Abstract
To identify new loci in abscisic acid (ABA) signaling, we screened a library of 35S∷cDNA Arabidopsis (Arabidopsis thaliana)-expressing lines for ABA-insensitive mutants in seed germination assays. One of the identified mutants germinated on 2.5 μm ABA, a concentration that completely inhibits wild-type seed germination. Backcrosses and F2 analyses indicated that the mutant exhibits a dominant phenotype and that the ABA insensitivity was linked to a single T-DNA insertion containing a 35S∷cDNA fusion. The inserted cDNA corresponds to a full-length cDNA of the AtPP2CA gene, encoding a protein phosphatase type 2C (PP2C). Northern-blot analyses demonstrated that the AtPP2CA transcript is indeed overexpressed in the mutant (named PP2CAox). Two independent homozygous T-DNA insertion lines, pp2ca-1 and pp2ca-2, were recovered from the Arabidopsis Biological Resource Center and shown to lack full-length AtPP2CA expression. A detailed characterization of PP2CAox and the T-DNA disruption mutants demonstrated that, whereas ectopic expression of a 35S∷AtPP2CA fusion caused ABA insensitivity in seed germination and ABA-induced stomatal closure responses, disruption mutants displayed the opposite phenotype, namely, strong ABA hypersensitivity. Thus our data demonstrate that the PP2CA protein phosphatase is a strong negative regulator of ABA signal transduction. Furthermore, it has been previously shown that the AtPP2CA transcript is down-regulated in the ABA-hypersensitive nuclear mRNA cap-binding protein mutant abh1. We show here that down-regulation of AtPP2CA in abh1 is not due to impaired RNA splicing of AtPP2CA pre-mRNA. Moreover, expression of a 35S∷AtPP2CA cDNA fusion in abh1 partially suppresses abh1 hypersensitivity, and the data further suggest that additional mechanisms contribute to ABA hypersensitivity of abh1.
The phytohormone abscisic acid (ABA), which regulates many agronomically important aspects of plant life, including seed development and dormancy, plays a critical role in plant stress responses such as drought, salinity, cold shock, wounding, and pathogen attack (Schroeder et al., 2001; Finkelstein et al., 2002; Hetherington and Woodward, 2003; Fan et al., 2004). These physiological responses to ABA are in large part due to changes in gene expression and a complex signal transduction network (Hoth et al., 2002; Seki et al., 2002; Leonhardt et al., 2004; Takahashi et al., 2004). Several transcription factors mediating ABA responses have been isolated (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Uno et al., 2000; Himmelbach et al., 2002; Abe et al., 2003).
Whereas the biosynthesis of ABA is well understood (Seo and Koshiba, 2002), the mechanisms by which ABA regulates multiple plant responses is beginning to be revealed through genetic and physiological analyses in Arabidopsis (Arabidopsis thaliana). To date, genetic screens for ABA-hypersensitive mutants have indicated that processes including farnesylation (era1; Cutler et al., 1996; Pei et al., 1998), inositol 1,4,5-triphosphate (IP3) dephosphorylation (fry1; Xiong et al., 2001b), and RNA metabolism (hyl1; Lu and Fedoroff, 2000; abh1, Hugouvieux et al., 2001; sad1, Xiong et al., 2001a) are required to attenuate the ABA signal. However, surprisingly few non-transcription factor-encoding genes have been identified as recessive ABA-insensitive disruption mutants, namely, the G-protein α-subunit GPA1 (Wang et al., 2001), the RCN1 protein phosphatase type 2A subunit (Kwak et al., 2002), the OST1/SnRK2E protein kinase (Mustilli et al., 2002; Yoshida et al., 2002), the AtRBOHD/F NADPH oxidases (Kwak et al., 2003), ABI8 (Brocard-Gifford et al., 2004), RPK1 (Osakabe et al., 2005), and GCA2 (Himmelbach et al., 1998).
Many gene families in the Arabidopsis genome have large numbers of homologs relative to other sequenced genomes (Arabidopsis Genome Initiative, 2000). Therefore, the relatively low number of recessive ABA-insensitive mutants is most likely due to redundancy in genes encoding ABA transducers, requiring analyses of double or multiple mutations in (partially) redundant genes (Kwak et al., 2003). In addition, the ABA signaling pathway is mediated by a network of events and interacts with many other signaling pathways including drought, salinity, cold, sugar, GA3, and ethylene (Finkelstein et al., 2002; Zhu, 2002; Himmelbach et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2005). Consequently, functional redundancy could also be explained by the fact that such complex networks have the ability to buffer a mutation's effects in a neighboring pathway (Cutler and McCourt, 2005).
To further enhance the chance of success in identifying new mutants in ABA signaling, we screened a library of 35S∷cDNA Arabidopsis-expressing lines (LeClere and Bartel, 2001) for ABA-insensitive mutants in seed germination assays. In theory, the 35S∷cDNA fusion can generate mutants due to (1) random insertional gene disruptions; (2) overexpression of the inserted full-length or truncated cDNA; or (3) silencing of the cDNA-corresponding endogenous gene (LeClere and Bartel, 2001). Consequently, this screen can identify both positive and negative regulators of a given signaling pathway. In this study, we report isolation of two strong ABA-insensitive mutants: a new insertional mutant of ABI5, coding for a basic Leu zipper transcription factor, a well-known positive regulator of ABA responses (Finkelstein and Lynch, 2000), as well as a constitutive overexpressor of AtPP2CA (named PP2CAox) encoding a protein phosphatase type 2C (PP2C).
Sixty-nine PP2Cs are encoded in the Arabidopsis genome (Kerk et al., 2002; Schweighofer et al., 2004), and a gene disruption phenotype has only been reported for one of these PP2Cs (Leonhardt et al., 2004; Saez et al., 2004). The protein phosphatase PP2CA belongs to group A of the Arabidopsis PP2C family, together with ABI1, ABI2, and AtP2C-HA (hereafter named AtP2C-HAB1; Schweighofer et al., 2004). Recently, reverse-genetics studies have demonstrated that AtP2C-HAB1 is a negative regulator of ABA signaling (Leonhardt et al., 2004; Saez et al., 2004). Isolation and characterization of the dominant negative abi1-1 and abi2-1 PP2C mutants and their intragenic revertants also support a negative role for these two PP2Cs (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998; Gosti et al., 1999; Merlot et al., 2001). However, in transgenic Arabidopsis plants overexpressing the ABI1 gene, no altered ABA sensitivity was found in seed germination or in suppression of ABA-mediated gene induction (Wu et al., 2003). To date, no allele of ABI1 and ABI2 in which the corresponding protein is not produced has been reported.
AtPP2CA has been shown to block ABA-induced gene induction when transiently overexpressed in protoplasts (Sheen, 1998). However, stable AtPP2CA overexpression in planta and corresponding knockout mutants have not been reported. In this study, the isolation and detailed phenotypic characterization of PP2CAox and two insertional mutants, pp2ca-1 and pp2ca-2, unequivocally demonstrate that PP2CA acts as a strong negative regulator of ABA signaling not only at the seed germination level but also in vegetative tissues. It has been previously shown that AtPP2CA transcripts are reduced in the ABA-hypersensitive mutant abh1, which encodes a nuclear mRNA cap-binding protein (Hugouvieux et al., 2001). We therefore also analyzed AtPP2CA mRNA splicing and the effects of the introduction of a 35S∷AtPP2CA fusion in the abh1 background to determine the role of AtPP2CA in mediating the ABA hypersensitivity of abh1.
RESULTS
Screening of a 35S∷cDNA-Expressing Line Library for ABA-Insensitive Mutants Identifies an Overexpressor of AtPP2CA
Because studies suggest genetic and network redundancy in ABA signal transduction, a screen was pursued that can include dominant ABA-insensitive mutants. Roughly one million seeds were screened from approximately 60,000 activation-tagged lines for ABA-insensitive seed germination at 5 μm ABA. Remarkably, after retesting putative mutants, no robust ABA-insensitive mutant line was isolated from the two activation-tagged populations that were tested twice independently. These findings may be attributed to the robustness of ABA signaling and the inherent limitations in the mutation rate of activation-tagged lines (Weigel et al., 2000).
To further enhance the frequency of obtaining dominant mutants in ABA signaling, we screened Arabidopsis lines expressing a library of random 35S∷cDNAs (LeClere and Bartel, 2001) for ABA-insensitive mutants in seed germination assays. This library is composed of approximately 400,000 T2 seeds coming from 33,000 different T1 lines in the ecotype Columbia (Col-0) background, each expressing a random 35S∷cDNA together with a Basta resistance gene inserted in the T-DNA of the 35SpBARN vector. From the original screen of 400,000 T2 seeds, 902 putative individual T2 seeds were able to germinate (radicle emergence plus expanded green cotyledons) 5 d after stratification on 2.5 μm ABA, a concentration that completely inhibits wild-type seed germination. These young ABA-resistant seedlings were then transferred to soil and allowed to self pollinate. The progeny of each of the T2 plants were then tested again for ABA insensitivity in germination assays. From this secondary screen, the progeny of two T2 plants, named 54.7 and 393.1, exhibiting strong ABA resistance were selected and further characterized.
Southern-blot analyses revealed that both mutants contained tandem T-DNA insertions at one locus each (data not shown). Segregation analyses of both ABA and Basta resistance in the next generations as well as in F1 and F2 populations from backcrosses indicated that both ABA insensitivities were linked to the corresponding T-DNAs. However, whereas 393.1 was heterozygous for the mutation and its ABA insensitivity was dominant, 54.7 was homozygous and its phenotype was due to a single recessive nuclear mutation.
Because the strong ABA insensitivity of the recessive mutant 54.7 was reminiscent of the loss-of-function phenotype of the classic positive regulators of ABA responses ABI3, ABI4, and ABI5, a PCR-based diagnosis was performed on 54.7 genomic DNA (see “Materials and Methods”). This analysis and sequencing showed that 54.7 has a tandem T-DNA insertion located in the first intron of ABI5 approximately 500 bp before the start codon (data not shown). Thus, considering its strong ABA insensitivity and the location of the T-DNA insertion, 54.7 is most likely a new allele of abi5.
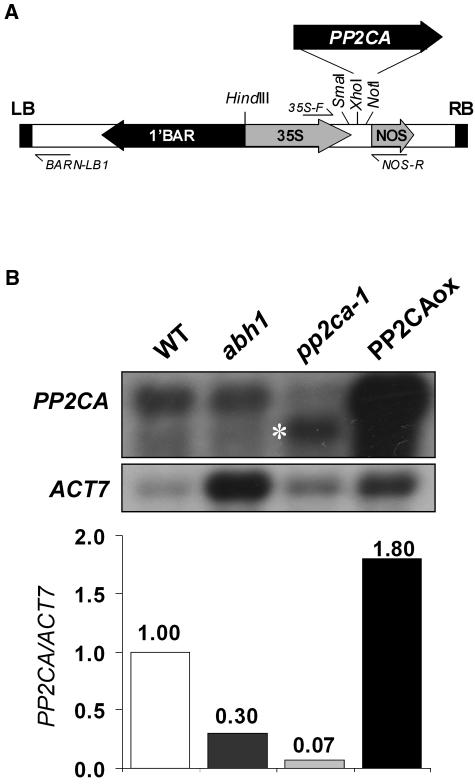
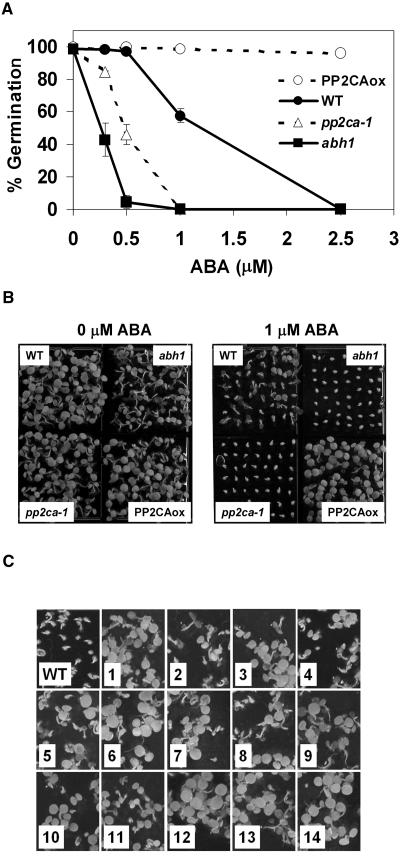
The cDNA contained within the T-DNA of the dominant 393.1 mutant was identified by PCR and perfectly matches the full-length cDNA of AtPP2CA, encoding a PP2C (Fig. 1A). The AtPP2CA gene belongs to group A of the Arabidopsis PP2C gene family (Schweighofer et al., 2004). A homozygous T3 line, PP2CAox (see below), was recovered from the progeny of the heterozygous 393.1 plant, and northern-blot analysis revealed that AtPP2CA was indeed overexpressed in PP2CAox compared to wild type (Fig. 1B). PP2CAox clearly exhibited ABA insensitivity in seed germination assays as it germinated almost completely on 2.5 μm ABA, whereas wild-type seeds do not germinate at all (Fig. 2A).
Figure 1.
The ABA-insensitive 393.1 mutant is an overexpressor of AtPP2CA encoding a PP2C. A, Schematic representation of the 393.1 mutant T-DNA containing the full-length cDNA of AtPP2CA. The AtPP2CA cDNA is inserted at the original XhoI and NotI restriction sites of the 35SpBARN vector. Arrows indicate the direction of transcription. 35S-F and NOS-R are the primers used for amplifying the cDNA. BARN-LB1 is the primer used for T-DNA left-border sequencing. Note that the XhoI restriction site no longer exists because it has been filled with T4 polymerase during the original cloning procedure (LeClere and Bartel, 2001). B, Northern-blot analysis of AtPP2CA in wild-type, abh1, pp2ca-1, and PP2CAox leaves. Hybridization signals with ACTIN7 cDNA (ACT7) were used for standardization of RNA and the value obtained from wild-type leaves was set to 1.
Figure 2.
Constitutive expression of AtPP2CA results in ABA insensitivity, while disruption of AtPP2CA causes ABA hypersensitivity during seed germination. A, Comparison of germination rates of wild-type (Col-0; black circles), pp2ca-1 (white triangles), abh1 (black squares), and PP2CAox (white circles) seeds exposed to 0, 0.3, 0.5, 1, and 2.5 μm ABA at 5 d. Data represent the mean ± sem of three independent experiments with 36 seeds per genotype and experiment. See Supplemental Figure 1A for similar data with pp2ca-2. Error bars are smaller than symbols, if not visible. B, Comparison of germination for wild-type, pp2ca-1, abh1, and PP2CAox seeds at 1 μm ABA or in the absence of ABA after 5 d. C, Germination of seeds from wild-type and 14 individual T1 lines with constitutive 35S∷AtPP2CA expression at 2.5 μm ABA 5 d after germination.
To further test whether the 35S∷AtPP2CA fusion caused the ABA-insensitive phenotype in PP2CAox, the full-length AtPP2CA cDNA was cloned back in the 35SpBARN binary vector (LeClere and Bartel, 2001; see “Materials and Methods”) and transformed into wild-type Arabidopsis plants. Fourteen independent T1 35S∷AtPP2CA lines were recovered as well as 10 T1 35SpBARN lines (empty-vector controls) and the germination rate of their T2 seeds was analyzed in the presence of 2.5 μm ABA. Whereas none of the 10 T1 35SpBARN lines exhibited ABA insensitivity in their progeny compared to untransformed wild-type Arabidopsis (data not shown), all 14 35S∷AtPP2CA lines displayed ABA resistance similar to the original heterozygous 393.1 mutant seeds (Fig. 2C). Thus, we conclude that ectopic expression of the 35S∷AtPP2CA fusion is responsible for the ABA insensitivity observed for PP2CAox seeds.
Seeds from Two Independent T-DNA Insertion Mutants for the AtPP2CA Gene Are Strongly ABA Hypersensitive
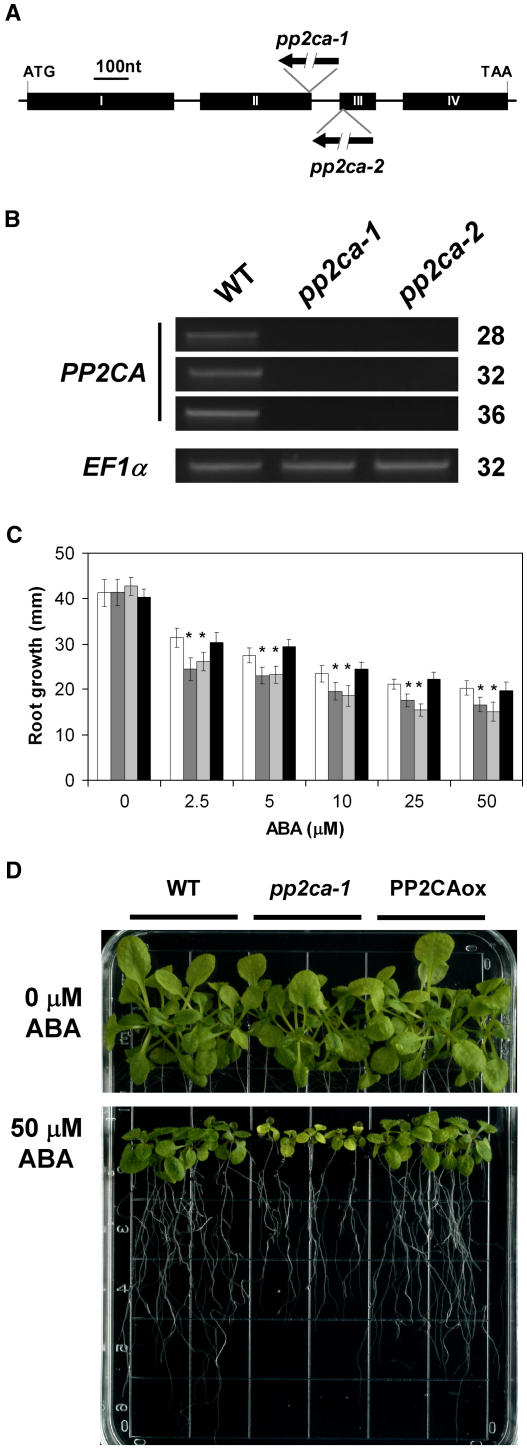
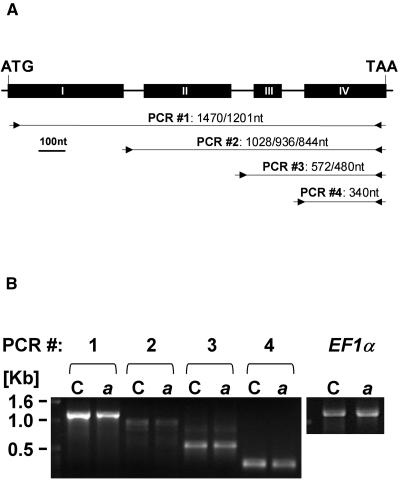
To further elucidate the role of AtPP2CA in ABA signaling, we isolated an insertion mutant from the Salk Institute Genomic Analysis Laboratory (SIGnAL; Alonso et al., 2003) database corresponding to donor stock number SALK_028132. Sequencing of the T-DNA flanking region indicated that the T-DNA is located at the end of exon II (position +864 [numbering refers to the ATG start codon]; Fig. 3A). Plants homozygous for the T-DNA insertion recovered by PCR genotyping were renamed pp2ca-1. Northern-blot analysis failed to detect a full-length AtPP2CA transcript in the pp2ca-1 mutant (Fig. 1B), although a shorter transcript could be detected (Fig. 1B, asterisk). In addition, reverse transcription (RT)-PCR analysis did not result in any product corresponding to a full-length cDNA in the T-DNA disruption allele pp2ca-1 (Fig. 3B).
Figure 3.
Disruption of AtPP2CA causes reduced root elongation in response to ABA. A, Schematic representation of the genomic organization of the AtPP2CA gene with four exons (black boxes). Positions of the pp2ca-1 and pp2ca-2 T-DNA insertions are indicated and orientation of the left-border sequence of the respective T-DNAs is represented by broken arrows. B, RT-PCR analysis shows no full-length transcript in AtPP2CA T-DNA disruption lines pp2ca-1 and pp2ca-2. PCR reactions were performed with oligonucleotides PP2CAEx1-F and PP2CAEx4-R (Table I) and samples were withdrawn from the reaction after 28, 32, and 36 cycles. Amplification of EF1α cDNA with primers EF1α-F and EF1α-R (Table I) was used for controls. C, Comparison of root elongation of wild-type (white bars), pp2ca-1 (dark gray bars), pp2ca-2 (light gray bars), and PP2CAox (black bars) seedlings; 6-d-old seedlings were transferred to plates supplemented with 0, 2.5, 5, 10, 25, and 50 μm ABA, and root elongation was monitored after 6 d. Each data point represents the mean of three independent experiments with eight seedlings each. Asterisks (*) indicate a significant change between wild-type and pp2ca-1 or pp2ca-2 plants (P < 0.001). D, Growth of wild type, pp2ca-1, and PP2CAox on 0.25× Murashige and Skoog medium supplemented with 50 μm ABA. The photographs show plants on plates with 0 (top) and 50 μm ABA (bottom) 18 d after transfer of 6-d-old seedlings from 0.25× Murashige and Skoog medium.
ABA germination assays were carried out with seeds from pp2ca-1 in parallel with seeds from the PP2CAox line, wild-type plants, and the ABA-hypersensitive mutant, abh1 (Fig. 2A). In the absence of exogenous ABA, pp2ca-1 mutant seeds germinated as well as wild-type seeds (Fig. 2, A and B). And in the presence of 1 μm ABA, a clear ABA-hypersensitive inhibition of seed germination was observed. Indeed, the ABA hypersensitivity of pp2ca-1 was almost as strong as that of abh1 (Fig. 2, A and B). Later, a second T-DNA insertion mutant line WiscDsLox341D03 was released by the Arabidopsis Biological Resource Center (ABRC; stock no. CS851888; Sussman et al., 2000). Plants homozygous for the T-DNA insertion recovered by PCR genotyping were renamed pp2ca-2 and sequencing of the T-DNA flanking region showed that the insertion lay at the beginning of exon III (position +966; Fig. 3A). As for pp2ca-1, RT-PCR failed to amplify a product corresponding to a full-length cDNA in the pp2ca-2 mutant (Fig. 3B). Seeds from pp2ca-2 were also tested in germination assays and displayed a strong ABA hypersensitivity very similar to that of pp2ca-1 (Supplemental Fig. 1A). Together, our data obtained from PP2CAox, as well as the pp2ca-1 and pp2ca-2 T-DNA disruption mutants, demonstrate an important role of AtPP2CA as a negative regulator of ABA signaling during seed germination.
AtPP2CA Affects ABA-Promoted Inhibition of Root Growth
To test whether AtPP2CA gene disruption or constitutive expression of AtPP2CA in plants could affect other ABA responses, and because AtPP2CA was shown to be expressed in the stele of the Arabidopsis root system (Cherel et al., 2002), we investigated ABA inhibition of root growth by transferring 6-d-old seedlings on 0.25× Murashige and Skoog plates with 0, 2.5, 5, 10, 25, and 50 μm ABA. Elongation of the primary root was measured 6 d after the transfer in three independent experiments (Fig. 3C). Disruption of AtPP2CA in pp2ca-1 and pp2ca-2 plants exhibits a moderate, but significant, increase in ABA sensitivity compared to wild type during root growth on 0.25× Murashige and Skoog media supplemented with 2.5, 5, 10, 25, and 50 μm ABA (Fig. 3C). Interestingly, ABA inhibition of root elongation in the originally isolated PP2CAox plants was the same as in wild type at all ABA concentrations measured (P > 0.04 to 0.36 in all conditions tested; Fig. 3C), suggesting that elevated AtPP2CA transcript levels have no dramatic effect on ABA regulation of root elongation. A possible explanation could be that during the ABA inhibition of root elongation, the expression of PP2CA interacting partners is the rate-limiting step rather than PP2CA levels. Moreover, due to the moderate ABA hypersensitivity of pp2ca-1 and pp2ca-2 in root assays compared to the strong seed germination phenotype, it is conceivable that partial redundancy with other PP2Cs is more pronounced in roots. When plants were exposed to 50 μm ABA for extended time periods (18 d), pp2ca-1 plants show ABA-hypersensitive inhibition of root elongation as well as retarded growth and chlorosis in aerial parts of plants (Fig. 3D), further exhibiting an enhanced sensitivity to ABA.
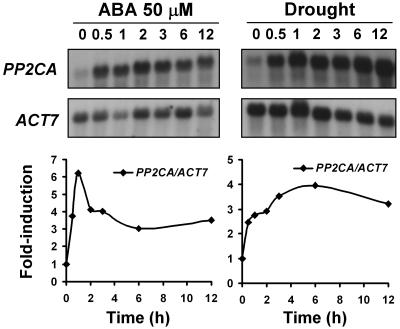
The AtPP2CA Transcript Level Is Up-Regulated by Both ABA and Drought Treatments
AtPP2CA is expressed ubiquitously in plant organs with the highest transcript levels in leaves and its expression is up-regulated by stresses, including ABA, cold, drought, and salt treatment (Tahtiharju and Palva, 2001; Cherel et al., 2002). However, the time courses of AtPP2CA gene induction in ABA or drought experiments have not yet been reported, to our knowledge. Thus, to assess the stress-related regulation of this gene, we studied the accumulation of AtPP2CA transcripts from wild-type plant leaves in response to ABA and drought treatments. The results clearly show that the AtPP2CA mRNA is highly and rapidly up-regulated by both treatments (Fig. 4). Significant transcript increases were detected within 30 min of exposure to ABA or drought (Fig. 4). However, whereas the peak of ABA induction occurred at 1 h, induction by drought increased rather progressively before reaching a peak at approximately 6 h.
Figure 4.
AtPP2CA transcripts are rapidly and highly up-regulated by both ABA and drought treatments. Northern-blot analyses of AtPP2CA in wild-type leaves either treated with ABA (50 μm; left) or excised and subjected to desiccation (right). Total RNA was extracted from leaves at times specified by the number above each lane. Hybridization signals with ACTIN7 cDNA (ACT7) were used for standardization of equal amounts of RNA. Values obtained prior to the indicated treatments were set to 1.
AtPP2CA Disruption Causes ABA Hypersensitivity in Stomatal Guard Cells
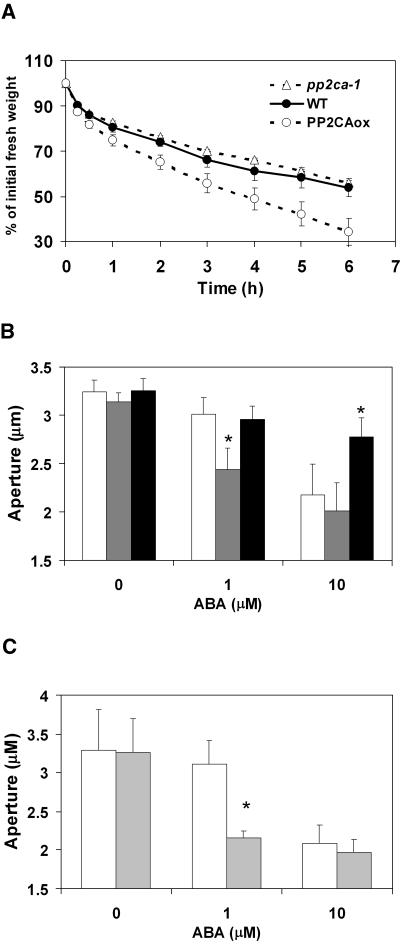
The control of water loss by ABA is a crucial survival mechanism for plants during drought periods. To investigate the role of AtPP2CA in regulating water homeostasis, we measured the loss of fresh weight of detached rosette leaves (Fig. 5A). Overexpression of the AtPP2CA cDNA in planta led to an approximately 1.5-fold increase in the water-loss rate (Fig. 5A). In contrast, the gene disruption line pp2ca-1 did not exhibit significant differences to transpiration rates of wild-type leaves (Fig. 5A). Similarly, some stomatal ABA response mutants show no detached-leaf water-loss phenotype, including earlier findings on mutant alleles of the PP2Cs ABI1, ABI2, and AtP2C-HAB1 (Gosti et al., 1999; Merlot et al., 2001; Saez et al., 2004). This lack of a phenotype in detached-leaf wilting assays may be attributable to the limited resolution of this method.
Figure 5.
AtPP2CA modulates the stomatal response to ABA. A, Ectopic expression of AtPP2CA in plants causes enhanced leaf evaporation rate compared to wild type. Loss of fresh weight of detached rosette leaves at the same developmental stages was measured for wild-type (black circles), pp2ca-1 (white triangles), and PP2CAox plants (white circles) at the indicated time points. Data represent the mean of three independent experiments ± sem. B, Stomatal closing is ABA hypersensitive in pp2ca-1 and ABA insensitive in PP2CAox plants. Stomatal aperture measurements of wild type (white bars), pp2ca-1 (shaded bars), and PP2CAox (black bars) in response to 0, 1, and 10 μm ABA. Data represent the mean of n = 4 independent experiments ± sem with 4 × 50 stomata per data point. Asterisks (*) indicate significant changes between the indicated genotype and wild type (P < 0.001). C, Stomatal closing is ABA hypersensitive in pp2ca-2. Stomatal aperture measurements of wild type (white bars) and pp2ca-2 (shaded bars) in response to 0, 1, and 10 μm ABA. Data represent the mean of two independent experiments ± sem with 2 × 50 stomata per data point. Asterisks (*) indicate significant changes between the indicated genotype and wild type (P < 0.001). See Supplemental Figure 1, B and C, for stomatal aperture ratios from experiments in Figure 5, B and C.
Therefore, we more directly analyzed stomatal movement responses to ABA in loss- and gain-of-function AtPP2CA plants (Fig. 5, B and C). Compared to wild type, guard cells from PP2CAox plants exhibit a clear insensitivity in ABA-induced stomatal closure analyses. Guard cells in these plants show a significantly reduced response to 10 μm ABA, which clearly results in stomatal closure in wild-type plants (Fig. 5B). In contrast, AtPP2CA gene disruption results in an ABA-hypersensitive stomatal closure response at 1 μm ABA (Fig. 5B for pp2ca-1, and 5C for pp2ca-2). These data show that AtPP2CA plays an important role in ABA signal transduction events and the regulation of stomatal aperture. Analyses of stomatal aperture responses (Fig. 5, B and C) and ratios of stomatal apertures to stomatal heights illustrate the same findings in AtPP2CA disruption and gain-of-function lines (Supplemental Fig. 1, B and C).
Analysis of AtPP2CA mRNA Splicing in the abh1 Mutant
Interestingly, the AtPP2CA mRNA was previously shown to exhibit a reduced mRNA level in the ABA-hypersensitive abh1 mutant (Hugouvieux et al., 2001). Transcript levels were analyzed in northern-blot analyses in abh1 and compared to those in wild-type controls. AtPP2CA transcripts normalized to Actin7 mRNA were 3.3-fold lower in abh1 compared to wild-type controls (Fig. 1B).
ABH1 is the Arabidopsis homolog of an 80-kD subunit of the dimeric mRNA cap-binding complex, which additionally consists of a 20-kD subunit, AtCBP20 (Hugouvieux et al., 2001; Kmieciak et al., 2002; Papp et al., 2004). In yeast (Saccharomyces cerevisiae) and human HeLa cells, the cap-binding complex was shown to participate in pre-mRNA splicing (Izaurralde et al., 1994; Lewis and Izaurralde, 1997; Fortes et al., 1999). We investigated whether the AtPP2CA transcript undergoes differential splicing in abh1 compared to wild type. Differential splicing could contribute to the down-regulation of the AtPP2CA mRNA in abh1 due to a reduced turnover of AtPP2CA transcript maturation (Clark et al., 2002). We designed an RT-PCR approach to analyze qualitative and quantitative differences in AtPP2CA transcript maturation (Fig. 6A). Forward primers were selected for amplification of the full reading frame or for amplification of intron sequences with the corresponding reverse primer being located at the 3′ part of the most downstream exon IV (Fig. 6A).
Figure 6.
Analysis of AtPP2CA intron splicing in wild type and abh1. A, Schematic representation of AtPP2CA genomic organization with exons (black boxes) and introns (lines between exons). Positions of primers (arrows) used for the RT-PCR analysis are indicated and resulting amplification products (dotted lines) are shown for each PCR reaction performed, numbered from 1 to 4. Product sizes for full-length amplification, corresponding to completely unspliced transcripts, or for smaller fragments, corresponding to fully or partially processed mRNAs, are given for each PCR reaction. B, DNA fragments of fully and partially spliced AtPP2CA mRNA amplification products (36 cycles) generated in PCR reactions 1 to 4 (Fig. 6A) for wild-type controls (C) and abh1 (a) are size fractionated on a 1.8% agarose gel. Amplification (28 cycles) of the EF1α was used as internal control. Sizes of coelectrophoresed DNA standard fragments are given in kilobase pairs.
RT-PCR on DNase-treated total RNA from leaves yielded similar amplification product qualities and quantities for all four reactions (Fig. 6B). PCR product 1 resulted in a single band corresponding to the full-length reading frame. PCR product 2, with the forward primer location in the first intron, showed faint bands with sizes corresponding to a fully unspliced pre-mRNA (1,028 nucleotides). PCR product 2 also resulted in splice intermediates emerging from intron I independent of removal of intron II or intron III (936 nucleotides) and even removal of both introns II and III (844 nucleotides) from the pre-mRNA without remarkable differences between wild type and abh1 (Fig. 6B). This holds equally true for PCR products 3 and 4 (Fig. 6B). Identical results were obtained for an analysis on RNA isolated from independently grown plants (data not shown). We conclude from these results that splicing of the AtPP2CA pre-mRNA is not affected in the abh1 background and down-regulation of AtPP2CA transcripts is more likely caused by other mechanisms.
To elucidate whether elevated transcript levels of AtPP2CA can restore normal ABA sensitivity or even cause ABA insensitivity in abh1, we transformed wild-type (Col-0) and abh1 plants with a AtPP2CA cDNA under the control of the cauliflower mosaic virus 35S promoter in a binary vector different from 35SpBARN (see “Materials and Methods”). Forty independent T1 plants were isolated each for wild type and for abh1 and tested for ABA responses. Individual lines with single-insertion segregation patterns and the strongest ABA insensitivity in seed germination were selected to obtain homozygous lines. ABA germination assays were performed in triplicate and confirmed earlier findings (Fig. 2C) that introduction of a 35S∷AtPP2CA fusion in the wild type always confers a strong ABA insensitivity in seed germination independent of the binary vector used (data not shown).
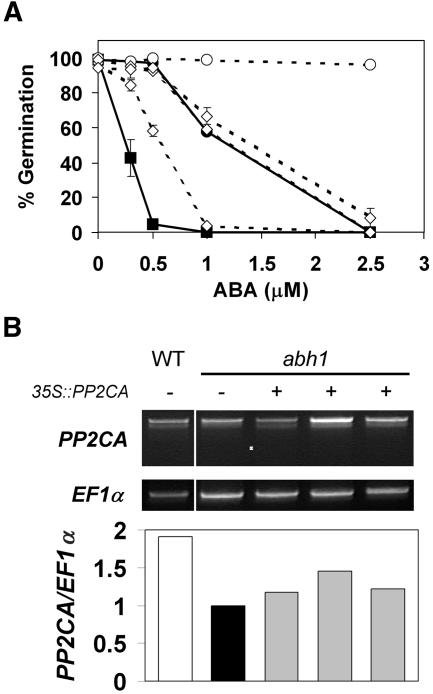
In the abh1 background, ectopic expression of the AtPP2CA cDNA had a much weaker effect than in the wild-type background, with only two homozygous single-insertion lines being able to restore wild-type-like ABA responses during germination (Fig. 7A, white diamonds). Most of the 40 abh1 mutant lines expressing the 35S∷AtPP2CA construct exhibited a range of ABA sensitivities between abh1 and wild type in T2 generation germination experiments (data not shown). Based on the hypothesis that abh1 might affect AtPP2CA transcripts in these constitutively cDNA-expressing lines, we tested AtPP2CA transcript integrity in RT-PCR experiments (Fig. 7B). Amplification of the full-length reading frame resulted in a single band in all lines investigated, indicating no detectable qualitative impact of abh1 on the full-length transcripts (Fig. 7B). Ratiometric analysis of the AtPP2CA transcript levels, with elongation factor 1α (EF1α) as a reference, indicated that AtPP2CA mRNA abundance in 35S∷AtPP2CA/abh1 lines was indeed increased up to 1.5-fold compared to abh1 (Fig. 7B). However, the transcript abundance was persistently lower than in wild-type plants (Fig. 7B). Northern analyses of AtPP2CA transcript levels confirmed these results (data not shown). Thus, elevated AtPP2CA transcript levels can partially suppress the ABA hypersensitivity of abh1 and the overall lower levels of AtPP2CA transcripts in abh1 compared to wild type indicate a continued down-regulation of AtPP2CA in abh1 (Fig. 7).
Figure 7.
Ectopic expression of AtPP2CA partially suppresses the ABA hypersensitivity of abh1. A, Comparison of germination rates of wild type (black circles), PP2CAox in wild type (white circles), abh1 (black squares), and seeds from three independent abh1 35S∷AtPP2CA (white diamonds) lines germinated on Murashige and Skoog plates containing 0, 0.3, 0.5, 1, and 2.5 μm ABA after 5 d. Data represent the mean ± sem of three independent experiments. Error bars are smaller than symbols, if not visible. Data for wild type, abh1, and PP2CAox in A are the same as in Figure 2A for reference purposes. B, RT-PCR analysis of AtPP2CA transcript levels in wild type, abh1, and the three independent abh1 35S∷AtPP2CA lines displayed in A from the more sensitive to the more ABA resistant. DNA fragments from RT-PCR analyses of EF1α were used for standardization of equal amplification rates and the value obtained for AtPP2CA quantity in abh1 was set to 1. RT-PCR reactions were performed for 20, 24, 28, and 32 cycles to quantify relative mRNA levels and representative images after 28 cycles are shown.
DISCUSSION
Here we report isolation of a strong dominant ABA response mutant overexpressing the AtPP2CA cDNA during an ABA-insensitive screen of a library of 33,000 35S∷cDNA-expressing Arabidopsis lines. We characterize AtPP2CA gene disruption and overexpression phenotypes in Arabidopsis. We show that T-DNA insertions in the AtPP2CA gene result in a strongly increased sensitivity to the phytohormone ABA during seed germination (Fig. 2, A and B; Supplemental Fig. 1A) and also render guard cells more sensitive to ABA during stomatal closure at 1 μm ABA (Fig. 5, B and C) and affect root elongation in the presence of exogenous ABA (Fig. 3, C and D). On the other hand, overexpression of AtPP2CA impairs stomatal closure in response to 10 μm ABA (Fig. 5B). Seed germination of PP2CAox lines displays a greatly decreased sensitivity to ABA (Fig. 2, A and C). Moreover, constitutive expression of AtPP2CA in the ABA-hypersensitive mutant abh1 is shown to partially restore ABA sensitivity in abh1 (Fig. 7A). Together, these results point to an important function of AtPP2CA as a negative regulator of ABA signal transduction events. The identification of a negative regulator in ABA signaling based on a cDNA overexpression screen shows that this approach can be used to isolate mutants in genes that modulate complex signaling networks in plants (Schroeder et al., 2001; Fedoroff, 2002; Finkelstein et al., 2002; Hetherington and Woodward, 2003; Himmelbach et al., 2003; Fan et al., 2004; Cutler and McCourt, 2005).
In an earlier study, AtPP2CA was shown to be linked to cold acclimation in Arabidopsis (Tahtiharju and Palva, 2001). It was shown that AtPP2CA is highly induced during cold acclimation. Plants with suppression of AtPP2CA transcripts by an antisense approach were shown to exhibit an ABA-dependent accelerated development of freezing tolerance (Tahtiharju and Palva, 2001) and an increased ABA sensitivity in seed germination was mentioned. As the antisense construct used shows nucleotide homology to five of the nine group A PP2Cs, and to avoid the possible effects of cosuppression of these related group A PP2C genes, we pursued analyses of T-DNA disruption lines of the AtPP2CA gene (Fig. 3A). Accordingly, northern-blot and RT-PCR analyses of the pp2ca-1 and the pp2ca-2 lines show absence of mature AtPP2CA transcripts (Figs. 1B and 3B).
AtPP2CA, a Negative Regulator of Physiological ABA Responses
We report that AtPP2CA gene disruption lines show a strongly increased sensitivity to ABA during seed germination, which appears to be more pronounced than in a AtP2C-HAB1 disruption line (Leonhardt et al., 2004; Saez et al., 2004). Thus, we investigated the effect of AtPP2CA gene disruption and ectopic expression at the whole-plant level.
Application of exogenous ABA is well established to affect root growth as an antagonist of auxin, impairing cell elongation and causing an arrest in mitotic cell cycle activity (Himmelbach et al., 1998). The ABA-insensitive dominant abi1-1 mutant and sustained expression of AtP2C-HAB1 in 35S∷AtP2C-HAB1 plants have been shown to exhibit less sensitivity to ABA inhibition of root growth (Beaudoin et al., 2000; Ghassemian et al., 2000; Saez et al., 2004). However, previous studies have not analyzed PP2C gene disruption lines for altered ABA-dependent root elongation. In our study, we show that AtPP2CA gene disruption results in a moderate hypersensitive response to ABA in root elongation compared to wild type under the conditions tested (Fig. 3, C and D).
Furthermore, we show that ectopic expression of AtPP2CA results in increased transpiration rates of detached rosette leaves (Fig. 5A). It has been shown previously that the dominant ABA-insensitive mutants abi1-1 and abi2-1 are sensitive to water stress conditions and impair early ABA signal transduction (Koornneef et al., 1984; Finkelstein, 1994; Leung et al., 1994, 1997; Meyer et al., 1994; Pei et al., 1997; Allen et al., 1999). More recently, plants overexpressing the AtP2C-HAB1 protein phosphatase were also shown to exhibit increased transpiration rates (Saez et al., 2004). However, we could not observe a difference in the transpiration rates of detached leaves from the pp2ca-1 plants (Fig. 5A), resembling findings on the abi1-1R1 to abi1-1R7 intragenic revertant lines and the T-DNA insertion line of AtP2C-HAB1 (Saez et al., 2004), even though these mutants show ABA-hypersensitive responses in stomatal movements (Gosti et al., 1999; Leonhardt et al., 2004). Previous leaf water loss analyses from detached leaves have shown that these assays can show phenotypic differences in mutations in which steady-state stomatal apertures already differ from wild-type controls prior to excising leaves (Kwak et al., 2001; Cominelli et al., 2005; Liang et al., 2005). More direct (double-blinded) analyses of ABA-induced stomatal closing show that AtPP2CA disruption causes ABA-hypersensitive stomatal closing (Fig. 5, B and C), which also appears to be more pronounced than in the AtP2C-HAB1 gene disruption line (Leonhardt et al., 2004). Together, these results emphasize the importance of AtPP2CA as a strong negative regulator of ABA signal transduction during seed germination and stomatal closure.
AtPP2CA Shows a Similar Expression Profile to Other Group A PP2C Genes
Sixty-nine PP2C genes are encoded in the Arabidopsis genome compared to 15 PP2C genes in humans (Kerk et al., 2002; Schweighofer et al., 2004). This implies genetic redundancy and a more specific role of plant PP2Cs during developmental stages and responses to environmental changes. With the exception of AtP2C-HAB1, no gene disruption phenotype for other plant PP2Cs has been reported (Leonhardt et al., 2004; Saez et al., 2004). AtP2C-HAB1 is highly expressed in guard cells in response to ABA and gene disruption causes ABA-hypersensitive stomatal closure (Leonhardt et al., 2004) and increased sensitivity during seed germination (Leonhardt et al., 2004; Saez et al., 2004).
AtPP2CA transcript levels are the second highest expressed of all group A PP2Cs in dry seeds of Arabidopsis and are significantly down-regulated upon imbibition (Supplemental Fig. 2; Zimmermann et al., 2004; Nakabayashi et al., 2005). A similar, but overall lower, expression pattern can be seen for AtP2C-HAB1 (Supplemental Fig. 2). Since other not-yet-characterized PP2Cs of group A also show high transcript levels in dry seeds and down-regulation during imbibition, an ABA-related function in seed germination can be anticipated for these genes. Moreover, AtPP2CA was found to be among the mRNAs with the highest levels of ABA-mediated induction in guard cells, showing an average 7-fold ABA induction (Leonhardt et al., 2004), again similar to the transcript abundance of AtP2C-HAB1 (Supplemental Fig. 3). Guard cell expression of AtPP2CA and AtP2C-HAB1 was also shown in independent studies, which investigated their promoter activities in planta with reporter gene fusions (Cherel et al., 2002; Saez et al., 2004).
Despite the large gene family of PP2Cs and similar expression patterns of AtP2C-HAB1 and AtPP2CA, the limited functional redundancy in single gene disruption lines during the process of ABA signal transduction in seeds and guard cells may imply a high degree of specificity toward downstream targets of these two PP2Cs. In plants, our knowledge about PP2C targets is still limited and no target has been identified for AtP2C-HAB1. However, the inward-rectifying potassium channel AKT2 was shown to interact with AtPP2CA in yeast and AKT2 channel activity is negatively modulated by AtPP2CA in heterologous expression systems (Cherel et al., 2002). Also, the ABI2 protein phosphatase has been found to interact in yeast with the protein kinase PKS3 (Guo et al., 2002). In addition, ABI1 can interact with the ABA-inducible homeodomain transcription factor AtHB6 (Himmelbach et al., 2002). Moreover, phospholipase Dα1 (PLDα1)-derived phosphatidic acid has been shown to bind and regulate ABI1 (Zhang et al., 2004).
Modulation of AtPP2CA in abh1
The abh1 mutation causes ABA hypersensitivity in seed germination and stomatal movements and modulates ion channel activities in guard cells (Hugouvieux et al., 2001, 2002). Down-regulation of the AtPP2CA transcript level was previously reported in the abh1 mutant (Hugouvieux et al., 2001). Because a protein homologous to ABH1, the 80-kD subunit of the dimeric nuclear cap-binding protein CBP80, has been shown to affect splicing during pre-mRNA maturation in yeast and mammalian cells (Izaurralde et al., 1994; Lewis and Izaurralde, 1997; Fortes et al., 1999; Clark et al., 2002), we investigated the hypothesis that the AtPP2CA transcript undergoes differential splicing in abh1 compared to wild type, therefore causing down-regulation of the transcript. An RT-PCR analysis specifically designed to amplify splice intermediates based on differential intron splicing efficiencies in abh1 and wild type did not reveal any differences in pre-mRNA splicing of AtPP2CA (Fig. 6). Therefore, pre-mRNA splicing seems unlikely to cause down-regulation of the AtPP2CA transcript, an effect that was observed for some transcripts in the yeast gcr3 mutant that encodes the CBP80 protein (Clark et al., 2002).
In this study, we investigated the hypothesis that AtPP2CA down-regulation in abh1 contributes to the ABA hypersensitivity in abh1. Constitutive expression of AtPP2CA in the abh1 mutant suppressed the ABA hypersensitivity of abh1 plants. Interestingly, however, overexpression did not render abh1 plants as ABA-insensitive as 35S∷AtPP2CA wild-type plants (Fig. 7A). Out of 40 abh1 plants harboring the 35S∷AtPP2CA construct, only two homozygous single-insertion lines resulted in ABA sensitivity similar to wild-type plants in seed germination experiments. The comparison of AtPP2CA transcript levels by RT-PCR in these gain-of-function lines revealed that the AtPP2CA transcript levels were up to 1.5-fold higher than the AtPP2CA transcript level in abh1, but still significantly lower than in wild-type plants (Fig. 7B). Because the identification of strong AtPP2CA gain-of-function phenotypes in the abh1 background proved substantially more difficult than in wild type, a negative feedback mechanism may limit AtPP2CA expression in abh1. With AtPP2CA gene disruption lines being less ABA hypersensitive in seed germination than abh1 (Fig. 2, A and B) and because AtPP2CA overexpression only partially restores wild-type-like ABA sensitivity in abh1, we conclude that additional mechanisms contribute to ABA hypersensitivity in abh1.
In conclusion, we demonstrate that the protein phosphatase AtPP2CA acts as a strong negative regulator of ABA signal transduction during seed germination (Fig. 2A) and the regulation of stomatal closure (Fig. 5B). Yoshida et al. (2006) have conducted an independent screen for ABA signaling components in Arabidopsis. They have characterized the same protein phosphatase AtPP2CA also showing ABA hypersensitivity in loss-of-function mutants and insensitivity in AtPP2CA overexpressors. Despite the large number of PP2C genes in the Arabidopsis genome, this study demonstrates that loss- and gain-of-function of AtPP2CA causes strong modulation of ABA responses. With the negative regulatory role of PP2CA in ABA signal transduction, a challenging question for future research will be to uncover the interacting proteins of PP2CA.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Transformation
Arabidopsis (Arabidopsis thaliana) plants (Col-0) were grown in a Conviron growth chamber (Controlled Environments Limited) in plastic pots filled with ready-to-use soil (Professional Blend). After sowing, pots were kept at 4°C for 4 to 7 d. Growing conditions were 22°C, 75% humidity, with a 16-h-light/8-h-dark photoperiod regime at approximately 75 μmol m−2 s−1. Seeds used for comparative studies were from plants grown and harvested in parallel.
Seeds of the activation-tagged lines for identification of ABA-insensitive mutants were kindly provided by D. Weigel (Max-Planck-Institute for Developmental Biology, Tuebingen, Germany; Weigel et al., 2000) and by W. Scheible (Carnegie Institution, Stanford, CA; Sedbrook et al., 2004). Seeds from the library of 35S∷cDNA Arabidopsis-expressing lines (LeClere and Bartel, 2001) were obtained from ABRC and correspond to the CS84450 stock number. The binary vectors constructed and described below were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, which was then used to transform wild-type (Col-0) or abh1 plants by floral dipping (Clough and Bent, 1998).
Molecular Characterization of a New abi5 Allele and pp2ca-1 and pp2ca-2 Insertional Mutants
Given the recessive nature of the strong ABA insensitivity in the homozygous 54.7 mutant, we hypothesized that its phenotype could be mediated by disruption of positive regulators such as ABI3, ABI4, and ABI5. To test this hypothesis, PCR on genomic DNA from 54.7 and wild-type plants with a set of specific primers for ABI3, ABI4, and ABI5 (Table I) was carried out. Only one PCR reaction with ABI5-F3/ABI5-R2 primers (Table I) did not lead to any products from 54.7 genomic DNA (data not shown). PCR genotyping reactions from 54.7 genomic DNA were then carried out with ABI5-F3/ABI5-R2 and with BARN-LB1 (Table I; Fig. 1A), a specific primer of the 35SpBARN T-DNA left border. This led to amplification of two PCR products whose sequencing showed that a tandem T-DNA insertion occurred in the first intron of ABI5 approximately 500 bp before the start codon.
Table I.
Oligonucleotide primers used in this study
| Primer Name | Sequence (5′-3′) |
|---|---|
| PP2CAEx1-F | CAAATGGCTGGGATTTGTTGC |
| PP2CAInt1-F | CTCATGTACGTTAATTTCCTCTGTTTAG |
| PP2CAInt2-F | CGAATTGGTTAGTATGAATCAAGATGGC |
| PP2CAInt3-F | CTGATGATTATTGTTTTTGTTTGTATAGGT |
| PP2CAEx4-R | AAGACGACGCTTGATTATTCCTC |
| SALK-LBa1 | TGGTTCACGTAGTGGGCCATCG |
| p745 | AACGTCCGCAATGTGTTATTAAGTTGTC |
| PP2CAEx1Sal1 | TACCGTCGACAAATGGCTGGGATTTGTTGC |
| PP2CAEx4Spe1 | TCCACTAGTTTAAGACGACGCTTGATTATTCC |
| EF1a-F | GGCCACGTCGATTCTGGAAA |
| EF1a-R | GGCTTGGTTGGAGTCATCTT |
| 35S-F | CGCACAATCCCACTATCCTTCGCAAG |
| NOS-R | GATAATCATCGCAAGACCGGCAACAGG |
| 1′BAR-F | GCGCAAGACGTGACGTAAGTATCCG |
| 1′BAR-R | CCCTCTAGAGTCGACCTGCAGGCATGC |
| BARN-LB1 | GGGCCAGGCGGTGAAGGGCAATC |
| ACTIN7-F | GGCCGATGGTGAGGATATTCAGCCACTTG |
| ACTIN7-R | TCGATGGACCTGACTCATCGTACTCACTC |
| ABI3F1 | CGGTTTTAGATTACTTATTAGC |
| ABI3R1 | CCACCGCCTAGTCTTCTTGCC |
| ABI3F2 | GGCGGTGGTAAAGAAGCGATGAAGC |
| ABI3R2 | CCGAGGTTACCCACGTCGC |
| ABI3F3 | GGGTAACCTCGGAAGGATCG |
| ABI3R3 | CCCATGCATGCACGAGAAG |
| ABI4F1 | GGGATGCTCATCGTATATAATATG |
| ABI4R1 | GGACCCTTTAGCTTCCCAAC |
| ABI5F1 | GGGAACACTAGTAAAGCAG |
| ABI5R1 | CCGCTTTGTAGGAAGACTGTTG |
| ABI5F2 | GCGGAGCTGGAAGTGTCAAAG |
| ABI5R2 | CCGCCTCCTACCCATTTATC |
| ABI5F3 | GCGCCATGGACGACTCTACTTCTCGC |
| ABI5R3 | GGTAACGAAAACTTTATTGG |
pp2ca-1 and pp2ca-2 mutants of the AtPP2CA gene (At3g11410) were obtained from ABRC and correspond to the SALK_028132 and WiscDsLox341D03 lines, respectively. Genotyping PCR reactions for pp2ca-1 and pp2ca-2 were performed with PP2CAEx1-F/PP2CAEx4-R primers and with PP2CAEx1-F/SALK-LBa1 primers (SALK_028132) or with PP2CAEx1-F/p745 primers (WiscDsLox341D03) and amplified products were sequenced (Table I).
Molecular Characterization of the PP2CAox Mutant and Generation of the Reconstructed 35S∷AtPP2CA Lines
The cDNA within the T-DNA of 393.1 plants was PCR amplified from genomic DNA with 35S-F/NOS-R primers (Table I; Fig. 1A) and purified with the QIAEX II kit (Qiagen). The amplified DNA was then cloned into pGEM-T Easy vector (Promega) and sequenced. Because the PCR fragment matched the full-length cDNA of AtPP2CA perfectly, it was then excised from pGEM-T Easy with SmaI and NotI restriction enzymes, subcloned into SmaI/NotI linearized 35SpBARN vector (Fig. 1A; LeClere and Bartel, 2001), and then used to transform Arabidopsis wild-type plants.
Vector PS173 (kindly provided by Professor Jeff F. Harper, University of Nevada, Reno) was used to constitutively express AtPP2CA cDNA in wild-type and abh1 plants. After amplification from total wild-type (Col-0) cDNA (first-strand cDNA synthesis kit; Amersham Biosciences) and cloning the AtPP2CA cDNA into the pGEM-T Easy vector (Promega), the sequenced cDNA was excised with SalI and SpeI restriction enzymes and subcloned into the PS173 vector previously digested with XhoI and XbaI restriction enzymes.
Northern-Blot and RT-PCR Analyses
Total RNA was extracted from leaves using TRIzol reagent (Life Technologies/Gibco-BRL) and quantified by absorption and migration of an aliquot on agarose gel. For ABA and drought treatments, rosette leaves of 3- to 4-week-old wild-type plants were either sprayed with 50 μm ABA or excised and subjected to desiccation for 0.5, 1, 2, 3, 6, or 12 h before extraction. Fifteen micrograms of total RNA were separated in a denaturing formaldehyde-agarose gel and blotted to a Hybond-N membrane (Amersham-Pharmacia). Blots were hybridized with random-priming 32P-labeled probes (Megaprime DNA labeling system; Amersham-Pharmacia). AtPP2CA and ACTIN7 probes were amplified by PCR from cDNA using PP2CAEx1-F/PP2CAEx4-R and ACTIN7-F/ACTIN7-R primers, respectively (Table I). PCR fragments were purified using the QIAEX II kit (Qiagen).
RT-PCR experiments were performed on total RNA isolated as described above after DNase I treatment (DNA-free; Ambion). Reverse transcription (first-strand cDNA synthesis kit, Amersham Biosciences) was performed on 2.5 μg of RNA and 2 μL were used for PCR reactions (Ex Taq DNA polymerase; TaKaRa Mirus Bio). Samples were withdrawn after 20, 24, 28, and 32 cycles (splicing) or 28, 32, and 36 cycles (T-DNA disruption lines) and products were analyzed by agarose gel electrophoresis. Hybridization/PCR signals were quantified using Adobe Photoshop 5.5 software (Adobe Systems) after subtraction of background levels. Expression levels for northern-blot and RT-PCR analyses were normalized against the corresponding ACTIN7 and EF1α RNA levels, respectively.
Root Growth and Germination Assays
For ABA germination assays, sterilized seeds were plated on minimal medium (0.25× Murashige and Skoog medium, no Suc) supplemented with increasing ABA concentrations. After stratification of 4 d at 4°C, plates were transferred to a Conviron growth chamber (Controlled Environments Limited). To score seed germination, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined in three independent experiments (36 seeds per genotype and experiment).
Root growth assays to assess ABA sensitivity were carried out by transferring 6-d-old seedlings onto minimal medium (0.25× Murashige and Skoog medium, no Suc) supplemented with the indicated ABA concentrations on 0.8% agar (Phytagel; Sigma) plates. Root growth was measured 6 d after the transfer in three independent experiments with eight individuals per genotype and experiment. t-Test (one-tailed, homoscedastic) P-values are as follows: pp2ca-1, P = 3.6E-07 (2.5 μm ABA); P = 3.3E-06 (5 μm ABA); P = 4.6E-04 (10 μm ABA); P = 1.6 E-07 (25 μm ABA); and P = 6.8E-06 (50 μm ABA). pp2ca-2, P = 9.7E-06 (2.5 μm ABA); P = 6.5E-06 (5 μm ABA); P = 7.2E-05 (10 μm ABA); P = 8.9E-13 (25 μm ABA); and P = 1.2E-07 (50 μm ABA).
Leaf Water Loss and Stomatal Closure Measurements
Time-dependent analyses of loss of fresh weight were performed with detached rosette leaves at the same developmental stage and size from single 3-week-old plants. Three leaves per genotype were excised, kept in the Conviron growth chamber (Controlled Environments Limited), and fresh weight was measured at the indicated periods of time in three independent experiments.
Double-blind stomatal movement assays were performed such that the genotype and applied ABA concentrations were unknown. Stomatal responses were analyzed in 3- to 4-week-old plants grown in a Conviron growth chamber. Leaves were floated for 2.5 h in stomatal opening solution (Pei et al., 1997) containing 50 mm KCl, 50 μm CaCl, and 10 mm MES (pH 6.15). After incubation in ABA for 2.5 h, leaves were blended and the stomatal aperture was measured. Control experiments were performed in parallel with no ABA added. t-Test (one-tailed, homoscedastic) P-values were calculated: P = 2.3E-08 for pp2ca-1 and P = 5.7E-13 for pp2ca-2 at 1 μm ABA, and P = 2.0E-09 for PP2CAox at 10 μm ABA.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_111974.
Supplementary Material
Acknowledgments
We thank David Waner and Majse Nafisi for screening the activation-tagging populations, Bonnie Bartel for providing the 35SpBARN vector as well as helpful discussions, Nadia Robert, Jean Colcombet, and Dongyul Sung for discussions, and Taehoun Kim and Jared Young for comments on the manuscript.
This work was supported by National Institutes of Health (R01GM060396) and National Science Foundation (MCB0417118) grants (to J.I.S.), and by a European Molecular Biology Organization fellowship (to J.M.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Julian I. Schroeder (julian@biomail.ucsd.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070318.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI (1999) Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR (2004) The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16: 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB (2002) Physical and functional interaction of the Arabidopsis K(+) channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Cutler S, McCourt P (2005) Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol 138: 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LM, Zhao Z, Assmann SM (2004) Guard cells: a dynamic signaling model. Curr Opin Plant Biol 7: 537–546 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (2002) Cross-talk in abscisic acid signaling. Sci STKE 2002: RE10. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol 105: 1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW (1999) Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol 19: 6543–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Iten M, Grill E (1998) Signalling of abscisic acid to regulate plant growth. Philos Trans R Soc Lond B Biol Sci 353: 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Murata Y, Young JJ, Kwak JM, Mackesy DZ, Schroeder JI (2002) Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol 130: 1276–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668 [DOI] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M (2002) The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 129: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmieciak M, Simpson CG, Lewandowska D, Brown JWS, Jarmolowski A (2002) Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene 283: 171–183 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI (2001) Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol 127: 473–485 [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Bartel B (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46: 695–703 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E (1997) The role of the cap structure in RNA processing and nuclear export. Eur J Biochem 247: 461–469 [DOI] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15: 1201–1206 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C (2004) A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol 55: 679–686 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) GUARD CELL SIGNAL TRANSDUCTION. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR (2004) The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26: 461–470 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56: 29–55 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sanchez JP, Lopez-Molina L, Himmelbach A, Grill E, Chua NH (2003) The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant J 34: 307–315 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001. a) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1: 771–781 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001. b) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis PP2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.