Abstract
5-Enolpyruvylshikimate 3-phosphate synthase (EPSPS) is a key enzyme in the shikimate pathway and is targeted by the wide-spectrum herbicide glyphosate. Here, we describe the use of a selection system based on directed evolution to select glyphosate-resistant mutants of EPSPS. Using this system, the rice (Oryza sativa) EPSPS gene, mutagenized by Error-Prone polymerase chain reaction, was introduced into an EPSPS-deficient Escherichia coli strain, AB2829, and transformants were selected on minimal medium by functional complementation. Three mutants with high glyphosate resistance were identified in three independent glyphosate selection experiments. Each mutant contained a C317→T transition within the EPSPS coding sequence, causing a change of proline-106 to leucine (P106L) in the protein sequence. Glyphosate resistance assays indicated a 3-fold increase in glyphosate resistance of E. coli expressing the P106L mutant. Affinity of the P106L mutant for glyphosate and phosphoenolpyruvate was decreased about 70-fold and 4.6-fold, respectively, compared to wild-type EPSPS. Analysis based on a kinetic model demonstrates that the P106L mutant has a high glyphosate resistance while retaining relatively high catalytic efficiency at low phosphoenolpyruvate concentrations. A mathematical model derived from the Michaelis-Menten equation was used to characterize the effect of expression level and selection conditions on kinetic (Ki and Km) variation of the mutants. This prediction suggests that the expression level is an important aspect of the selection system. Furthermore, glyphosate resistance of the P106L mutant was confirmed in transgenic tobacco (Nicotiana tabacum), demonstrating the potential for using the P106L mutant in transgenic crops.
The enzyme 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS; 3-phosphoshikimate 1-carboxyvinyl-transferase; EC2.5.1.19) catalyzes the reaction between phosphoenolpyruvate (PEP) and shikimate-3-phosphate (S3P) to form 5-enolpyruvylshikimate 3-phosphate (EPSP) and inorganic phosphate (Pi; Haslam, 1974). EPSPS is the sixth key enzyme on the shikimate pathway, which is essential for production of the aromatic amino acids (l-Phe, l-Tyr, and l-Trp) and chorismate-derived secondary metabolites. Because the shikimate pathway is present in plants, bacteria, and fungi but absent from animals, EPSPS has been an attractive target for developing effective herbicide and antimicrobial agents (Coggins et al., 2003; Priestman et al., 2005a). One of the widely used EPSPS inhibitors, glyphosate (N-[phosphonomethyl]-glycine) is a broad-spectrum, nonselective, postemergence herbicide (Amrhein et al., 1980, 1983) with low risk to humans and the environment due to its low toxicity, lack of residual soil activity, and rare occurrence of resistant weeds (Smith and Oehme, 1992; Giesy et al., 2000).
EPSPS has attracted considerable attention since it was identified as the primary target of glyphosate in the 1980s (Steinrucken and Amrhein, 1980; Amrhein et al., 1983). Two types of EPSPS have been identified. Class I EPSPS, generally identified from plants and bacteria, has been the subject of most studies of EPSPS enzyme kinetics and active site analysis. Class II EPSPS was identified from some bacteria, e.g. Pseudomonas sp. strain PG2982 (Fitzgibbon and Braymer, 1990), Agrobacterium tumefaciens sp. strain CP4 (Barry et al., 1992), and Staphylococcus aureus (Priestman et al., 2005b). These two types of EPSPS share less than 50% amino acid sequence identity.
Structural analysis and mutagenesis have revealed details of the enzymatic mechanism of EPSPS catalysis. EPSPS from Escherichia coli (Mr 46,000), a class I EPSPS, is a two-domain enzyme that closes upon binding of S3P to form the active site in the interdomain cleft. Glu-341 of EPSPS acts as a proton donor to the methylene group of PEP. The 5′ hydroxyl of S3P is deprotonated by Asp-313 and attacks the C2 oxocarbenium ion of PEP. A tetrahedral intermediate is formed and Lys-22 protonates the oxygen, cleaving the scissile bond to release inorganic phosphate and EPSP (Schonbrunn et al., 2001). Glyphosate acts as a competitive inhibitor of PEP and appears to occupy the PEP binding site, mimicking an intermediate state of the ternary enzyme-substrate complex (Schonbrunn et al., 2001). Thus, structural changes of EPSPS that reduce glyphosate affinity often impair PEP binding, as demonstrated when studying the structure and enzyme kinetics of an E. coli G96A mutant (Eschenburg et al., 2002).
Studies of glyphosate-resistant EPSPS mutants have not only illustrated the mechanism of glyphosate inhibition but also provided tools for engineering glyphosate-resistant crops. Several glyphosate-resistant mutants of class I EPSPS have been identified. The first known glyphosate-resistant EPSPS mutant was identified with a single substitution of P101S from Salmonella typhimurium (Comai et al., 1983; Stalker et al., 1985). Recently, a corresponding substitution (P106S) in EPSPS was also found to be responsible for glyphosate resistance of a weed, goosegrass (Eleusine indica). This P106S mutant is the only reported example of spontaneously occurring glyphosate-resistant EPSPS (Baerson et al., 2002). An E. coli EPSPS mutant containing a G96A substitution was isolated following the selection of E. coli B for growth in the presence of glyphosate (Kishore et al., 1986), and subsequent kinetic assays indicated that this substitution dramatically decreased its affinity for glyphosate. The same substitution of the corresponding site, using site-directed mutagenesis, conferred high glyphosate tolerance to plant EPSPS enzymes (Padgette et al., 1991). Overexpression of a herbicide-sensitive form of EPSPS was demonstrated to be responsible for glyphosate resistance in a petunia (Petunia hybrida) cell line (MP4-G; Steinrucken et al., 1986).
Unfortunately, transgenic plants that overexpress wild-type EPSPS or the EPSPS mutants described above failed to show sufficient glyphosate resistance for commercial utilization (Bradshaw et al., 1997). Most of the class I EPSPS mutants that display increased glyphosate resistance also have an elevated PEP Km, indicating an effect on the interaction between EPSPS and PEP, thereby reducing the efficiency of reaction rate, especially at low PEP concentrations in chloroplasts (Bradshaw et al., 1997). Accordingly, EPSPS with higher Ki (glyphosate) and normal Km (PEP) are considered to be more useful for engineering glyphosate resistance in crops. A class II EPSPS from A. tumefaciens sp. strain CP4 was then explored and successfully used in glyphosate-tolerant crops due to its high glyphosate resistance and near normal Km (Barry et al., 1992; Padgette et al., 1996).
Directed evolution has emerged as a powerful alternative to rational engineering of biocatalysts in the past few years. Prerequisites for directed evolution are functional expression in a suitable microbial host, and rapid and efficient selection for the desired feature(s) of the target protein (Kuchner and Arnold, 1997). As the only case of directed evolution targeting the glyphosate resistance of class I EPSPS, He et al. reported a T42M mutant that was identified from the random-hybridized EPSPS gene of S. typhimurium and E. coli based on a directed evolution strategy. The T42M mutant was reported to have increased glyphosate resistance and enzyme activity (He et al., 2003).
In this study, we have created a glyphosate-resistant P106L mutant of a rice (Oryza sativa) EPSPS using a directed evolution strategy (Arnold et al., 2001). The rice EPSPS gene was randomly mutagenized by error-prone PCR, and glyphosate selection was carried out in an E. coli expression system absent of EPSPS background. Interestingly, this P106L substitution repetitively occurred in independent processes. The repeated occurrence of this mutant provoked us to explore the intrinsic relationship between the P106L mutation and glyphosate resistance.
RESULTS
Identification of OsEPSPS Mutant Conferring Glyphosate Resistance
A pBR322-derived vector, pBREP, was constructed to functionally express the mature form of wild-type rice EPSPS (OsEPSPS; Xu et al., 2002) in E. coli under the control of the bla promoter (Russell and Bennett, 1981). Random mutagenesis via error-prone PCR must be performed on sequences less than 1 kb (Cadwell and Joyce, 1994), thus the OsEPSPS coding sequence was separated into two regions (36–542 bp, 542–1,335 bp) to perform error-prone PCR. The error-prone PCR product was recombined into pBREP to replace the corresponding sequences of the wild-type OsEPSPS gene. The reconstructed product was transformed into an auxotrophic E. coli strain, AB2829. Consequently, more than 2 × 105 AB2829 clones were obtained on M9 plates for each of the two mutagenized regions. Glyphosate selection was then performed by replica plating the clones onto M9 plates supplemented with 5 mm glyphosate and resistant clones were isolated after 48 h of culture at 37°C. E. coli AB2829 (pBREP) that express wild-type OsEPSPS grow slowly on M9 medium containing 1 mm glyphosate but not on M9 medium containing more than 2 mm glyphosate.
Thirteen resistant clones were isolated and classified into four types based on the results of sequencing the OsEPSPS coding region. Type I, including three clones identified from independent processes, comprised a C-T transition at position 317 (C317→T) on the coding strand, which caused a single amino acid substitution of Pro to Leu at position 106 (P106L) in the protein sequence. Type II consists of six clones with nucleotide sequences containing synonymous mutations (two clones with G12→A and four clones with G246→A, A468→T, C954→T, and A1050→T, respectively), whereas no amino acid sequences were changed. Type III contains a single clone in which two nucleotides (GT) were inserted just before the initial codon but no other changes were detected, either in nucleotide or amino acid sequence. Type IV contains three clones with no change detected in the sequenced region. Since the P106L mutant was the only type with an amino acid substitution in the protein sequence, it was further studied and is discussed systematically in this article.
Plasmid DNA of the resistant clones was isolated and transformed into AB2829. Glyphosate resistance of all types of the mutants was confirmed again to rule out self-mutation of the host E. coli. Furthermore, the mutant OsEPSPS coding sequence was recombined to pBREP to confirm that the original glyphosate resistance mutant was essential. Only mutant types I and II retained glyphosate resistance after reconstruction.
Refined Analysis of Glyphosate Resistance in E. coli
Glyphosate resistance was analyzed in vivo by growth of E. coli harboring pBREP-P106L (expressing P106L mutant) in minimal medium containing increased glyphosate concentrations. E. coli harboring pBREP was used as a wild-type control. E. coli harboring pBREP-G101A, a vector expressing OsEPSPS with a site-directed G101A substitution, was also used as a control due to the reported high glyphosate resistance conferred by a homologous G101A substitution in EPSPS from various sources (Padgette et al., 1991).
The increase in OD600 over the original dilution was compared between bacterium strains with increasing glyphosate concentration in the medium (Fig. 1A). At glyphosate concentrations below 1.0 mm, E. coli (pBREP-P106L) grew more rapidly than the other two clones. However, growth was gradually reduced at higher glyphosate concentrations. In medium containing 0.4 mm glyphosate, growth of this clone was about 60%, which was only 15% lower than its growth in glyphosate-free medium and 3-fold higher than the other two clones. Growth of all three clones decreased more than 80% at glyphosate concentrations above 1.0 mm, with E. coli (pBREP-G101A) having a slightly higher growth rate (2%–5%) than the others.
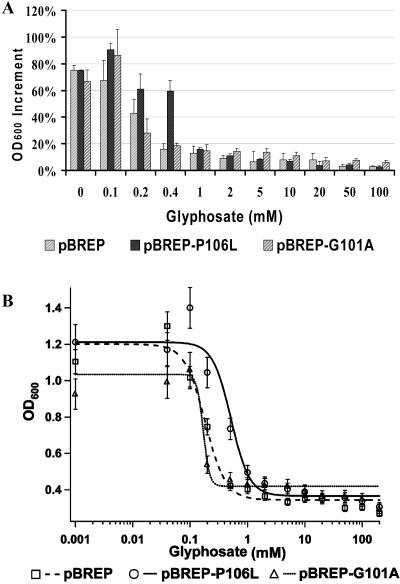
Figure 1.
Glyphosate resistance analysis of EPSPS mutants via growth of E. coli transformant. DH5α was transformed with pBREP (wild type), pBREP-P106L, and pBREP-G101A. The transformants were cultured in liquid MOPS medium containing glyphosate in increased concentration gradients. Aliquots of culture sample were measured for OD600 before (ODb) and after (ODa) 12 h of incubation in 37°C by vigorous vibration. Error bars represent the sd resulting from three repetitions. A, Bar chart; OD600 increment was calculated as (ODa − ODb)/OD0, in which OD0 is the final value of OD600 absent glyphosate. B, Curve chart; the curve was determined by fitting data of ODb to f(x) = C + (D − C)/(1 + (x/LD50)b) (Seefeldt et al., 1995; Igor Pro 4.00), where C = lower limit, D = upper limit, LD50 = 50% lethal dose, and b = slope of the curve at LD50. The first and last points on the curve represent the data of OD0 and ODb.
A log-logistic model (Seefeldt et al., 1995) was used to analyze the data to predict the trend of glyphosate resistance (Fig. 1B). In this model, the equation y = f(x) = C + (D − C)/(1 + (x/LD50)b) was used to fit the data (LD50 = 50% lethal dose, b = slope of the curve at LD50, C = lower limit, and D = upper limit). LD50 was determined to be 0.517 mm, 0.183 mm, and 0.169 mm for E. coli (pBREP-P106L), E. coli (pBREP), and E. coli (pBREP-G101A), respectively (Table I), suggesting a 3-fold increase in glyphosate resistance of the P106L mutant compared to wild-type OsEPSPS. Curves for E. coli (pBREP-P106L) and E. coli (pBREP) have similar shapes, with nearly the same values of C, D, and b, suggesting that the P106L mutant has a similar mode of catalysis as wild-type OsEPSPS. Although the value of C increased 20% in the E. coli (pBREP-G101A) curve, the lower LD50 and D and the approximately 3-fold increase in b implies that the G101A mutant has a different mode of catalysis and is much more sensitive to glyphosate than the P106L mutant in this experiment.
Table I.
Fitting coefficients of the resistance curves
Fitting parameters of the OD600 values of two different mutant types, P106L and G101A, and the wild-type rice EPSPS. The equation used to fit the data is a general dose response curve: y = f(x) = C + (D − C)/(1 + (x/LD50)b), where C is the lower base line, D is the upper limit, LD50 is the 50% lethal dose, and b is the slope of the curve at LD50. Standard deviation for each parameter is also listed in the table.
| Coefficients | pBREP | pBREP-P106L | pBREP-G101A |
|---|---|---|---|
| C | 0.344 ± 0.015 | 0.366 ± 0.031 | 0.419 ± 0.018 |
| D | 1.200 ± 0.059 | 1.211 ± 0.087 | 1.034 ± 0.061 |
| LD50 | 0.183 ± 0.014 | 0.517 ± 0.046 | 0.169 ± 0.044 |
| b | 2.429 ± 0.287 | 2.698 ± 0.567 | 8.038 ± 1.764 |
Kinetic Characterization of the P106L Mutant
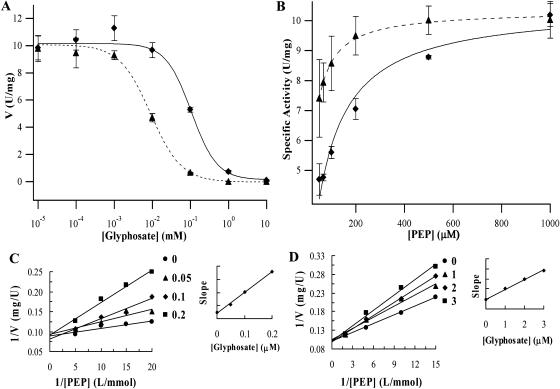
The coding sequence of the P106L mutant and OsEPSPS was cloned into a pET-28a inducible expression vector, and crude protein extract was prepared for enzyme assays. The kinetic constants of the P106L mutant and the wild-type OsEPSPS were determined as shown in Table II. At 1.0 mm substrate, the specific activity was 10.032 units (U)/mg and 10.225 U/mg for the P106L mutant and wild-type OsEPSPS, respectively. The IC50 (50% inhibition concentration of glyphosate) was 104.606 μm for the P106L mutant, which is about 11-fold greater than the IC50 determined for wild-type OsEPSPS, 8.982 μm (Fig. 2A). The Km (PEP) of the P106L mutant was 88.260 μm (Fig. 2B), which is about 4.6-fold greater than the Km (PEP) of wild-type OsEPSPS (19.940 μm). The Ki (glyphosate) of the P106L mutant and wild-type OsEPSPS were 4.020 μm and 0.057 μm, respectively, indicating an approximately 70-fold decreased affinity of the P106L mutant for glyphosate compared to the wild-type OsEPSPS. If the value of Ki/Km (PEP) is taken as a measure of the selectivity for PEP over glyphosate binding, the P106L mutant has an approximately 15-fold greater Ki/Km than wild-type OsEPSPS.
Table II.
Kinetic constants of E. coli-expressed OsEPSPS and P106L mutant
Kinetic constants of wild-type EPSPS and P106L were obtained from the least-squares fitting that was carried out by using Igor Pro 4.00 software. Kinetic parameters are expressed as mean value ± sd derived from the best fit.
| Kinetic Constants | OsEPSPS | OsEPSPS-P106L |
|---|---|---|
| Specific activity (U/mg) | 10.032 ± 0.613 | 10.225 ± 0.599 |
| IC50 (glyphosate; μm) | 8.982 ± 0.799 | 104.606 ± 38.948 |
| Km (PEP; μm) | 19.940 ± 0.822 | 88.260 ± 9.480 |
| Ki (glyphosate; μm) | 0.057 ± 0.021 | 4.020 ± 0.087 |
| Vmax (U/mg) | 10.562 ± 0.490 | 10.663 ± 0.346 |
| Ki/Km (PEP) | 0.003 | 0.045 |
Figure 2.
Enzyme assay results. Km (PEP) and IC50 were obtained from the least-squares fitting of the data to appropriate equations (Igor Pro 4.00). Black triangles and diamonds represent the measured data points of wild-type OsEPSPS and P106L EPSPS, respectively. Error bars represent the sd resulting from three repetitions. Dashed and solid curves represent the fitting curves of OsEPSPS and P106L mutant, respectively. S3P concentration was fixed at 1 mm. A, Km (PEP) was determined by fitting the data to V = Vmax[S]/(Km + [S]), where V is the velocity of the reaction (expressed in U/mg), Vmax is the maximum velocity, and [S] is the concentration of the substrate assayed for the Km. B, The curve was determined by fitting data to V = Vmin + (Vmax − Vmin)/(1 + ([I]/IC50)s), where [I] is concentration of glyphosate and s is the slope of the curve at the IC50. The first point on the curve represents the data obtained with no glyphosate. Ki value was determined by the method as described (Copeland, 1996). C, Ki (glyphosate) of OsEPSPS was determined by fixed PEP concentration of 0.05, 0.067, 0.1, and 0.2 mm, respectively, and glyphosate concentration was 0, 0.05, 0.1, and 0.2 μm. D, Ki (glyphosate) of P106L mutant was determined by fixed PEP concentration of 0.067, 0.1, 0.2, and 0.5 mm, respectively, and glyphosate concentration was 0, 1, 2, and 3 μm.
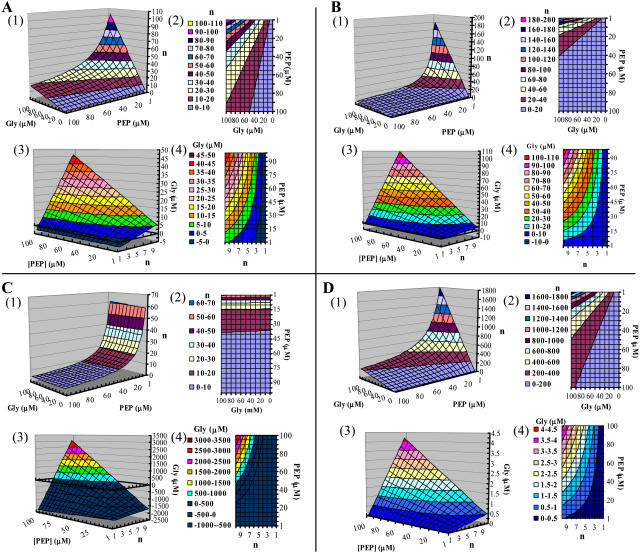
Construction and Analysis of the RSO Model
A Resistance-Substrate-Overexpression (RSO) model (Fig. 3) was constructed to analyze the relationship between glyphosate resistance, substrate (PEP) concentration, and level of P106L mutant expression (Fig. 3A). As controls, two EPSPS mutants, P106S mutant from goosegrass (Fig. 3B; Baerson et al., 2002) and G101A mutant from petunia (Fig. 3C; Padgette et al., 1991), and the wild-type OsEPSPS (Fig. 3D) were compared with the RSO model based on the kinetic data from the references.
Figure 3.
RSO model. Model construction was based on Michaelis-Menten equation. A to D represent the RSO model of P106L mutant, P106S mutant from goosegrass (Baerson et al., 2002), G101A mutant from petunia (Padgette et al., 1991), and wild-type OsEPSPS, respectively. Calculations for A and D were carried out using the kinetic parameters shown in Table II. Calculations for B and C were carried out using the kinetic parameters reported previously (Padgette et al., 1991; Baerson et al., 2002). Sections 1 and 3 represent the surface charts generated by Excel from Equations 1 and 2, respectively. Sections 2 and 4 represent the contour charts of 1 and 3, respectively. Gly, Abbreviation of glyphosate.
Using the Michaelis-Menten equation, the wild-type OsEPSPS has a normal catalytic reaction velocity defined as 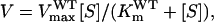 in which [S] represents PEP concentration,
in which [S] represents PEP concentration, 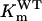 is the Michaelis constant of PEP for wild-type OsEPSPS, and
is the Michaelis constant of PEP for wild-type OsEPSPS, and 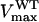 is the maximum velocity of wild-type OsEPSPS. When glyphosate is present, the velocity of enzyme has the form
is the maximum velocity of wild-type OsEPSPS. When glyphosate is present, the velocity of enzyme has the form 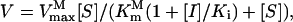 in which
in which 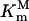 is the Michaelis constant of PEP for P106L mutant and
is the Michaelis constant of PEP for P106L mutant and 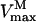 is the maximum velocity of P106L mutant, Ki is the inhibitor constant of glyphosate, and [I] is the glyphosate concentration. To achieve the normal catalytic velocity, the P106L mutant must be expressed n-fold over wild-type OsEPSPS, thus:
is the maximum velocity of P106L mutant, Ki is the inhibitor constant of glyphosate, and [I] is the glyphosate concentration. To achieve the normal catalytic velocity, the P106L mutant must be expressed n-fold over wild-type OsEPSPS, thus:
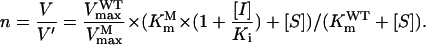 |
(1) |
From Equation 1, we can then derive:
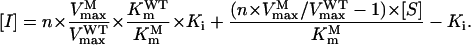 |
(2) |
The RSO model was constructed of four parts. As shown in Figure 3, (1) is a three-dimensional (3-D) surface chart and (2) is the corresponding contour chart created by defining the z axis as n of Equation 1, thus the surface in A represents the lowest overexpression ratio (n) of the enzyme needed to achieve the normal catalytic velocity when PEP concentration ranges from 1 to 100 μm and glyphosate concentration ranges from 0 to 100 μm. Similarly, (3) is a 3-D surface chart and (4) is the corresponding contour chart created by defining the z axis as [I] of Equation 2, and the surface in (3) represents the highest concentration of glyphosate at which the enzyme can maintain the normal catalytic velocity when PEP concentration ranges from 1 to 100 μm and overexpression ratio ranges from 1- to 10-fold. Note that some z axis data of (3) are less than zero (denoted in dark blue) from theoretical calculation, indicating that the enzyme fails to achieve normal catalytic velocity even when no glyphosate is present.
The overexpression ratio of P106L mutant necessary to achieve normal catalytic velocity is increased when PEP concentration is decreased and glyphosate concentration is increased (Fig. 3A, 1 and 2). Approximately 100-fold overexpression of P106L mutant is needed to tolerate 100 μm glyphosate at 1 μm PEP. The glyphosate resistance conferred by P106L mutant decreased as the PEP concentration and overexpression ratio decreased (Fig. 3A, 3 and 4). Ten-fold overexpression of the P106L mutant can confer glyphosate resistance of approximately 45 μm at 100 μm PEP.
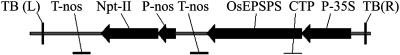
Figure 4.
T-DNA cassette containing encoding region of OsEPSPS on pTi-OsEPSPS for plant transformation. Npt-II, Neomycin phosphotransferase gene; P-nos, nopaline synthase promoter; T-nos, nopaline synthase terminator; TB(L) and TB(R), left and right border of T-DNA; P-35S, cauliflower mosaic virus 35S promoter; OsEPSPS, coding sequence of premature form of OsEPSPS; CTP, coding sequence of chloroplast transit peptide.
In the RSO model, the P106S mutant (Fig. 3B) behaves similar to the P106L mutant. The overexpression ratio is lower compared to the P106L mutant in high PEP concentrations, while at PEP concentrations lower than 30 μm and glyphosate concentrations higher than 50 μm the overexpression ratio increases rapidly (Fig. 3B, 1 and 2). Approximately 180-fold overexpression of the P106S mutant is needed to tolerate 100 μm glyphosate at 1 μm PEP. Ten-fold overexpression of the P106S mutant can confer glyphosate resistance of approximately 110 μm at 100 μm PEP (Fig. 3B, 3 and 4).
In the RSO model, the G101A mutant (Fig. 3C) behaves very different from the P106L and P106S mutants. The overexpression ratio is elevated at approximately 35 μm PEP and rapidly increases as PEP concentration is reduced (Fig. 3C, 1 and 2). Glyphosate concentration had a slight effect on the overexpression ratio. Approximately 62-fold overexpression of the G101A mutant is needed to tolerate 100 μm glyphosate at 1 μm PEP. The large area in dark blue represents glyphosate resistance values below zero, which means the G101A mutant cannot achieve normal catalytic velocity even in the absence of glyphosate (Fig. 3C, 3 and 4). However, when the threshold value is surpassed (more than 35 μm for PEP concentration and more than a 5-fold overexpression ratio, approximately), glyphosate resistance increased dramatically. Ten-fold overexpression of the G101A mutant can confer glyphosate resistance of approximately 2,500 μm at 100 μm PEP.
The RSO model of wild-type OsEPSPS appears almost the same as that of the P106L mutant except that the z axis scale differs by approximately one order of magnitude (Fig. 3D). Approximately 1,800-fold overexpression of OsEPSPS is needed to tolerate 100 μm glyphosate at 1 μm PEP. Ten-fold overexpression of OsEPSPS can only confer glyphosate resistance of approximately 4 μm at 100 μm PEP.
The RSO model (Fig. 3) indicates that EPSP synthesis is a comprehensive effect of kinetic characterizations, level (n) of EPSPS expression, and concentrations of PEP and glyphosate, i.e. concentrations of the substrate and the competitive inhibitor. Data from the RSO model are shown at the fixed criteria of 10-fold overexpression, 50 μm glyphosate, and 50 μm PEP, respectively (Table III). The results indicate that P106L and P106S mutants display similar behavior of glyphosate resistance and range in the same order of magnitude. The P106L and P106S mutants can survive greater glyphosate concentrations than the G101A mutant in relatively low PEP (0∼50 μm) concentrations and at low expression level (2∼5). In contrast, the G101A mutant needs relatively high expression (n ≥ ∼5) and PEP concentrations (≥∼35 μm) to achieve normal catalytic activity, regardless of glyphosate presence, although significant glyphosate resistance was conferred when the threshold conditions were satisfied.
Table III.
RSO model comparison of several mutants
Data were summarized according to the RSO model when taking 10-fold overexpression, 50 μm PEP, and 50 μm glyphosate as fixed parameters, respectively.
| Mutant Type
|
n = 10 1 μm ≤ PEP ≤ 100 μm
|
PEP = 50 μm 2 ≤ n ≤ 10
|
Glyphosate = 50 μm 1 μm ≤ PEP ≤ 100 μm
|
|---|---|---|---|
| Glyphosate | Glyphosate | Overexpression | |
| μm | μm | n | |
| P106L mutant | 0∼40 | 0∼20 | 10∼50 |
| P106S mutant | 0∼100 | 0–60 | 5∼90 |
| G101A mutant | 0∼3,000 ([PEP] > ∼35) | 0∼500 (n > ∼8) | 8∼90 |
| Wild-type OsEPSPS | 0∼4 | 0∼2 | 100∼900 |
Glyphosate Resistance of P106L Mutant in Transgenic Tobacco
A mini-Ti plasmid, pTi-P106L, containing the P106L mutant coding sequence, was constructed for plant transformation. In pTi-P106L, the chloroplast transit peptide sequence derived from the premature OsEPSPS was fused to P106L (Fig. 4) to direct the mutant to chloroplasts, the intracellular location of EPSPS catalysis in plants (Della-Cioppa et al., 1986). As a wild-type control, pTi-OsEPSPS, containing the OsEPSPS coding sequence, was constructed in the same way.
Subsequently, pTi-P106L was introduced into tobacco plants (Nicotiana tabacum cv Xanthi) via agrobacterium-mediated transformation. Transgenic plants were obtained from plantlets regenerated on medium containing 50 mg/L kanamycin and further confirmed by PCR analysis and northern blotting (data not shown). The glyphosate resistance of transgenic plants was then compared at different growth stages. Callus differentiation of transgenic plants on glyphosate-containing media was inspected (Fig. 5A). Actively growing callus tissue appeared on the periphery of all leaf discs on the control plate lacking glyphosate. On media containing 0.02 mm glyphosate, little difference was detected between the control discs and the pTi-P106L-transformed tissue, whereas growth of nontransformed tissue was affected severely. On medium containing 0.1 mm glyphosate, substantial calli grew from pTi-P106L-transformed discs, whereas very little callus grew from pTi-OsEPSPS-transformed tissue and growth of the nontransformed control was inhibited completely. Seedlings of transgenic plants also exhibited various levels of resistance on medium containing glyphosate (Fig. 5B). P106L-A seedlings, the most glyphosate-resistant plant line transformed with pTi-P106L, exhibited near normal growth on medium containing 0.1 mm glyphosate and stayed green on medium containing up to 10 mm glyphosate, with little growth. However, WT-5 seedlings, the most glyphosate-resistant line transformed with pTi-OsEPSPS, showed chlorosis at 5 mm glyphosate, and seedlings of nontransformed plants showed chlorosis at 0.1 mm glyphosate. The six- to eight-leaf-stage transgenic plants were sprayed with a 1% (v/v) solution of the herbicide Roundup (isopropylamine glyphosate salt as active ingredient, 41.0%) at a dose of 2 L/ha. P106L-A plants grew well with normal morphology, whereas WT-5 plants showed severe chlorosis on most leaves and nontransformed plants showed very rapid chlorosis and bleaching, then wilted and died (Fig. 5C). None of the plants survived after glyphosate treatment at a dose of 4 L/ha. These results indicated that pTi-P106L-transformed plants are more resistant to glyphosate exposure than pTi-OsEPSPS-transformed and nontransformed plants.
Figure 5.
Glyphosate resistance of transgenic tobacco. A, Photograph was taken after 30 d of culture on medium containing 0 to 0.5 mm glyphosate. (Columns 1–7 contained 0, 0.01, 0.02, 0.05, 0.1, 0.2, and 0.5 mm glyphosate, respectively.) The top line shows the leaf discs of nontransformed plants, the middle line shows the leaf discs transformed with pTi-OsEPSPS, and the bottom line shows the leaf discs of plants transformed with pTi-P106L. B, Photograph was taken at 1 month after the transplant. Seedlings were germinated and selected on medium containing 100 mg/L kanamycin for 7 to 10 d to eliminate the negative plants; the green seedlings with two to four true leaves were transplanted onto medium containing 0 to 10 mm glyphosate. (Columns 1–7 contained 0, 0.05, 0.1, 1, 2, 5, and 10 mm glyphosate, respectively.) The top line shows the seedlings of nontransformed plants, the middle line shows the seedlings of transgenic plant line CEP5 expressing wild-type OsEPSPS, and the bottom line shows the seedlings of plant line P106L-A expressing P106L mutant OsEPSPS. C, Photograph was taken at 2 weeks after the spray treatment. The top line was control plants without treatment of Roundup. The bottom line was sprayed with Roundup at a dose of 0.2 mL/m2 in 1% concentration. Numbers 1 and 4 represent nontransformed plants. Numbers 2 and 5 represent transgenic plant line CEP5 expressing wild-type OsEPSPS. Numbers 3 and 6 represent transgenic plant line P106L-A expressing P106L mutant OsEPSPS.
DISCUSSION
In this study, a selection system based on directed evolution was designed to select an EPSPS mutant conferring high glyphosate resistance and retaining high catalytic activity. Because the E. coli host AB2829 is deficient in endogenous EPSPS, only those transformants containing complementary EPSPS that provide adequate enzyme activity can grow on minimal medium without extra aromatic amino acid supplements. In addition, resistant clones that survive subsequent glyphosate selection should contain a mutant with enough EPSPS catalytic activity in the presence of glyphosate. The low copy number vector pBR322 (Mayer, 1995) as well as the weak and constitutive bla promoter (Liang et al., 1999) were chosen to provide relatively low EPSPS expression, which not only ensures a low selection background of glyphosate but also selects for a mutant enzyme that exhibits relatively high catalytic activity. It is expected that no mutants, such as the frequently reported G101A mutant (Padgette et al., 1991), will occur in our selection conditions due to the significantly decreased catalytic activity, despite low glyphosate affinity. The host in our study, AB2829, cannot grow on M9 medium, with or without glyphosate. AB2829 expressing wild-type OsEPSPS cannot grow on medium containing more than 1 mm glyphosate due to the low EPSPS expression. Correspondingly, 5 mm glyphosate was enough to select glyphosate-resistant mutants from wild-type transformants in our system. In another similar study, a randomly mutagenized EPSPS gene was generated by recombination of the aroA genes from S. typhimurium and E. coli through StEP (Zhao et al., 1998), and then expressed in E. coli BL21 under control of a modified isopropylthio-β-galactoside-inducible T7 promoter (He et al., 2001). In their system, the BL21 host was reported to spontaneously survive 1 mm glyphosate on M9 medium due to an endogenous EPSPS gene. After introduction of the exogenous wild-type EPSPS, the BL21 grew on medium containing 10 mm glyphosate because of high levels of EPSPS expression. Consequently, they selected the glyphosate-resistant mutant on medium containing 50 to 60 mm glyphosate. Compared with early methods to identify glyphosate-resistant mutants, another advantage of this directed evolution system is the ease of genetic manipulation of the mutant EPSPS gene on the plasmid, which is independent from the chromosome, as well as subsequent identification and analysis.
A glyphosate-resistant mutant strain of S. typhimurium generated by ethyl methanesulfonate mutagenesis, which confers glyphosate resistance due to the substitution of Pro-106 with Ser (P106S), was reported previously (Comai et al., 1983). The corresponding P106S mutant is also responsible for the glyphosate resistance of goosegrass, a weed found in orchards where glyphosate was used intensively as a herbicide for 8 to 10 years in Malaysia (Baerson et al., 2002). In this study, it is surprising that, although we cloned 30 μg of error-prone PCR product for each mutation region, which is about 6-fold greater than the quantity sufficient for a mutant library (Cadwell and Joyce, 1994), no mutations similar to the reported results were obtained, except the P106L substitution, which occurred repetitively in our three independent experiments. Although Baerson et al. (2002) have concluded that goosegrass might be predisposed to this mutation due to species-specific genetic or physiological characteristics, the high frequency of the Pro-106 substitution occurring in bacteria- or plant-derived EPSPS from artificial (Comai et al., 1983) or natural selection (Baerson et al., 2002) indicates that Pro-106 might be a universal mutation hot spot for glyphosate resistance. These results also imply that our selection system simulated the natural conditions for wild-type EPSPS to spontaneously evolve glyphosate resistance.
The crystal structure of E. coli EPSPS, with about 50% sequence identity to OsEPSPS, provides a structural basis to understand the roles of Pro-106 and Gly-101 (see supplemental files). Glyphosate directly forms a hydrogen bond with the main-chain nitrogen atom of Gly-101 in the native enzyme, making Gly-101 critical for glyphosate binding. In G101A mutant, the Cβ atom seems too close to glyphosate phosphor oxygen (1.9 Å versus 2.8 Å) and may push glyphosate away from the binding site. Pro-106 is not directly involved in glyphosate binding. However, Pro-106 is located in an α-helix starting from Gly-101 and bends this helix slightly, which is unusual due to the unstable conformation (Yun et al., 1991). The large hydrophobic side chain of Leu-106 in the P106L mutant probably affects the conformation of this α-helix or the orientation of the side chain of Arg-105, resulting in a movement of Gly-101. As a consequence, the affinity to glyphosate decreases. The essential relationship between glyphosate resistance and Pro-106 substitution should be further studied by protein crystallography, which will help the rational design of EPSPS engineering and promote our understanding of the interaction between EPSPS, ligands, and inhibitor.
Kinetic characterization of the P106L mutant and the wild-type OsEPSPS was determined by in vitro enzyme assays. Although the results of enzyme assays in different studies vary based on experimental conditions, it is useful to compare the kinetic parameters of different EPSPS mutants. Compared to wild-type EPSPS, the P106L mutant showed a 70-fold increase in Ki, and the P106S substitution resulted in a 22-fold increase in Ki in goosegrass. The G101A substitution in petunia EPSPS showed a 5,000-fold increase in Ki. These results indicate that the P106L and P106S mutants had similar glyphosate affinities that were about one order of magnitude lower than wild type, whereas the G101A mutant has a significantly lower glyphosate affinity (three orders of magnitude). As a popular criterion of glyphosate resistance, Ki/Km (PEP) is approximately 11-fold greater in the P106L mutant and similarly 12-fold increased in the P106S mutant. However, Ki/Km (PEP) for the G101A mutant was increased 119-fold. The IC50 was 10-fold greater in the P106L mutant and 6-fold greater in the P106S mutant, whereas IC50 increased 567-fold in the G101A mutant. These data demonstrate the utility of the G101A mutant as a glyphosate-resistant mutant. However, the G101A mutant has a 40% lower Vmax and a 42-fold greater Km (PEP). Conversely, the P106L and P106S mutants showed only 4.6- and 2-fold greater Km (PEP), respectively, and no obvious Vmax decrease detected in the P106L mutant. Such results indicate that the Pro-106 substitution had a less severe impact on EPSPS catalytic activity than the G101A mutant.
In this study, an RSO model was constructed to illustrate the relationship among the expression level, PEP concentration, and glyphosate resistance for a specific mutant enzyme. This model indicates that survival of the host organism in the presence of glyphosate relies on the synthesis velocity of EPSP, which is determined by the expression level (n) and catalytic velocity of EPSPS. Catalytic velocity of the mutant EPSPS is determined by Km (PEP), Vmax, Ki (glyphosate), and the concentrations of glyphosate and PEP presented in the system. Among these factors, PEP concentration in the host is relatively constant and can hardly be adjusted. The level of EPSPS expression and the glyphosate concentration in the selection system are the only two adjustable parameters that, if changed, will affect the selection system, resulting in different mutant types with different Km (PEP), Vmax, and Ki (glyphosate), i.e. different catalytic activity and glyphosate affinity. From Equation 1, a derivation of the Michaelis-Menten equation, we can deduce Equation 3:
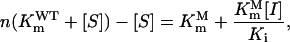 |
(3) |
where Vmax of the mutant and the wild-type EPSPS were assumed to be equal. Actually, compared to the wild-type EPSPS, Vmax of the mutants generally showed only a very small change or was in the same order of magnitude. Let 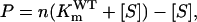 then Equation 3 can be rewritten as follows:
then Equation 3 can be rewritten as follows:
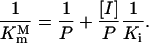 |
(4) |
This equation is nominated as Double-Reciprocal Affinity (DRA) model. If we plot 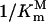 versus
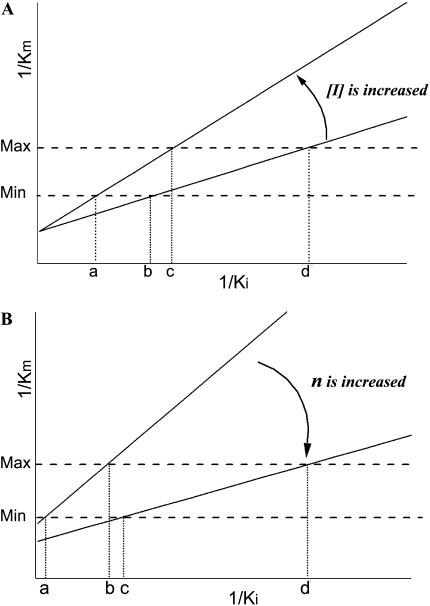
versus 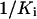 schematically, a linear relationship will be obtained with a slope of [I]/P and an intercept of 1/P (Fig. 6). This schematic plot illustrates the relationship between glyphosate affinity (1/Ki) and PEP affinity (1/Km) when the normal catalytic velocity has to be achieved by the mutant EPSPS that is expressed at a constant level (n) and at a constant intracellular PEP concentration. Therefore, the Km and Ki of the mutant are determined by the slope and the intercept, respectively, which are related to the glyphosate concentration and expression level, the only two adjustable parameters in the selection system. 1/Km is proportional to 1/Ki because of the positive slope, which is consistent with competitive binding between glyphosate and PEP. We can theoretically assume a maximum and minimum value of 1/Km, i.e. theoretically, the EPSPS has a maximum catalytic activity with a maximum PEP affinity and a minimum PEP affinity to sustain metabolism for survival. When expression level n is assumed to be a constant while [I] is increased such that the slope is increased, the range of 1/Ki will be decreased (b–d to a–c), whereas the change of 1/Km becomes more pronounced (Fig. 6A). Therefore, the glyphosate affinity of the mutant will be decreased when selection concentration is increased; however, PEP affinity (1/Km) will change dramatically when the selection concentration is increased too much. In contrast, when [I] is assumed to be a constant while the expression level n increases, such that the slope ([I]/P) and the intercept (1/P) both decrease (Fig. 6B), then the range of glyphosate affinity (1/Ki) will increase (a–b to c–d), indicating that higher expression is adverse for selection of high glyphosate-resistant mutants. Therefore, the selection must be carried out with a moderate selection concentration and low expression level if higher PEP affinity and lower glyphosate affinity, i.e. high glyphosate resistance, are expected. In the directed evolutions under a series of gradient concentrations or with several rounds of mutagenesis/selection, mutants that survived lower or moderate concentration of inhibitor (e.g. glyphosate) were generally ignored in subsequent research (Comai et al., 1983; Stalker et al., 1985; Stemmer, 1994; He et al., 2001, 2003). However, our analysis above indicates that the moderate-resistant mutants may have some advantages or unique features that the high-resistant mutants do not have. This analysis also suggests that the selection system of directed evolution can be regulated by changing expression level n and selection concentration [I]. At the same time, the kinetic characterization (Ki and Km) of the mutants selected under specific conditions can be predicted theoretically. The selection system in our study might be coincidentally suitable for the P106L mutant to evolve, and the repetitive occurrence of the P106L mutant probably suggests the evolutionary aptitude of the class I EPSPS under glyphosate selection.
schematically, a linear relationship will be obtained with a slope of [I]/P and an intercept of 1/P (Fig. 6). This schematic plot illustrates the relationship between glyphosate affinity (1/Ki) and PEP affinity (1/Km) when the normal catalytic velocity has to be achieved by the mutant EPSPS that is expressed at a constant level (n) and at a constant intracellular PEP concentration. Therefore, the Km and Ki of the mutant are determined by the slope and the intercept, respectively, which are related to the glyphosate concentration and expression level, the only two adjustable parameters in the selection system. 1/Km is proportional to 1/Ki because of the positive slope, which is consistent with competitive binding between glyphosate and PEP. We can theoretically assume a maximum and minimum value of 1/Km, i.e. theoretically, the EPSPS has a maximum catalytic activity with a maximum PEP affinity and a minimum PEP affinity to sustain metabolism for survival. When expression level n is assumed to be a constant while [I] is increased such that the slope is increased, the range of 1/Ki will be decreased (b–d to a–c), whereas the change of 1/Km becomes more pronounced (Fig. 6A). Therefore, the glyphosate affinity of the mutant will be decreased when selection concentration is increased; however, PEP affinity (1/Km) will change dramatically when the selection concentration is increased too much. In contrast, when [I] is assumed to be a constant while the expression level n increases, such that the slope ([I]/P) and the intercept (1/P) both decrease (Fig. 6B), then the range of glyphosate affinity (1/Ki) will increase (a–b to c–d), indicating that higher expression is adverse for selection of high glyphosate-resistant mutants. Therefore, the selection must be carried out with a moderate selection concentration and low expression level if higher PEP affinity and lower glyphosate affinity, i.e. high glyphosate resistance, are expected. In the directed evolutions under a series of gradient concentrations or with several rounds of mutagenesis/selection, mutants that survived lower or moderate concentration of inhibitor (e.g. glyphosate) were generally ignored in subsequent research (Comai et al., 1983; Stalker et al., 1985; Stemmer, 1994; He et al., 2001, 2003). However, our analysis above indicates that the moderate-resistant mutants may have some advantages or unique features that the high-resistant mutants do not have. This analysis also suggests that the selection system of directed evolution can be regulated by changing expression level n and selection concentration [I]. At the same time, the kinetic characterization (Ki and Km) of the mutants selected under specific conditions can be predicted theoretically. The selection system in our study might be coincidentally suitable for the P106L mutant to evolve, and the repetitive occurrence of the P106L mutant probably suggests the evolutionary aptitude of the class I EPSPS under glyphosate selection.
Figure 6.
Schematic diagrams of the relationship between PEP affinity 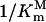 and glyphosate affinity
and glyphosate affinity 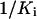 as shown in Equation 4. Two horizontal dash lines represent minimum and maximum values of
as shown in Equation 4. Two horizontal dash lines represent minimum and maximum values of 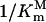 . Four vertical dotted lines represent the corresponding values of
. Four vertical dotted lines represent the corresponding values of 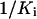 ; A is obtained when the glyphosate concentration [I] is increased, and B is obtained when the expression level n is increased.
; A is obtained when the glyphosate concentration [I] is increased, and B is obtained when the expression level n is increased.
The RSO model also indicates that the resistance level conferred by the EPSPS mutant is highly dependent on the intracellular environment, especially the physiological PEP concentration. The P106L mutant in E. coli displayed only a 3-fold LD50 increase, whereas an 11-fold IC50 increase was obtained in in vivo assays. This is most likely to be a consequence of higher PEP concentration (1 mm) in the assay compared to physiological PEP concentration in E. coli. Contrary to the reported results, high sensitivity of the G101A mutant to glyphosate in the E. coli glyphosate resistance assay might result from relatively low expression and/or low PEP concentration. Physiological PEP concentration in some plant species has been reported to fluctuate diurnally between 5 to approximately 60 nmol g−1 fresh weight (Chen et al., 2002), which is equal to roughly 5 to approximately 60 μm PEP. At such a low PEP concentration, much higher expression of the G101A mutant might be needed to confer complete glyphosate resistance compared to the P106L and P106S mutants.
Tobacco plants transformed with the P106L mutant showed higher glyphosate resistance compared to the wild-type control. The P106L-A transgenic plant line survived treatment with 2 L/ha glyphosate. A plant-derived EPSPS mutant is likely to be more efficiently engineered for glyphosate tolerance in crops due to optimal codon usage and protein stability in plants. Furthermore, genetically modified crops expressing an endogenous plant protein will be more acceptable to consumers than those expressing bacterial proteins.
SUMMARY
In this study, a strategy based on directed evolution was developed to select glyphosate-resistant EPSPS mutants. Repeated occurrence of the Pro-106 substitution suggests a possible evolutionary advantage of this substitution for class I EPSPS under glyphosate selection. Analysis of enzyme kinetics revealed that the P106L mutant confers high glyphosate resistance while retaining relatively high catalytic activity. The RSO model and the DRA model derived from the Michaelis-Menten equation could be useful for evaluating glyphosate resistance and the kinetic characterizations of an EPSPS mutant and for rationally designing a directed evolution system. We speculate that the repetitive occurrence of the P106L mutant in our experiments is related to the level of EPSPS expression and the selection conditions, e.g. the promoter, the copy number of the vector, as well as the selection concentration, and that different mutant types would be obtained if the above conditions were changed. Therefore, we believe this directed evolution strategy, as well as kinetic analysis of the RSO model and the DRA model, are not only applicable to EPSPS but also to other enzymes susceptible to competitive inhibition.
MATERIALS AND METHODS
Strains and Reagents
Escherichia coli AB2829 (aroA354, supE42; Pittard and Wallace, 1966) came from the E. coli Genetic Stock Center. Glyphosate and PEP were from Sigma-Aldrich. Herbicide Roundup was from Monsanto. Taq DNA polymerase was from Tianwei Biotech. S3P was prepared by a simplified protocol as described (Weiss and Mingioli, 1956; Knowles and Sprinson, 1970).
Mutagenesis via Error-Prone PCR
DNA fragment encoding rice (Oryza sativa) EPSPS mature enzyme was obtained by reverse transcription (RT)-PCR with the primers 5′-ATT GAA AAA GGA AGA GTA TGA GGC GGA GGA GAT C and 5′-AAA CTG CAG CTC ACT CTT TTA AAA from reverse transcriptional product of total RNA of rice. After digestion with PstI, the PCR product was inserted into the vector pBR322 and replaced the coding sequence of the bla gene at the sites of SspI and PstI to construct pBREP vector.
Error-prone PCR was performed as described (Cadwell and Joyce, 1994). Two regions of OsEPSPS coding sequence used as the template were obtained by restriction digestion and purified by low-melt agarose gel with a standard lab method. The oligonucleotide primers used for region I were 5′-TTT AGA TCT CCG GGG CGG TT and 5′CAA GTA CTG ACT GCT GAT GG, and primers for region II were 5′TTA GTA CTT GAG TGC CTT GC and 5′-AAA CTG CAG CTC ACT CTT TTA AAA.
Isolation of the Resistance Clones
Digested by BglII and ScaI, the error-prone PCR product of region I was inserted into the same restriction sites of pBREP to replace the wild-type sequence. The product of region II was processed similarly except that the restriction sites used were ScaI and PstI. For 0.2 μg of ligated DNA product, 40 μL of competent cells were used for AB2829 transformation by electroporation as a standard method. The resultant cells were washed with M9 liquid medium and spread on M9 plates at approximately 3 clones cm−2 after 37°C incubation for 48 h. Clones were then transplanted by a velvet replica-plating tool (Lederberg and Lederberg, 1952) onto M9 plates containing 5 mm glyphosate at 37°C incubated for 48 h. Plasmids were purified from resistant clones, and nucleotide sequences of the recombinant epsps gene were determined with a model ABI 377 DNA sequencer (Applied Biosystems).
Glyphosate Resistance Assay in E. coli
Site-directed mutation was performed by PCR (Fisher and Pei, 1997) to generate a G101A mutation on OsEPSPS (confirmed by sequencing). Vectors containing coding sequence of mutant and wild-type OsEPSPS were transformed into E. coli DH5α; subsequent transformants were placed in a shaking incubator (200 rpm) overnight in Luria-Bertani broth at 37°C. The overnight culture was collected by centrifugation and washed twice by MOPS liquid medium. The bacteria were diluted to approximately OD600 0.4 with MOPS medium containing glyphosate. After another 12 h of shaking incubation at 37°C, the OD600 of the culture was measured again. The OD600 values were averaged from three repeats of the experiment. Glyphosate concentration ranged from 0 to 100 mm in liquid MOPS medium.
Enzyme Assay
Coding sequences of OsEPSPS of wild type and P106L mutant were reconstructed into pET-28a (Novagen) and expressed in strain BL21/DE3. Transformants were grown in 50 mL of Luria-Bertani broth containing 50 mg/L kanamycin to OD600 0.75, 1 mm isopropylthio-β-galactoside was added, and the bacteria were further incubated for 3 h at 37°C. Small aliquots of the final culture were analyzed by SDS-PAGE to determine the induced expression of EPSPS, as described.
Cell pellets of the culture were washed and suspended in extraction buffer (5 mL of 5 mm Tris-HCl, pH 7.8, 1 mm EDTA, 1 mm NaCl, and 1 mm dithiothreitol). After sonication lysis and centrifugation, supernatant of extraction buffer containing the crude extracted enzyme was added to glycerin to 40% (w/v) and stored at −20°C. This crude extraction was diluted to approximately 0.05 mg/mL protein with dilution buffer (20 mm HEPES/NaOH, 1 mm dithiothreitol, 5% [w/v] glycerin, pH 7.0) just before the assay.
Specific activity was determined in 100 μL reaction buffer of 50 mm HEPES/NaOH, 1 mm PEP, 1 mm S3P, and 5 μL of diluted crude enzyme extraction, pH 7.0. Reactions were started by adding the enzyme after all the reaction contents were kept at 28°C for 5 min; reaction velocity was deduced by measuring the production velocity of Pi after 2 min of reaction at 28°C as described (He et al., 2001). Enzyme activity was expressed as increased amount of Pi (μmol) (min reaction time)−1 (mg protein)−1 [U/mg]. Protein concentration was determined by the method of Bradford (Bradford, 1976) using bovine γ-globulin as a standard.
Kinetic characterization was performed in reaction buffer containing different concentrations of PEP or glyphosate. The data were fitted to appropriate equations to analyze the kinetic constants using program Igor Pro 4.00 (Wavemetrics). To obtain Km (PEP), data were fitted to V = Vmax [S]/(Km + [S]), in which V is the velocity of the reaction (expressed in U/mg), Vmax is the maximum velocity, and [S] is the concentration of the substrate assayed for the Km. Ki value was determined by the method as described (Copeland, 1996), S3P concentration was fixed at 1 mm, PEP concentration ranged from 50 μm to 200 μm, and glyphosate concentration ranged from 0 μm to 3 μm. IC50 was determined by fitting the data to V = Vmin + (Vmax − Vmin)/(1 + ([I]/IC50)s), where [I] is concentration of glyphosate, s is the slope of the curve at the IC50 (Eschenburg et al., 2002), and V was determined at 1 mm PEP and S3P with glyphosate concentration ranging from 0.0001 mm to 10 mm.
Construction of the RSO Model
Calculation was based on the kinetic constants and formulas described above. Overexpression data series were calculated at a fixed concentration of PEP within 1 to 100 μm and glyphosate within 0 to 100 μm. Data points were calculated at every 5 μm PEP concentration and 10 μm glyphosate concentration. With glyphosate concentration as x axis, PEP concentration as y axis, and overexpression ratio as z axis, 3-D surface section 1 and its corresponding contour section 2 were generated by Microsoft Excel 2000. Similarly, data points of glyphosate resistance were calculated at every 5 μm PEP concentration and 1-fold of overexpression ratio, and with overexpression ratio as x axis, PEP concentration as y axis, and glyphosate concentration as z axis, 3-D surface section 3 and its corresponding contour section 4 were generated by Microsoft Excel 2000.
Tobacco Transformation and Glyphosate Resistance Analysis of Transgenic Plants
Transformation vectors containing coding sequence of wild type and mutant OsEPSPS were constructed based on pCAMBIA-2300 (CAMBIA; Hajdukiewicz et al., 1994). The vectors were transferred into Agrobacterium tumefaciens LBA4404 by electroporation according to the instruction of the E. coli Pulser apparatus (Bio-Rad). The tobacco (Nicotiana tabacum cv Xanthi) was transformed by the ameliorated leaf discs method via the Agrobacterium-mediated transformation procedure described by Gao et al. (1998). Shoots were rooted on medium containing 50 mg/L kanamycin, then transferred into soil and grown in a greenhouse.
Sterile leaf discs of the transgenic tobacco from individual clones were grown on Murashige and Skoog (MS) medium containing 1.0 mg/L 6-benzyl adenine, 0.1 mg/L indole-3-acetic acid, and 0 to 4.0 mm glyphosate for 30 d with 16 h of light (100 μmol photons m−2 s−1) and 8 h of dark. Seedlings of T2 generations were germinated on half-strength MS medium containing 100 mg/L of kanamycin for 7 to 10 d, then green and active growing seedlings with two true leaves were selected and transplanted on half-strength MS medium containing different amounts of glyphosate and cultured for 30 d. After being transplanted into soil, the six- to eight-leaf-stage transgenic plants were sprayed with the herbicide Roundup (isopropylamine salt of glyphosate as active ingredient, 41.0%) at a dose equal to 2 L/ha. Injury was observed visually 2 weeks after the spray.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF413081.
Supplementary Material
Acknowledgments
We thank Mary Berlyn and the staff of the E. coli Genetic Stock Center for providing E. coli strain AB2829.
This work was supported by the China National 863 Program (grant nos. 2001AA212041 and 2004AA22180), the National Program on Key Basic Research Projects (grant no. 2004CB720406), and the Program Strategic Scientific Alliances and the Program for Strategic Scientific Alliances between China and The Netherlands (KNAW-PSA 04–PSA–BD–04 for Dr. P.B.F. Ouwerkerk [Leiden University], and KNAW-CEP 04CDP022 for Y. Xiao [KNAW-CEP]).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhen Zhu (zhuzhen@cashq.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068577.
References
- Amrhein N, Deus B, Gehrke P, Steinrucken HC (1980) The site of the inhibition of the shikimate pathway by glyphosate. II. Interference of glyphosate with chorismate formation in vivo and in vitro. Plant Physiol 66: 830–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrhein N, Johanning D, Schab J, Schulz A (1983) Biochemical basis for glyphosate-tolerance in a bacterium and a plant tissue culture. FEBS Lett 157: 191–196 [Google Scholar]
- Arnold FH, Wintrode PL, Miyazaki K, Gershenson A (2001) How enzymes adapt: lessons from directed evolution. Trends Biochem Sci 26: 100–106 [DOI] [PubMed] [Google Scholar]
- Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM (2002) Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Kishore G, Padgette SR, Taylor ML, Kolacz K, Weldon M, Re DB, Eichholtz DA, Fincher K, Hallas L (1992) Inhibitors of amino acid biosynthesis: strategies for imparting glyphosate tolerance to crop plants. In BK Singh, HE Flores, JC Shannon, eds, Biosynthesis and Molecular Regulation of Amino Acids in Plants. American Society of Plant Physiologists, Madison, WI, pp 139–145
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bradshaw L, Padgette SR, Kimball SL, Wells BH (1997) Perspectives on glyphosate resistance. Weed Technol 11: 189–198 [Google Scholar]
- Cadwell RC, Joyce GF (1994) Mutagenic PCR. PCR Methods Appl 3: S136–S140 [DOI] [PubMed] [Google Scholar]
- Chen LS, Lin Q, Nose A (2002) A comparative study on diurnal changes in metabolite levels in the leaves of three crassulacean acid metabolism (CAM) species, Ananas comosus, Kalanchoe daigremontiana and K. pinnata. J Exp Bot 53: 341–350 [DOI] [PubMed] [Google Scholar]
- Coggins JR, Abell C, Evans LB, Frederickson M, Robinson DA, Roszak AW, Lapthorn AP (2003) Experiences with the shikimate-pathway enzymes as targets for rational drug design. Biochem Soc Trans 31: 548–552 [DOI] [PubMed] [Google Scholar]
- Comai L, Sen LC, Stalker DM (1983) An altered aroA gene product confers resistance to the herbicide glyphosate. Science 221: 370–371 [DOI] [PubMed] [Google Scholar]
- Copeland RA (1996) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. VCH Publishers, New York
- Della-Cioppa G, Bauer SC, Klein BK, Shah DM, Fraley RT, Kishore GM (1986) Translocation of the precursor of 5-enolpyruvylshikimate 3-phosphate synthase into chloroplasts of higher plants in vitro. Proc Natl Acad Sci USA 83: 6873–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenburg S, Healy ML, Priestman MA, Lushington GH, Schonbrunn E (2002) How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 216: 129–135 [DOI] [PubMed] [Google Scholar]
- Fisher CL, Pei GK (1997) Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23: 570–571, 574 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon JE, Braymer HD (1990) Cloning of a gene from Pseudomonas sp. strain PG2982 conferring increased glyphosate resistance. Appl Environ Microbiol 56: 3382–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YF, Zhu Z, Xiao GF, Zhu Y, Wu Q, Li XH (1998) Isolation of soybean kunitz trypsin inhibitor gene and its application in plant insect-resistant genetic engineering. Acta Bot Sin 40: 405–411 [Google Scholar]
- Giesy JP, Dobson S, Solomon KR (2000) Ecotoxicological risk assessment for Roundup herbicide. Rev Environ Contam Toxicol 167: 35–120 [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Haslam E (1974) The Shikimate Pathway. John Wiley and Sons, New York
- He M, Nie YF, Xu P (2003) A T42M substitution in bacterial 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) generates enzymes with increased resistance to glyphosate. Biosci Biotechnol Biochem 67: 1405–1409 [DOI] [PubMed] [Google Scholar]
- He M, Yang ZY, Nie YF, Wang J, Xu P (2001) A new type of class I bacterial 5-enopyruvylshikimate-3-phosphate synthase mutants with enhanced tolerance to glyphosate. Biochim Biophys Acta 1568: 1–6 [DOI] [PubMed] [Google Scholar]
- Kishore G, Brundage L, Kolk K, Padgette SR, Rochester D, Huynh QK, Della-Cioppa G (1986) Isolation, purification, and characterization of a glyphosate-tolerant mutant E. coli EPSP synthase. Fed Proc 45: 1506 [Google Scholar]
- Knowles PF, Sprinson DB (1970) Preparation of shikimate 5-phosphate. Methods Enzymol 17A: 351–354 [Google Scholar]
- Kuchner O, Arnold FH (1997) Directed evolution of enzyme catalysts. Trends Biotechnol 15: 523–530 [DOI] [PubMed] [Google Scholar]
- Lederberg J, Lederberg EM (1952) Replica plating and indirect selection of bacterial mutants. J Bacteriol 63: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Bipatnath M, Xu Y, Chen S, Dennis P, Ehrenberg M, Bremer H (1999) Activities of constitutive promoters in Escherichia coli. J Mol Biol 292: 19–37 [DOI] [PubMed] [Google Scholar]
- Mayer MP (1995) A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163: 41–46 [DOI] [PubMed] [Google Scholar]
- Padgette SR, Re DB, Barry G, Eichholtz DA, Delannay X, Fuchs RL, Kishore G, Fraley RT (1996) New weed control opportunities: development of soybeans with a Roundup Ready gene. In SO Duke, ed, Herbicide Resistant Crops: Agricultural, Economic, Environmental, Regulatory, and Technological Aspects. CRC Press, Boca Raton, FL, pp 53–84
- Padgette SR, Re DB, Gasser CS, Eichholtz DA, Frazier RB, Hironaka CM, Levine EB, Shah DM, Fraley RT, Kishore GM (1991) Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J Biol Chem 266: 22364–22369 [PubMed] [Google Scholar]
- Pittard J, Wallace BJ (1966) Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol 91: 1494–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestman MA, Funke T, Singh IM, Crupper SS, Schonbrunn E (2005. b) 5-Enolpyruvylshikimate-3-phosphate synthase from Staphylococcus aureus is insensitive to glyphosate. FEBS Lett 579: 728–732 [DOI] [PubMed] [Google Scholar]
- Priestman MA, Healy ML, Becker A, Alberg DG, Bartlett PA, Lushington GH, Schonbrunn E (2005. a) Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail. Biochemistry 44: 3241–3248 [DOI] [PubMed] [Google Scholar]
- Russell DR, Bennett GN (1981) Characterization of the beta-lactamase promoter of pBR322. Nucleic Acids Res 9: 2517–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JN, Kabsch W (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci USA 98: 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt SS, Jensen JE, Fuerst EP (1995) Log-logistic analysis of herbicide dose-response relationships. Weed Technol 9: 218–227 [Google Scholar]
- Smith EA, Oehme FW (1992) The biological activity of glyphosate to plants and animals: a literature review. Vet Hum Toxicol 34: 531–543 [PubMed] [Google Scholar]
- Stalker DM, Hiatt WR, Comai L (1985) A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J Biol Chem 260: 4724–4728 [PubMed] [Google Scholar]
- Steinrucken HC, Amrhein N (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun 94: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Steinrucken HC, Schulz A, Amrhein N, Porter CA, Fraley RT (1986) Overproduction of 5-enolpyruvylshikimate-3-phosphate synthase in a glyphosate-tolerant Petunia hybrida cell line. Arch Biochem Biophys 244: 169–178 [DOI] [PubMed] [Google Scholar]
- Stemmer WP (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370: 389–391 [DOI] [PubMed] [Google Scholar]
- Weiss U, Mingioli ES (1956) Aromatic biosynthesis. XV. The isolation and identification of shikimic acid 5-phosphate. J Am Chem Soc 78: 2894–2898 [Google Scholar]
- Xu J, Feng D, Song G, Wei X, Chen L, Wu X, Li X, Zhu Z (2002) Cloning of genomic DNA of rice 5-enolpyruvylshikimate 3-phosphate synthase gene and chromosomal localization of the gene. Sci China C Life Sci 46: 561–569 [DOI] [PubMed] [Google Scholar]
- Yun RH, Anderson A, Hermans J (1991) Proline in alpha-helix: stability and conformation studied by dynamics simulation. Proteins 10: 219–228 [DOI] [PubMed] [Google Scholar]
- Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol 16: 258–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.