophoresis for fractionation of Arabidopsis membranes. Electrophoresis 19: 1145–1153 [DOI] [PubMed] [Google Scholar]
- Barre A, Hervé C, Lescure B, Rougé P (2002) Lectin receptor kinases in plants. CRC Crit Rev Plant Sci 21: 379–399 [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Taylor A, Brownlee C (1994) Cell fate determination by the cell wall in early Fucus development. Science 263: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal AJ, Willats WGT (2004) Plant science in the age of phage. Trends Plant Sci 9: 465–468 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Abergel C, Cambillau C, Frey M, Rougé P, Fontecilla-Camps JC (1990. a) X-ray crystal structure determination and refinement at 1.9 A resolution of isolectin I from the seeds of Lathyrus ochrus. J Mol Biol 214: 571–584 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Roussel A, Frey M, Rougé P, Fontecilla-Camps JC, Cambillau C (1990. b) Three-dimensional structures of complexes of Lathyrus ochrus isolectin I with glucose and mannose: fine specificity of the monosaccharide-binding site. Proteins 8: 365–376 [DOI] [PubMed] [Google Scholar]
- Canut H, Carrasco A, Galaud JP, Cassan C, Bouyssou H, Vita N, Ferrara P, Pont-Lezica R (1998) High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana links the cell wall. Plant J 16: 63–71 [DOI] [PubMed] [Google Scholar]
- Erb L, Liu J, Ockerhausen J, Kong Q, Garrad RC, Griffin K, Neal C, Krugh B, Santagio-Perez LI, Gonzalez FA, et al (2001) An RGD sequence in the P2Y(2) receptor interacts with alpha(V)beta(3) integrins and is required for G(o)-mediated signal transduction. J Cell Biol 153: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ, Mc Queen-Mason S, Mandel T, Kuhlemeier C (1997) Induction of leaf primordia by the cell wall protein expansin. Science 276: 1415–1420 [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP (1987) Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett 224: 149–155 [DOI] [PubMed] [Google Scholar]
- Henry CA, Jordan JR, Kropf DL (1996) Localized membrane-wall adhesions in Pelvetia zygotes. Protoplasma 190: 39–52 [Google Scholar]
- Hervé C, Serres J, Dabos P, Canut H, Barre A, Rougé P, Lescure B (1999) Characterization of the Arabidopsis lecRK-a genes: members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol Biol 39: 671–682 [DOI] [PubMed] [Google Scholar]
- Kohorn BD (2000) Plasma membrane-cell wall contacts. Plant Physiol 124: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD (2001) WAKs; cell wall associated kinases. Curr Opin Cell Biol 13: 529–533 [DOI] [PubMed] [Google Scholar]
- Koivunen E, Gay DA, Ruoslahti E (1993) Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem 268: 20205–20210 [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lee-Stadelmann OY, Bushnell WR, Stadelmann EJ (1984) Changes in plasmolysis form in epidermal cells of Hordeum vulgare infected by Erysiphe graminis: evidence for increased membrane-wall adhesion. Can J Bot 62: 1714–1723 [Google Scholar]
- Levitt J (1983) Plasmolysis shape in relation to freeze hardening of cabbage plants and to the effect of penetrating solutes. Plant Cell Environ 6: 465–470 [Google Scholar]
- Lintilhac PM, Vesecky TB (1984) Stress-induced alignment of division plane in plant tissues grown in vitro. Nature 307: 363–364 [Google Scholar]
- Mas MT, Smith KC, Yarmush DL, Aisaka K, Fine RM (1992) Modeling the anti-CEA antibody combining site by homology and conformational search. Proteins Struct Func Genet 14: 483–498 [DOI] [PubMed] [Google Scholar]
- Mc Cabe PF, Valentine TA, Forsberg LS, Pennell RI (1997) Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 9: 2225–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh DG, Heath MC (2001) Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Timmers CJ, Niebel A, Hervé C, Boutet E, Bono JJ, Imberty A, Cullimore JV (2003) Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula. Structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol 133: 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GK, Ouzounis C, Eliopoulos E (1998) RGD sequences in several receptor proteins: novel cell adhesion function of receptors? Int J Biol Macromol 22: 51–57 [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM (1987) Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol 193: 775–791 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Haigler CH (1989) Rise in chlorotetracycline fluorescence accompanies tracheary element differentiation in suspension cultures of Zinnia. Protoplasma 152: 37–45 [Google Scholar]
- Roussel A, Cambillau C (1989) TURBO-FRODO. Silicon Graphics Geometry Partners Directory (Committee, S.G., Ed.). Silicon Graphics, Mountain View, CA
- Schindler M, Meiners S, Cheresh DA (1989) RGD-dependent linkage between plant cell wall and plasma membrane: consequences for growth. J Cell Biol 108: 1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchou V, Weide R, Carrasco A, Bouyssou H, Pont-Lezica R, Govers F, Canut H (2004) High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell Mol Life Sci 61: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DJ (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 15: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU (2000) Receptor kinase activation and signal transduction in plants: an emerging picture. Curr Opin Plant Biol 3: 361–367 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P (1998) Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. CRC Crit Rev Plant Sci 17: 575–692 [Google Scholar]
- Wyatt SE, Carpita NC (1993) The plant cytoskeleton-cell-wall continuum. Trends Cell Biol 3: 413–417 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Shi J, Singh U, Wyatt SE, Bressan RA, Hasegawa PM, Carpita NC (1993) Enrichment of vitronectin- and fibronectin-like proteins in NaCl-adapted plant cells and evidence for their involvement in plasma membrane-cell wall adhesion. Plant J 3: 637–646 [PubMed] [Google Scholar]