Abstract
Ran is an evolutionarily conserved eukaryotic GTPase. We previously identified a cDNA of TaRAN1, a novel Ran GTPase homologous gene in wheat (Triticum aestivum) and demonstrated that TaRAN1 is associated with regulation of genome integrity and cell division in yeast (Saccharomyces cerevisiae) systems. However, much less is known about the function of RAN in plant development. To analyze the possible biological roles of Ran GTPase, we overexpressed TaRAN1 in transgenic Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). TaRAN1 overexpression increased the proportion of cells in the G2 phase of the cell cycle, which resulted in an elevated mitotic index and prolonged life cycle. Furthermore, it led to increased primordial tissue, reduced number of lateral roots, and stimulated hypersensitivity to exogenous auxin. The results suggest that Ran protein was involved in the regulation of mitotic progress, either in the shoot apical meristem or the root meristem zone in plants, where auxin signaling is involved. This article determines the function of RAN in plant development mediated by the cell cycle and its novel role in meristem initiation mediated by auxin signaling.
Ran is one of the important small G-protein families in organisms. In animals and yeast (Saccharomyces cerevisiae), it functions in many aspects, including nuclear transport, cell cycle control, postmitotic nuclear assembly, and spindle assembly (for review, see Dasso, 2001). The core biochemistry of Ran is similar to that of many Ras-related GTPases (for review, see Görlich and Kutay, 1999; Sazer and Dasso, 2000). Ran's intrinsic rates of nucleotide exchange and hydrolysis are slow. In vivo, these reactions require a nucleotide exchange factor (RCC1) and a GTPase-activating protein (RanGAP1) to achieve physiological rates. The Ran binding protein 1 (RanBP1) binds to RanGTP with high affinity and acts as an essential accessory factor to increase RanGAP1-mediated nucleotide hydrolysis (Sazer and Dasso, 2000). During interphase, RCC1 is a chromatin-associated nuclear protein, while RanBP1 and RanGAP1 are largely cytosolic. The asymmetric distribution of nucleotide exchange and hydrolysis enzymes across the nuclear envelope suggests that RanGTP should be largely nuclear and RanGDP largely cytosolic. This distribution plays a key role in determining the directionality of nuclear transport (Dasso, 2001). The requirement for Ran in nuclear transport has been extensively studied in animals (for review, see Görlich and Kutay, 1999). Established functions of Ran have been reported in transport of RNA and proteins across the nuclear pore (Görlich and Kutay, 1999), mitotic spindle organization (Hetzer et al., 2002), and nuclear envelope assembly (Zhang and Clarke, 2000).
In plants, although Ran's function has been identified, it has not been elucidated broadly (Yang, 2002). The tomato (Lycopersicon esculentum) Ran protein was functionally homologous with a yeast Ran-like protein in suppressing the effects of a mutation in a yeast homolog of RCC1 in mitosis (Ach and Gruissem, 1994). A RanBP1 homolog in Arabidopsis (Arabidopsis thaliana) alters the cell cycle progression in yeast (Xia et al., 1996). Haizel et al. (1997) isolated RanBP genes in Arabidopsis, characterized their binding specificity to Ran, and demonstrated the expression pattern of its transcripts. Recent intriguing findings have shown that the N terminus of RanGAPs is responsible for its targeting to the plant nuclear rim, and RanBP transgenic Arabidopsis is hypersensitive to auxin and shows mitotic progress arrested at metaphase (Kim et al., 2001; Rose and Meier, 2001).
The basic mechanism and logic of cell cycle control are highly conserved in eukaryotes and so are the key genes that mediate cell cycle progression (Nasmyth, 1996; Novak et al., 1998). Compared with what is known about mitosis in animals or yeast, our molecular-level knowledge of mitosis in higher plants is still in its infancy (Criqui and Genschik, 2002). During the past decade, the key molecules associated with cell proliferation have been identified experimentally in plants (Potuschak and Doerner, 2001). Most of these molecules are homologs of or contain domains that are homologous to yeast and animal genes known to have a role in the regulation of cell division. But the function of Ran in the plant cell cycle has not been elucidated (Potuschak and Doerner, 2001).
Wheat (Triticum aestivum) is an important crop characterized to date as having a large genome size (approximately 17,000 Mb). Until now, the technique of gene transformation and plantlet regeneration in wheat has been difficult for use in functional genomics, thus challenging researchers in creating successful transgenic wheat and analyzing gene function (Chong et al., 1998; Vasil and Vasil, 1999). To investigate the function of genes in wheat, investigators use tractable model systems, Arabidopsis and rice (Oryza sativa), to analyze the possible biological roles. In this article, we also took this strategy to identify the function of wheat RAN1 (TaRAN1), a wheat Ran GTPase. Our previous result has shown that TaRAN1 was associated with regulation of genome integrity and cell division in yeast systems (Wang et al., 2004). Much less is known about the process Ran is involved in. Here we show that overexpressed TaRAN1 in transgenic Arabidopsis and rice increased the proportion of cells in the G2 phase of the cell cycle and resulted in elevation of the mitotic index and prolongation of the life cycle. Furthermore, overexpression of TaRAN1 led to an increased amount of primordial meristem, a decreased number of lateral roots, and stimulated hypersensitivity to exogenous auxin. This article indicates that Ran protein is involved in regulation of the mitotic progress in the shoot apical meristem or root meristem zone in plants.
RESULTS
Molecular Characterization of TaRAN1-Overexpressed Transgenic Lines
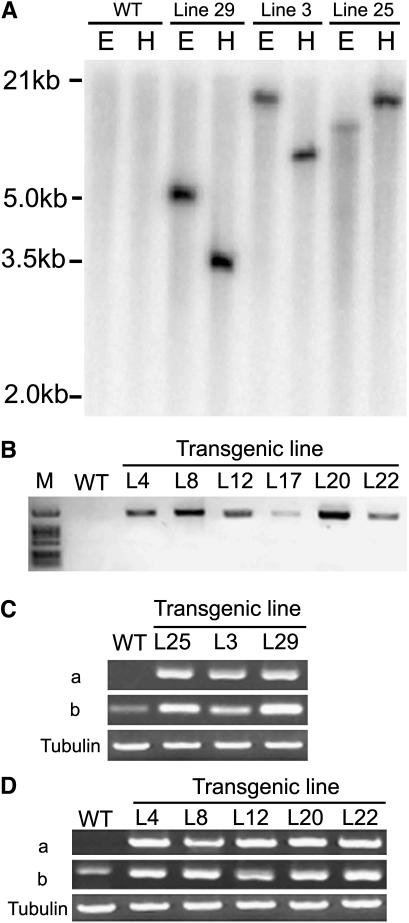
To analyze the role of TaRAN1 in plants, we overexpressed TaRAN1 in transgenic Arabidopsis and rice under the control of a constitutive cauliflower mosaic virus (CaMV) 35S promoter and a ubiquitin promoter of maize (Zea mays), respectively. Transformed lines of rice were confirmed by Southern blotting (Fig. 1A). The specific sequence of the reporter β-glucuronidase (GUS) gene revealed a single band on digestion with either HindIII or EcoRI in transgenic rice lines and different lines showed diverse hybridization maps. In the wild type, however, no signals occurred under the same conditions (Fig. 1A). Therefore, the transgenic lines of rice could be independent. In Arabidopsis, the 5′-end specific primer of the 35S promoter and the 3′-end specific primer of TaRAN1 were used to check the existence of TaRAN1 driven by a CaMV 35S promoter in genomes of transgenic Arabidopsis lines. PCR product was detected in the independent transformed lines, but not in wild-type Arabidopsis under the same conditions (Fig. 1B). Thus, TaRAN1 was integrated into the genome of transformed plants and highly expressed in transgenic rice and Arabidopsis.
Figure 1.
Molecular characterization of TaRAN1 transgenic plants of rice and Arabidopsis. A, Southern-blot assay for rice transgenic plants. Genomic DNA isolated from the transformed line or wild type (WT) was digested either with EcoRI (E) or HindIII (H). The blot was hybridized with the encoded region of the GUS gene labeled with 32P-dCTP and washed as described in “Materials and Methods.” B, Identification of independent transformed Arabidopsis lines by tissue PCR analysis. Total DNAs were isolated from WT or transgenic Arabidopsis plants. Transgene of TaRAN1 driven by the CaMV 35S promoter was examined by tissue PCR analysis as described in “Materials and Methods.” C and D, An examination of different transgenic plant lines of rice and Arabidopsis by semiquantitative RT-PCR. The specific primers (a) and the common primers (b) were used for RT-PCR. Tubulin RT-PCR was included as a loading control. The experiment was repeated five times. TaRAN1 expression in rice (C) and Arabidopsis (D).
To examine the expression of exogenous TaRAN1 in transgenic lines, we performed semiquantitative reverse transcription (RT)-PCR with either the specific primers, which could detect TaRAN1 (Fig. 1, Ca and Da), or the common primers, which could amplify the conserved domain of the Ran family (Fig. 1, Cb and Db). TaRAN1 was highly expressed at the transcriptional level in transgenic plants in both rice and Arabidopsis (Fig. 1, C and D), but not at all expressed in wild-type plants (Fig. 1, Ca and Da).
Overexpression of RAN1 Increased Primordia, Later Flowering, and Reduced Apical Dominancy
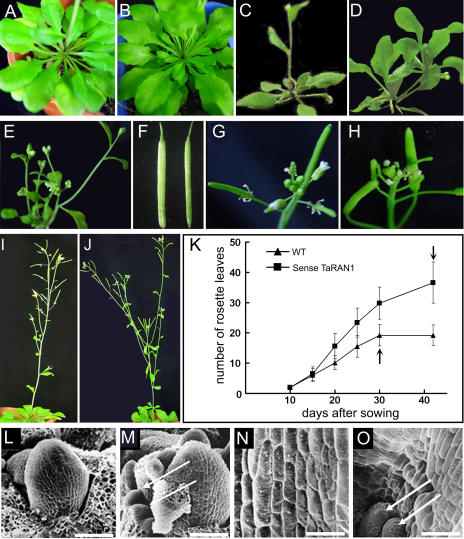
Transgenic plants of Arabidopsis produced seedlings with a range of phenotypes. Compared with the wild type, transgenic Arabidopsis showed distinct phenotypes, such as increased tiller number, weak apical dominance, abnormal root development, excess rosette leaves, and wider siliques (Fig. 2, A–F). The flower stalk emerged about 10 d later in RAN1-overexpressed plants than in wild-type plants under long-day conditions (Fig. 2K). The floral stalk of TaRAN1-overexpressed Arabidopsis plants was shorter and had more lateral floral branches than wild-type plants. In other words, the apical dominance of transgenic Arabidopsis was reduced (Fig. 2, G–J). Similarly, in transgenic rice plants, the tiller number reached 14.8 per plant on average. In contrast, wild-type rice had fewer tillers, about 5.6 per plant (Table I). These results suggest that TaRAN1 expression affected rosette leaf or tiller initiation in the meristematic region and subsequent growth.
Figure 2.
The phenotypes of the T2 generation of different lines of overexpressed TaRAN1 transgenic Arabidopsis. A, Rosette leaves of wild-type Arabidopsis grown for 3 weeks. B, Increased rosette leaves of transgenic Arabidopsis grown for 3 weeks. C, Normal phenotype of wild-type Arabidopsis. D, Two branches with rosette leaves in a transgenic plant. E, More tillery number and more buds in a transgenic plant. F, Mature silique morphology. Left, Transgenic plant; right, wild type. G, Apical inflorescence of wild-type Arabidopsis. H, Apical inflorescence including partial abortion of transgenic Arabidopsis. I, Normal floral apical dominance of wild-type Arabidopsis. J, Transgenic Arabidopsis with reduced apical dominance. K, Time curve of development of rosette leaves in transgenic Arabidopsis plants. Time of flowering is marked with arrows. Data are presented as mean ± se from three experiments (n = 15). WT, Wild type. L, Normal shoot apical point region of wild-type Arabidopsis on scanning electron microscopy. M, Additional new organ primordia around the shoot apical point of transgenic Arabidopsis. N, Axillary cells of wild type. O, Axillary cells of transgenic plants. Some hyperplastic cells and new primordial meristems emerged on the side of axillary cells of transgenic plants. The new primordia are marked with arrows. Arrows in M and O indicate the new primordia. Bar = 60 μm.
Table I.
Increased tillery number in 35S-sense TaRAN1 transgenic mature rice
se of the mean is based on 10 mature plants.
| Rice Plant Line | Tillery No. | Plant Height |
|---|---|---|
| cm | ||
| Wild type | 5.56 ± 1.64 | 55.6 ± 2.43 |
| Transgenic line 20 | 14.8 ± 5.22a | 44.8 ± 3.67a |
| Transgenic line 25 | 13.5 ± 4.3a | 43.7 ± 4.69a |
| Transgenic line 29 | 13.5 ± 2.12a | 41.9 ± 3.42a |
Significant differences, P < 0.01.
Previous results suggested that expression of TaRAN1 at the transcriptional level was higher in young stems and inflorescences in wheat (Wang et al., 2004). To investigate a possible role for TaRAN1 in the control of cell division and differentiation, we examined its effects on the shoot apex in transgenic Arabidopsis. In the wild type, the shoot apex contains the meristem itself, which consists of a central zone with fewer rapidly dividing stem cells and a surrounding peripheral region where new organ primordia are initiated. The growth of primordia is driven initially by cell division, and further rosette leaf and tiller development resulted from a combination of cell division and cell expansion accompanied by progressive differentiation. Compared with the wild type, transgenic Arabidopsis showed additional new organ primordia around the shoot apical point. Furthermore, some hyperplastic cells and new primordia also emerged at the axil of transgenic Arabidopsis (Fig. 2, L–O). Therefore, TaRAN1 protein might be involved in the regulation of cell division (Wang et al., 2004).
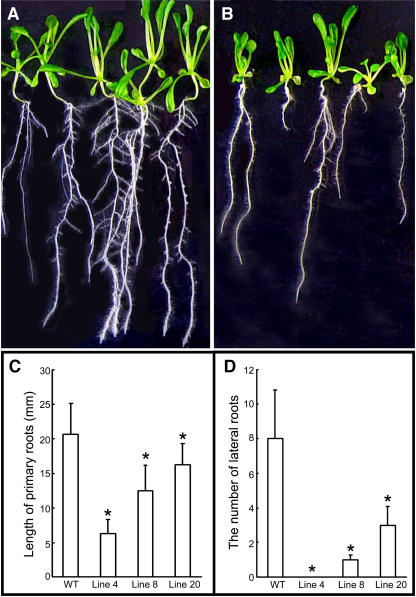
Transgenic Lines Displayed Inhibition of Primary Root Growth and Reduced Lateral Root Initiation
The striking and common phenotypes among TaRAN1 transgenic Arabidopsis seedlings were observed in root development (Fig. 3, A and B). Various lines of transgenic seedlings showed a similar phenotype, whereby the number of lateral roots was reduced and the growth of primary roots was suppressed (Fig. 3, C and D). The number of lateral roots was only 1.3 per plant on average in the transgenic seedlings. In contrast, the wild type showed eight per plant under the same conditions (Fig. 3, C and D).
Figure 3.
Root development of T2 generation of transgenic Arabidopsis. A, Wild-type Arabidopsis. B, Transgenic Arabidopsis. C, Length of primary roots in transgenic Arabidopsis lines. D, Number of lateral roots in transgenic Arabidopsis lines. C and D, Plants were allowed to grow for 10 d to determine the root length and lateral root production from the primary root after vernalization. The number of transgenic plants and wild type is more than 12 in each experiment. Results are presented as average values ± se from three experiments. Asterisk (*), Significant difference, P < 0.01.
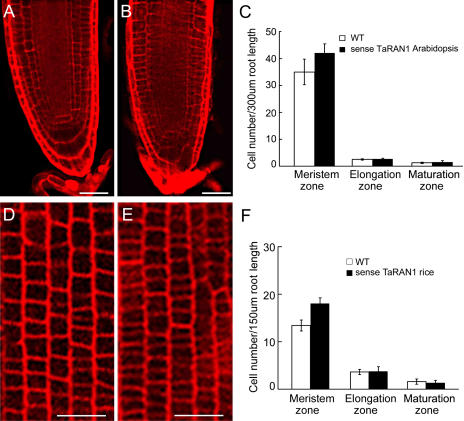
Roots of transgenic Arabidopsis showed no difference in cell size at both the elongation and maturation zones compared with those of wild-type Arabidopsis (Fig. 4C), but the number of cells in the meristem was greatly increased over that in the wild type (Fig. 4, A–C). The meristem cells of transgenic Arabidopsis were smaller and more tightly arrayed than those of the wild type. In transgenic plants of rice, all smaller and tightly arrayed cells were totally reemerged in the meristem zone (Fig. 4, D–F). The results suggest that TaRAN1 was involved in meristem cell proliferation in root development.
Figure 4.
Comparison of cell numbers between transgenic plants and wild-type Arabidopsis and rice. Cells of the meristem zone of primary roots of wild-type (A) and transgenic (B) Arabidopsis stained with PI. The number of transgenic Arabidopsis cells in the meristem was greatly increased over those in the wild type. Bar = 30 μm. C, Cell number of primary roots of Arabidopsis at the meristem, elongation, and maturation zones. Results are presented as mean ± se from three experiments (n = 10). Cells of the meristem zone of primary roots of wild-type (D) and transgenic (E) rice stained with PI. The number of transgenic rice cells in the meristem was greatly increased over those in the wild type. Bar = 30 μm. F, Cell number in primary roots of rice at the meristem, elongation, and maturation zones. Results are presented as mean ± se from three experiments (n = 10).
Hypersensitive Response to Indoleacetic Acid for Induction of Lateral Root Initiation
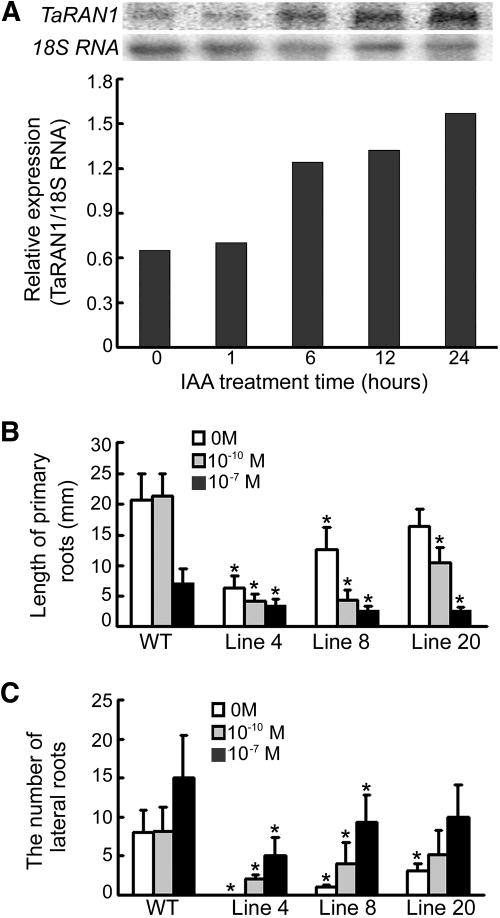
Multiple root and shoot phenotypes are commonly associated with many auxin-related mutants, and parallel phenotypes are often observed among different classes of auxin-related mutants (Berleth et al., 2000; Rogg and Bartel, 2001). To test whether root development was affected by auxin in transgenic plants, wheat seedlings were treated with 10−7 m indoleacetic acid (IAA) and showed substantial increase in TaRAN1 expression 24 h after treatment (Fig. 5A). Transgenic plants of Arabidopsis treated with 10−10 m IAA showed stimulated lateral root formation as compared with wild-type plants, which showed no lateral root formation (Fig. 5, B and C). The data suggest that sensitivity to auxin was increased in transgenic Arabidopsis compared with the wild type.
Figure 5.
Transcriptional response of TaRAN1 to IAA treatment in wheat and effect of auxin on root development in transgenic Arabidopsis plants. A, RNA gel-blot analysis of 10−7 m/L IAA-responsive expression of TaRAN1 in wheat. The bars represent the relative amount of the normalized expression of TaRAN1 after IAA treatment for different times. B, Effect of IAA (10−10 or 10−7 m) on growth of primary roots in transgenic plants of Arabidopsis. The transgenic plants treated with IAA showed suppressed primary root growth as compared with wild-type plants. C, Effect of IAA (10−10 or 10−7 m) on lateral root numbers in transgenic Arabidopsis plants. The number of lateral roots of transgenic plants was increased compared with wild-type plants after IAA (10−10 or 10−7m) treatment. B and C, All plates were allowed to grow for 10 d to determine the effect of various concentrations of auxin on root length and lateral root production after vernalization. Results are presented as average values ± se from three experiments. More than 12 roots were used in each experiment. Asterisk (*), Significant difference, P < 0.01.
Transgenic Lines Increased the Proportion of the G2 Phase in the Cell Cycle in Yeast and Rice
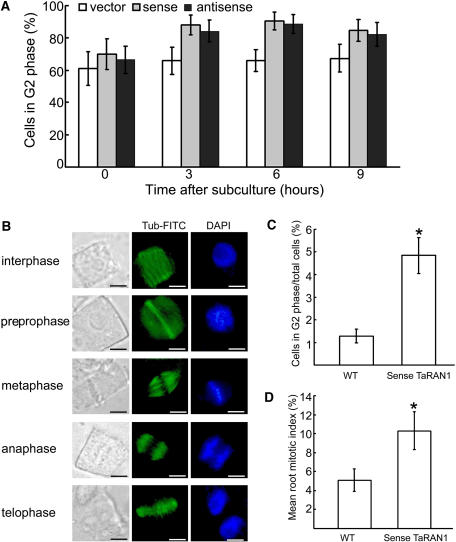
Fluorescence-activated cell sorter (FACS) was used to determine whether the DNA had replicated in the arrest cells of yeast by measuring rather than sorting (Liang et al., 1999). Fission yeast (Schizosaccharomyces pombe) cells transformed with sense or antisense TaRAN1, which has been shown previously to express foreign TaRAN1 or suppress the expression of yeast RAN1 (Wang et al., 2004), were grown overnight at 30°C. The cells were collected after subculture at 0, 3, 6, and 9 h, respectively. Cells were exposed to a fluorescent dye (propidium iodide [PI]) that binds to DNA so that the amount of fluorescence was directly proportional to the amount of DNA in each cell. Then the DNA content was analyzed by flow cytometry. In mammals and yeast, the G1 phase of the cell cycle is the major period of cellular growth (Neufeld and Edgar, 1998; Polymenis and Schmidt, 1999), but in fission yeast the G2 phase is the major period of cellular growth (Forsburg, 2001). Wild-type cells transformed with an empty expression vector displayed the normal distribution of cells in the G2, M, G1, and S phases. At all times, however, the average G2-phase populations were increased significantly (61.9% to 90.5%) in TaRAN1-transformed cells compared with wild-type cells (65.9% to 67.4%; Fig. 6A). TaRAN1-transformed cells were long or round large cells with duplicated DNA. Many cells showed a lag in their G2 phase, which is consistent with the abnormal chromosome segregation observed previously (Wang et al., 2004). No changes in the proportion of another phase were observed. These results suggest that the TaRAN1 transgenic cells are defective in the G2-to-mitosis transition.
Figure 6.
Effect of TaRAN1 transgene on the cell cycle phases of yeast and rice cells. A, Yeast cells transformed with pESPM-TaRAN1 analyzed by FACS at 0, 3, 6, and 9 h after subculture. The number of cells in the G2 phase was calculated as a proportion of the total cell population. Results are mean ± se from different lines analyzed in three independent experiments. B, Examination of the cell cycle phases of the nuclei of primary root cells by fluorescence microscopy in rice. Microtubules are stained green and chromosomes are stained blue with 4,6-diamidino-2-phenylindole. Bar = 4 μm. C, Proportion of cells in the G2 phase in wild-type (WT) and transgenic rice. D, Mitotic index in tips of primary roots of wild-type (WT) and transgenic rice. Results are presented as mean ± se from three experiments (n = 10). Asterisk (*), P < 0.01.
We examined the distribution of nuclei in transgenic rice root tips using fluorescence microscopy. At some points in the G2 phase, the cortical microtubules rearranged to form a band that encircles the cell, just below the plasma membrane (Fig. 6B). This preprophase band (PPB) of microtubules can be a marker of the G2 phase of the cell (Hoshino et al., 2003). A total of 4.8% of transgenic cells were in preprophase as compared with 1.3% of the wild type. Immunofluorescence observations of rice root-tip cells also showed an increased PPB of microtubules in the G2 phase. A striking increase of 3.6% in the proportion of G2 nuclei was observed in transgenic plants as compared with the wild type (Fig. 6C). These results suggest that the G2 phase of the transgenic root cells was delayed and the number of root cells in the G2 phase was increased. These results are consistent with the effects of the TaRAN1 protein seen in the yeast cell cycle (Wang et al., 2004) and suggest that TaRAN1 had a primary effect of increasing the tendency of cells to exit from the G1 phase, resulting in their accumulation in the G2 phase, and promoting the G2-M transition. The mitotic index in the primary roots of transgenic plants increased greatly as compared with that of wild-type plants (Fig. 6D). The proportion of mitotic cells in the transgenic roots were metaphase, 43%; anaphase, 23%; and telophase, 34%; whereas those in wild-type primary roots had a more even phase distribution (metaphase, 37%; anaphase, 25%; and telophase, 37%). The number of mitotic cells at the metaphase of transgenic plants was increased slightly. Indeed, the effect of TaRAN1 expression on cell phase distribution in the root tip suggested that TaRAN1 possibly plays a primary role in cell cycle progression.
DISCUSSION
Overexpression of TaRAN1 in Arabidopsis Increases Primordial Meristem Number
In the past years, three roles of Ran have been identified in animals: RNA and protein nuclear transportation (Görlich and Kutay, 1999), nuclear envelope reconstitution at the mitosis-to-interphase transition (Zhang and Clarke, 2000; Hetzer et al., 2002), and aster and spindle formation in mitosis (Zhang and Clarke, 2000). The RanGTP/RanGDP gradient controls the trafficking of molecules exceeding the diffusion limit of the nuclear pore across the envelope in animal cells. Molecular genetic evidence suggests cross talk between the organization of actin cytoskeleton and Ran-mediated nuclear transport in Drosphila (Minakhina et al., 2005). In Xenopus egg extracts, RanGTP induces aster and spindle assembly even in the absence of centrosome and DNA (Carazo-Salas et al., 1999; Kalab et al., 1999; Wilde and Zheng, 1999; Zhang and Clarke, 2000). Overexpression of plant Ran cDNA suppresses the phenotype of the pim46-1 cell cycle mutant in yeast. RanBPs interacted with the GTP-bound forms of the Ran1, Ran2, and Ran3 proteins of Arabidopsis. Both the AtRan and the AtRanBP genes are expressed coordinately and predominantly in meristematic tissues (Haizel et al., 1997). Overexpression of TaRAN1 causes increased primordial number, reduced apical dominancy and delayed flowering in Arabidopsis (Fig. 2), and increased tiller number in rice (Table I). This evidence supports a hypothesis that Ran protein may be involved in the regulation of shoot apical meristem and apical dominancy in plants.
Functional Analysis of TaRAN1 Reveals a Role in the Cell Cycle Regulation of Meristematic Root Cells
The cell cycle consists of alternating phases of DNA replication (S phase) and mitosis (M phase) that result in the formation of two daughter cells. These phases are usually separated by gaps: G1 and G2, which represent the interval between the M and S phases and between the S and M phases, respectively. To ensure that each daughter cell receives the correct hereditary material, controls must operate during the G1-S and G2-M transitions. Cyclin-dependent kinases (CDKs) play a central role in mediating cell cycle progression (Potuschak and Doerner, 2001; Inzé, 2005). In higher eukaryotes, the CDKs have evolved into gene families whose individual members have specialized functions during cell cycle progression. In contrast to animals, plants have two different classes of CDKs (A-type and B-type CDKs) that both seem to be involved in mitotic entry and progression. A-type CDKs are involved in both the G1-S and G2-M transitions (Hemerly et al., 1995). Expressing a dominant-negative mutant version of B-type CDK kinase protein in transgenic tobacco (Nicotiana tabacum) plants delays the G2-M transition (Mironov et al., 1999). Microinjection of affinity-purified active mitotic CDK complexes into stamen hair cells significantly accelerated chromosome condensation and the progression of prophase, and produced a rapid destabilization of the PPB in plant cells (Huch et al., 1996). Furthermore, a member of the A-type CDK class and a B1-type cyclin have been reported to bind to chromosomes (Mews et al., 1997; Stals et al., 1997). During interphase, the B1-type cyclins are found predominantly in the cytoplasm. During G2-M, these cyclins move to the nucleus, where they accumulate on nuclear material. Interestingly, these cyclins also accumulate around the nuclear envelope, which suggests that they may be involved in the breakdown of the nuclear envelope (Mews et al., 1997). Subcellular localization of active TaRAN1 protein is nuclear predominant (Wang et al., 2004). The overexpression of TaRAN1 in transgenic plants may result in a disturbed rate of delivery of proteins required for mitotic cell cycle progression to the nucleus and abnormal cell cycle completion. So we hypothesized that plant Ran protein may be involved in the transportation of these key proteins involved in the mitotic cell cycle across the nuclear envelope and regulate the cell cycle events and nuclear envelope assembly.
Revealing both conserved and plant-specific peculiarities in comparison with the mammalian system, the players and functions of the core machinery of the plant cell cycle are beginning to be clarified. Detailed knowledge of particular processes during cell cycle progression, however, is still missing, and many questions still remain to be answered (Rossi and Varotto, 2002). In mammals and yeast, the G1 phase of the cell cycle is the major period of cellular growth, and commitment to division during the G1 phase generally is subject to cell size control (Neufeld and Edgar, 1998; Polymenis and Schmidt, 1999). Plant meristematic cells do not undergo cell expansion and are relatively uniform in size. The meristematic cells of the primary root tip were reduced in average cell size in our TaRAN1-overexpressed plants. We speculate that TaRAN1 may promote precocious G1 exit and increase the number of G2-phase cells. So, cells in the root-tip meristem of TaRAN1-overexpressed plants are smaller than those in the wild type. Thus, TaRAN1 expression overrides the normal size control for cell division and results in a shift in the cell cycle distribution of meristem cells in the G1-to-G2 phase. For increased mitotic index in TaRAN1-overexpressed plants, the total number of the meristematic cells was also increased.
TaRAN1 Is Involved in Auxin Signal Transduction, and Overexpressed TaRAN1 Renders Arabidopsis Hypersensitive to Auxin
Heterotrimeric G proteins and Ras-related small GTPases in animals and yeast are prominent signaling molecules that mediate a wide variety of external stimuli to intracellular signaling pathways (Bourne et al., 1991; Bar-Sagi and Hall, 2000; Hur and Kim, 2002). Previous biochemical studies have suggested the involvement of GTP-binding proteins in the transduction of the auxin signal in rice coleoptiles (Zaina et al., 1990) and wheat mesophyll protoplasts (Bossen et al., 1991). Recent genetic analysis of a heterotrimeric G-protein mutants in Arabidopsis has suggested a role for this protein in modulating several hormonal signals, including auxin (Ullah et al., 2001; Wang et al., 2001; Assmann, 2002). Ran GTPases are emerging as important molecular switches that regulate the signaling of diverse cellular processes in plants. The results presented here show that Ran GTPases, in particular the wheat TaRAN1, play a pivotal role in auxin signaling. These results are consistent in part with previous findings that antisense expression of AtRanBP1c renders transgenic roots hypersensitive to auxin (Kim et al., 2001), which provides compelling evidence that these small GTPases play important roles in auxin-modulated signaling transduction.
In higher plants, where organogenesis occurs continuously, most cells maintain their ability to reenter and regulate the cell cycle in response to molecular signals. The mitotic cyclins comprise the A- and B-type cyclins involved in the regulation of the cell cycle from the S-to-M phases. The cycA2;2, a gene of alfalfa A2-type cyclin, is regulated by auxin and is involved in meristem formation. In the case of cycA2;2, auxin affects the spatial expression pattern of this cyclin by shifting the cycA2;2 expression from the phloem to the xylem poles where lateral roots initiate (Roudier et al., 2003). The G1-to-S checkpoint was also suggested to be a target for auxin-mediated lateral root initiation. In addition, a CDK-inhibitory protein (KRP2) was shown to be regulated transcriptionally by auxin and to prevent lateral root initiation by blocking the G1-to-S transition (Himanen et al., 2002).
Exogenous IAA (10−9 m) typically promotes the growth of lateral roots in wild-type Arabidopsis (Knee and Hangarter, 1996). If the concentration of exogenous IAA is lower than 10−9 m, the growth of lateral roots will not be promoted. In contrast, this response is induced at 10−10 m IAA in TaRAN1 transgenic plants. The roots of transgenic plants are hypersensitive to IAA on lateral root initiation. RanBP transgenic Arabidopsis is hypersensitive to auxin and arrests mitotic progress at the metaphase (Kim et al., 2001; Rose and Meier, 2001). Roots in transgenic plants accumulate higher levels of endogenous IAA (data not shown) and thus may require less exogenous IAA to respond (Kim et al., 2001). So, transgenic roots were hypersensitive to auxin. In both the suppression and induction of auxin responses, suppressors of auxin action must be delivered to the nucleus to block the expression of auxin-induced genes (Ulmasov et al., 1999). Overexpression of TaRAN1 protein might result in an abnormal or reduced rate of delivery of these key modulated proteins to the nucleus; in other words, they might cause diverse changes similar to the observed effects of TaRAN1 on regulation of cell cycle, gene expression, growth, and development and might provide an alternative explanation that the hypersensitivity to auxin is caused by abnormal distribution of auxin suppressors. The evidence that RanBP is predominantly expressed in meristerms and transgenic Arabidopsis (Kim et al., 2001; Rose and Meier, 2001) and that RAN1-overexpressed transgenic plants are hypersensitive to auxin supports a hypothesis that Ran is regulated by auxin and is involved in auxin-mediated modulation of the meristem. The relation between Ran and cyclins is worthy of investigation for understanding the molecular mechanism of the cell cycle in the meristem.
Ran GTPases in animals and yeast relay multiple signals to elicit multiple downstream responses (Dasso, 2001). Plant Ran and RanBP/RanGAP are known to be predominantly expressed in meristems and involved broadly in various cellular and developmental processes (Kim et al., 2001; Rose and Meier, 2001). Ran protein may participate in multiple signaling pathways. Our overexpression of RAN1 causes greatly increased primordial meristems, tillering, and number of cells in the meristem zone of roots, which is a novel function in the Ran family. Overexpressed Ran transgenic lines increased the proportion of cells in the G2 phase. We propose that TaRAN1, like several other RanBP/RanGAPs, also signals cellular activities that regulate cell division and auxin responses (Kim et al., 2001; Rose and Meier, 2001) and suggest that TaRAN1 is involved in auxin signaling pathways. Further work will be needed to reveal the participation of these signaling molecules as a class or as individual GTPases, such as RAN1, in mediating stimuli during plant growth and development.
MATERIALS AND METHODS
Plant Material
Overexpression of TaRAN1 was in Arabidopsis (Arabidopsis thaliana ecotype C24) and rice (Oryza sativa L. cv Zhonghua 10).
Generation of TaRAN1-Overexpressing Transgenic Plants
Transgenic Arabidopsis Plants
TaRAN1 was amplified for construction of the overexpressed vector with oligonucleotide 5′-GCTCTAGAATGGCGCTGCCGA-3′ and 5′-CGAGCTCCTCGATCAGATCG-3′ as primers. The PCR fragment was cloned into the pBI121 vector. The RAN1 gene was driven by the CaMV 35S promoter. This construct was verified by sequencing and electroporated into the Agrobacterium tumefaciens GV3101 and used for Arabidopsis transformation as described (Clough and Bent, 1998).
For plant transformation, Arabidopsis plants were grown in a greenhouse under long-day conditions (16-h light/8-h dark) for 4 weeks before a floral-dip procedure (Clough and Bent, 1998). Briefly, Agrobacterium cells were grown in Luria-Bertani broth for 24 h at 30°C. The cells were collected by centrifugation and resuspended in infiltration medium (one-half-strength Murashige Skoog medium, 5% Suc, 1× Gamborg's vitamins, 0.044 μm benzylaminopurine, and 0.04% Silwet L77) to an OD600 of 1.5 to 2.0. Plants were dipped into this suspension for 10 min and transferred to a greenhouse.
Seeds from plants treated by Agrobacterium were harvested and screened on selection medium (one-half-strength Murashige and Skoog medium, 1× Gamborg's vitamins, and 50 μg/mL kanamycin) for transformants. The putative transformants (defined as T1) were rescued from plates and grown in a greenhouse under long-day conditions (16-h light/8-h dark). The T3 generation was used for further experiments.
Transgenic Rice Plants
The digestion product TaRAN1 from pTripEX2-TaRAN1 was directionally cloned into the KpnI-SacI sites of a UN1301 vector to create UN1301-TaRAN1, which carried a gene of GUS as a marker. The coding sequence of TaRAN1 in the construct was verified by sequencing. The UN1301-TaRAN1 was used to transform the A. tumefaciens EHA105.
Rice embryonic calli were induced on scutella from germinated seeds and transformed with A. tumefaciens EHA105 containing the desired binary vector, as described (Ge et al., 2004; Xu et al., 2005). Transgenic plants were selected in one-half-strength Murashige and Skoog medium containing 75 mg/L hygromycin (Sigma). Hygromycin-resistant plants from calli, defined as transgenic plants of the T0 generation, were transplanted into soil and grown in a greenhouse at 28°C. For analysis of root phenotypes of transgenic plants, seeds of the T1 generation were germinated in one-half-strength Murashige and Skoog medium containing 75 mg/L hygromycin and confirmed by GUS staining.
RNA Gel Blots and RT-PCR for Gene Expression Analysis
Winter wheat (T. aestivum L. cv Jingdong No. 1) seeds were sown on culture plates after being surface sterilized in 2% (v/v) NaOCl for 0.5 h. The plumules excised from seeds were collected and stored in liquid N2 for isolation of total RNA. Total RNA of rice and Arabidopsis leaves was isolated by use of the Qiagen RNeasy plant mini kit (Qiagen). Total RNA of 15 μg was loaded on each lane for electrophoresis. RNA transfer and cross-linking onto a nylon membrane (Hybrid N+; Amersham) was as described (Sambrook et al., 1989; Ge et al., 2000). The probe of TaRAN1 cDNA was labeled with [32P]dCTP (China Isotope). After hybridization for 20 h at 68°C, the membrane was washed once with 2× SSC plus 0.1% SDS at 68°C for 20 min, then washed with 1× SSC plus 0.1% SDS at 37°C for 30 min. The membrane was exposed to x-ray film (Eastman-Kodak) at −70°C for 3 to 7 d.
RT-PCR was performed according to the manual of the RT-PCR kit (TaKaRa). The conserved region of Ran family primers was 5′-GAGAACATCCCCATTGTCC-3′ and 5′-CAAACAGTTTGCAGCCCACCA-3′. To ensure TaRAN1 gene-specific amplification, PCR primers were designed according to the sequence of no conservative untranslated regions of the 3′ terminus. TaRAN1 primers were 5′-TGCCAAGAGCAACTACAA-3′ and 5′-ATGATCCACATATTGAGCC-3′. Tubulin primers were 5′-TCAGATGCCCAGTGACAGGA-3′ and 5′-TTGGTGATCTCGGCAACAGA-3′ from the tubulin gene sequence. RT-PCR reactions were repeated five times.
Electron Microscopy of Shoot Apex Cells
Shoot apexes were immediately fixed in formaldehyde acetic acid solution (3.7% formaldehyde, 50% ethanol, and 5% acetic acid) for 12 h and dehydrated in a graded ethanol series. The dehydrated materials were critical-point dried in liquid CO2 and mounted on metallic stubs. The mounted material was shadowed with gold before scanning electron microscopy (Hitachi S-800).
Imaging of Root Cell Size
To examine cell arrangement and size, root tips were stained with 100 μg/mL PI solution and observed under a confocal laser scanning microscope (Zeiss) with an argon laser.
Flow Cytometric Analysis
The fission yeast (Schizosaccharomyces pombe) Leu− strain SPQ-01 was used. Expression vectors with the sense and the antisense TaRAN1 were constructed in pESPM. The transcriptional expression and phenotype analysis of TaRAN1 in fission yeast was as described (Wang et al., 2004). Transgenic sense and antisense TaRAN1 cells were grown overnight in minimal medium containing thiamine at 30°C. The cells were washed three times with the minimal medium without thiamine to derepress the NMT1 promoter and then diluted to 2 × 106 cells/mL in Edinburgh minimal medium. The cells were incubated at 30°C and samples were taken at 0, 3, 6, and 9 h. The DNA content of individual transgenic cells was determined by flow cytometry. Cells were prepared for the FACS by staining with PI (Sazer and Sherwood, 1990). Briefly, cells were fixed in ethanol overnight at 4°C, washed, and resuspended in 0.5 mL of 50 mm sodium citrate, pH 7.0, containing 0.1 mg/mL RNase A for 2 h at 37°C followed by incubation in 4 μg/mL PI (final concentration). Each sample was analyzed with use of a FACS caliber cytometer (B-D Corporation).
Immunolabeling of Root Cells
Excised 1- to 2-mm-long root tips were fixed for 1 h in PEM buffer (50 mm PIPES, pH 6.9, 5 mm MgSO4, 5 mm EGTA) containing 4% formaldehyde, then washed in PEM (3 × 10 min). Cell walls were digested (50 min, 28°C) with 1% (w/v) cellulase “Onozuka” R-10 and 0.1% pectolyase Y23 (Yakult) in PEM. Root tips were washed in PEM (3 × 10 min), and then squashed with use of a pencil eraser between two multiwell slides (ICN) coated with 0.1% polyethyleneimine (Sigma). Cells were treated at −20°C (5 min), dried at room temperature for 2 min, then extracted with 1% Triton X-100 at room temperature for 30 min. Cells were washed in phosphate-buffered saline (PBS; 131 mm NaCl, 5.1 mm Na2HPO4, 1.56 mm KH2PO4, pH 7.2) for 10 min three times before being blocked in incubation buffer (PBS containing 1% bovine serum albumin, 20 min). Incubations of primary and secondary antibodies, diluted in incubation buffer, were for either 1 h at 20°C or overnight at 4°C, with antibodies in multiple labeling experiments applied concurrently. Washes with PBS (3 × 10 min) were run after each antibody incubation. Cells were stained with 0.1 μg/mL 4,6-diamidino-2-phenylindole (Sigma) in PBS (5 min), rinsed briefly in PBS, and mounted with 1,4-diazabicyclo[2,2,2]octane (triethylenediamine; Sigma). Coverslips were sealed with nail polish. Slides were viewed under a Zeiss epifluorescence microscope. For each root tip, all mitotic figures and total numbers of cells in the meristematic zone were counted. Mitotic indices were calculated with use of the following formula: number of cells in mitosis/total number of cells.
Analyses of Auxin Effects
Seeds were surface sterilized in 70% ethanol for 1 min, then in 50% (v/v) NaClO solution for 8 min, and rinsed in sterile water. Seeds were then placed on plates and vernalized for 72 h to synchronize germination. After vernalization, all plates were placed in the same growth chamber and allowed to grow for 10 d to determine the effect of various concentrations of auxin (IAA) on root length and lateral root production (Kim et al., 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF488730.
Acknowledgments
We thank Yuan Cheng for assisting with microscope techniques and Huili Liu for assistance with the constructs.
This work was supported by the Major State Basic Research Program of the People's Republic of China (2005CB120806), the National Natural Science Foundation of China (30470157 and 30470167), and the Innovation Grant of the Chinese Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kang Chong (chongk@ibcas.ac.cn).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071670.
References
- Ach RA, Gruissem W (1994) A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc Natl Acad Sci USA 91: 5863–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell (Suppl) 14: S355–S373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Hall A (2000) Ras and Rho GTPases: a family reunion. Cell 103: 227–235 [DOI] [PubMed] [Google Scholar]
- Berleth T, Mattsson J, Hardtke CS (2000) Vascular continuity and auxin signals. Trends Plant Sci 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Bossen M, Tretyn A, Kendrick RE, Vredenberg WJ (1991) Comparison between swelling of etiolated wheat (Triticum aestivum L.) protoplasts induced by phytochrome and α-naphthal-leneacetic acid, benzylaminopurine, gibberellic acid, abscisic acid and acetylcholine. J Plant Physiol 137: 706–710 [Google Scholar]
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanisms. Nature 349: 117–127 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW (1999) Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400: 178–181 [DOI] [PubMed] [Google Scholar]
- Chong K, Bao S, Xu T, Liang T, Huang H, Zeng J, Xu J, Xu Z (1998) Functional analysis of ver gene using antisense transgenic wheat plant. Physiol Plant 102: 87–92 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Genschik P (2002) Mitosis in plants: how far we have come at the molecular level? Curr Opin Plant Biol 5: 487–493 [DOI] [PubMed] [Google Scholar]
- Dasso M (2001) Running on Ran: nuclear transport and the mitotic spindle. Cell 104: 321–324 [DOI] [PubMed] [Google Scholar]
- Forsburg SL (2001) Fission yeast. In SL Forsburg, ed, 2002 Yearbook of Science and Technology. McGraw-Hill, New York, pp 108–110
- Ge L, Chen H, Jiang J, Zhao Y, Xu M, Xu Y, Tan K, Xu Z, Chong K (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135: 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Liu JZ, Wong WS, Hsiao WLW, Chong K, Xu ZK, Yang SF, Kung SD, Li N (2000) Identification of a novel multiple environmental factor-responsive 1-aminocyclopropane-1-carboxylate synthase gene, NT-ACS2, from tobacco. Plant Cell Environ 23: 1169–1182 [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Haizel T, Merkle T, Pay A, Fejes E, Nagy F (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J 11: 93–103 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler Jde A, Bergounioux C, Van Montagu M, Engler G, Inze D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Gruss OJ, Mattaj IW (2002) The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol 4: E177–E184 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inze D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino H, Yoneda A, Kumagai F, Hasezawa S (2003) Roles of actin-depleted zone and preprophase band in determining the division site of higher-plant cells, a tobacco BY-2 cell line expressing GFP-tubulin. Protoplasma 222: 157–165 [DOI] [PubMed] [Google Scholar]
- Huch J, Wu L, John PCL, Hepler LH, Hepler PK (1996) Plant mitosis promoting factor disassembles the microtubule preprophase band and accelerates prophase progression in Tradescantia. Cell Biol Int 20: 275–287 [DOI] [PubMed] [Google Scholar]
- Hur EM, Kim KT (2002) G protein-coupled receptor signaling and cross-talk: achieving rapidity and specificity. Cell Signal 14: 397–405 [DOI] [PubMed] [Google Scholar]
- Inzé D (2005) Green light for the cell cycle. EMBO J 24: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M (1999) The ran GTPase regulates mitotic spindle assembly. Curr Biol 9: 481–484 [DOI] [PubMed] [Google Scholar]
- Kim SH, Arnold D, Lloyd A, Roux SJ (2001) Antisense expression of an Arabidopsis Ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell 13: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee EM, Hangarter RP (1996) Differential IAA dose response of the axr1 and axr2 mutants of Arabidopsis. Physiol Plant 98: 320–324 [Google Scholar]
- Liang DT, Hodson JA, Forsburg SL (1999) Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J Cell Sci 112: 559–567 [DOI] [PubMed] [Google Scholar]
- Mews M, Sek FJ, Moore R, Volkmann D, Gunning BES, John PCL (1997) Mitotic cyclin distribution during maize cell division: implications for the sequence diversity and function of cyclins in plants. Protoplasma 200: 128–145 [Google Scholar]
- Minakhina S, Myers R, Druzhinina M, Steward R (2005) Crosstalk between the actin cytoskeleton and Ran-mediated nuclear transport. BMC Cell Biol 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inze D (1999) Cyclin-dependent kinases and cell division in plants—the nexus. Plant Cell 11: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (1996) At the heart of the budding yeast cell cycle. Trends Genet 12: 405–412 [DOI] [PubMed] [Google Scholar]
- Neufeld P, Edgar BA (1998) Connections between growth and the cell cycle. Curr Opin Cell Biol 10: 784–790 [DOI] [PubMed] [Google Scholar]
- Novak B, Csikasz-Nagy A, Gyorffy B, Nasmyth K, Tyson J (1998) Model scenarios for evolution of the eukaryotic cell cycle. Philos Trans R Soc Lond B Biol Sci 353: 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV (1999) Coordination of cell growth with cell division. Curr Opin Genet Dev 9: 76–80 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Bartel B (2001) Auxin signaling: derepression through regulated proteolysis. Dev Cell 1: 595–604 [DOI] [PubMed] [Google Scholar]
- Rose A, Meier I (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 98: 15377–15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V, Varotto S (2002) Insights into the G1/S transition in plants. Planta 215: 345–356 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fedorova E, Lebris M, Lecomte P, Gyorgyey J, Vaubert D, Horvath G, Abad P, Kondorosi A, Kondorosi E (2003) The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol 131: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sazer S, Dasso M (2000) The ran decathlon: multiple roles of Ran. J Cell Sci 113: 1111–1118 [DOI] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW (1990) Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci 97: 509–516 [DOI] [PubMed] [Google Scholar]
- Stals H, Bauwens S, Traas J, Van Montagu M, Engler G, Inze D (1997) Plant CDC2 is not only targeted to the pre-prophase band, but also co-localizes with the spindle, phragmoplast, and chromosomes. FEBS Lett 418: 229–234 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im K-H, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil IK, Vasil V (1999) Transgenic cereals: Triticum aestivum (wheat). In IK Vasil, ed, Molecular Improvement of Cereal Crops. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 137–147
- Wang X, Xu W, Xu Y, Chong K, Xu Z, Xia G (2004) Wheat RAN1, a nuclear small G protein, is involved in regulation of cell division in yeast. Plant Sci 167: 1183–1190 [Google Scholar]
- Wang X-Q, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y (1999) Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284: 1359–1362 [DOI] [PubMed] [Google Scholar]
- Xia G, Ramachandran S, Hong Y, Chan YS, Simanis V, Chua NH (1996) Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J 10: 761–769 [DOI] [PubMed] [Google Scholar]
- Xu ML, Jiang JF, Ge L, Xu YY, Chen H, Zhao Y, Bi YR, Wen JQ, Chong K (2005) FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep 24: 79–85 [DOI] [PubMed] [Google Scholar]
- Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaina S, Reggiani R, Bertani A (1990) Preliminary evidence for involvement of GTP-binding protein(s) in auxin signal transduction in rice (Oryza sativa L.) coleoptile. J Plant Physiol 136: 653–658 [Google Scholar]
- Zhang C, Clarke PR (2000) Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288: 1429–1432 [DOI] [PubMed] [Google Scholar]