Abstract
Profilin is a small actin-binding protein that regulates cellular dynamics of the actin cytoskeleton. In Arabidopsis (Arabidopsis thaliana), five profilins were identified. The vegetative class profilins, PRF1, PRF2, and PRF3, are expressed in vegetative organs. The reproductive class profilins, PRF4 and PRF5, are mainly expressed in pollen. In this study, we examined the role of the first intron in the expression of the Arabidopsis profilin gene family using transgenic plants and a transient expression system. In transgenic plants, we examined PRF2 and PRF5, which represent vegetative and reproductive profilins. The expression of the PRF2 promoter fused with the β-glucuronidase (GUS) gene was observed in the vascular bundles, but transgenic plants carrying the PRF2 promoter-GUS with its first intron showed constitutive expression throughout the vegetative tissues. However, the first intron of PRF5 had little effect on the reporter gene expression pattern. Transgenic plants containing PRF5 promoter-GUS fusion with or without its first intron showed reproductive tissue-specific expression. To further investigate the different roles of the first two introns on gene expression, the first introns were exchanged between PRF2 and PRF5. The first intron of PRF5 had no apparent effect on the expression pattern of the PRF2 promoter. But, unlike the intron of PRF5, the first intron of PRF2 greatly affected the reproductive tissue-specific expression of the PRF5 promoter, confirming a different role for these introns. The results of a transient expression assay indicated that the first intron of PRF1 and PRF2 enhances gene expression, whereas PRF4 and PRF5 do not. These results suggest that the first introns of profilin genes are functionally distinctive and the first introns are required for the strong and constitutive gene expression of PRF1 and PRF2 in vegetative tissues.
Diverse actin-binding proteins regulate cellular dynamics of the actin cytoskeleton to perform various biological processes, such as locomotion, elongation, shape change, cytoplasmic streaming, division, and development of the cell (Bamburg et al., 1999; Kost et al., 1999; Meagher et al., 1999; Balŭska et al., 2001). Among these proteins, the small actin-binding protein, profilin (12–15 kD), plays an important role in actin dynamics (Gibbon and Staiger, 2000; Kovar et al., 2000; McKinney et al., 2001). In plants, profilin was originally identified in birch pollen as a potent allergen (Valenta et al., 1991), and it has been isolated and characterized in various plants, such as maize (Zea mays), tobacco (Nicotiana tabacum), bean (Phaseolus vulgaris), Arabidopsis (Arabidopsis thaliana), tomato (Lycopersicon esculentum), and castor bean (Ricinus communis; Staiger et al., 1993; Mittermann et al., 1995; Vidali et al., 1995; Christensen et al., 1996; Huang et al., 1996; Yu et al., 1998; Guillen et al., 1999; Schobert et al., 2000). Plant profilins have been shown to bind G-actin in vitro, and they are biochemically similar to nonplant profilins (Valenta et al., 1993; Guillen et al., 1999; Kovar et al., 2000). Arabidopsis profilin complemented both a budding yeast (Saccharomyces cerevisiae) profilin deletion mutant and a fission yeast (Schizosaccharomyces pombe) cdc3-124/profilin mutant, indicating that plant profilin is functionally equivalent with yeast profilin in vivo (Christensen et al., 1996). Like many other plant cytoskeletal genes, profilins exist as a multigene family and fall into either vegetative or reproductive classes, as shown in maize and Arabidopsis (Staiger et al., 1993; Christensen et al., 1996; Huang et al., 1996). In Arabidopsis, three vegetative and two reproductive profilins have been isolated and characterized. Profilins expressed in vegetative tissues are encoded by PRF1, PRF2, and PRF3. Both PRF4 and PRF5 encode reproductive class profilins that are preferentially expressed in pollen (Christensen et al., 1996; Huang et al., 1996). Analysis of transgenic plants harboring PRF1 and PRF2 promoter-β-glucuronidase (GUS) fusion constructs showed that these two profilins are mainly expressed in the vascular bundles of vegetative tissues (Christensen et al., 1996; Ramachandran et al., 2000). Recently, Kandasamy et al. (2002) investigated the spatial and developmental expression of profilins in Arabidopsis using vegetative and reproductive profilin-specific antibodies. They examined the root tip, sepal, and pistil using a monoclonal antibody that reacts strongly with PRF1 and PRF2, and weakly with PRF3. Based on the results of biochemical and structural localization analysis, the authors concluded that vegetative profilins are expressed in all vegetative organs and tissues. The discrepancy with previous work on profilin gene expression may reflect the presence of regulatory sequences in profilin genes. However, such regulatory elements have not yet been identified in profilin genes. Recently, it has been shown that some introns are involved in the regulation of spatial or temporal expression in potato (Solanum tuberosum) Sus3 (Fu et al., 1995), bean PsaD (Bolle et al., 1996), tobacco Ubi.U4 (Plesse et al., 2001), petunia (Petunia hybrida) PhADF1 (Mun et al., 2002), and Arabidopsis PAT1 (Rose and Last, 1997), AGAMOUS (Deyholos and Sieburth, 2000), and FLC (Sheldon et al., 2002). Particularly, in the case of genes that are known to express constitutively, such as PAT1, PhADF1, and Ubi.U4, introns were required for their strong and constitutive expression (Rose and Last, 1997; Plesse et al., 2001; Mun et al., 2002).
In this study, to identify the regulatory elements controlling profilin gene expression, we examined the effect of an intron of the profilin genes. Although genes encoding profilins are interrupted by two introns, we only tested the first intron, because introns that regulate gene expression are generally located near the 5′ region of the gene (Clancy and Hannah, 2002; Rose, 2002), and the first intron was about 4 times longer in length than the second intron. Our results show that the first intron of vegetative profilins (PRF1 and PRF2) plays an important role in strong and constitutive expression in vegetative tissues both in the transgenic and/or transient assay and enhances expression both at the mRNA and at the protein level. We also tested the first intron of reproductive profilins (PRF4 and PRF5) to determine whether a regulatory role of introns is conserved in reproductive profilin. In contrast with PRF1 and PRF2, the first intron of PRF4 and PRF5 did not alter expression patterns significantly. Furthermore, the first intron of PRF2 was able to alter spatial expression patterns of both PRF2 and PRF5 promoters.
RESULTS
The First Intron of PRF2 Is Required for Strong and Constitutive PRF2 Expression in Vegetative Tissues
Although introns are removed during the mRNA maturation process, some introns are known to enhance or regulate gene expression in various ways. To characterize the role of introns in vegetative profilin gene expression, we have cloned the PRF2 gene and generated two PRF2 promoter-GUS fusion constructs. The first construct, pPRF2-5′, includes the 1.6-kb promoter, 5′-untranslated region (UTR), and the start codon fused with the GUS gene in the pBI101 vector. The second construct, pPRF2e-p2i, contains the promoter, the entire first exon including 5′-UTR, the first intron, and 24 bp of the second exon (Fig. 1A). The short second exon was included for proper intron splicing. Three independent transgenic plants carrying single copies of the pPRF2-5′ or pPRF2e-p2i constructs were examined to analyze the quantitative effect of the first intron of PRF2 on gene expression.
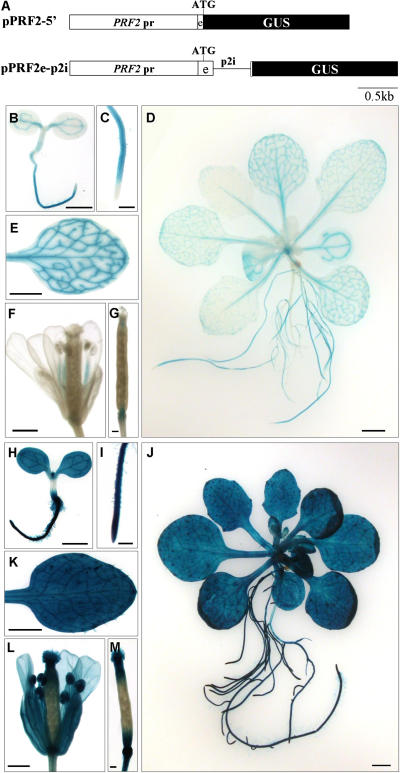
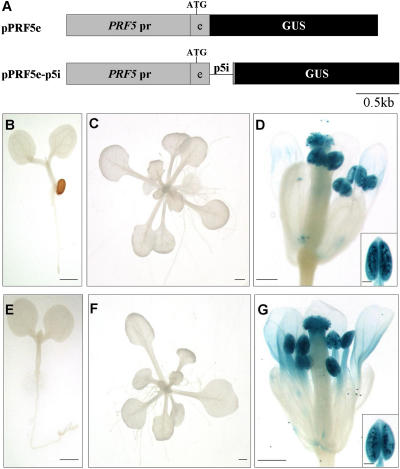
Figure 1.
Histochemical analysis of GUS expression in transgenic Arabidopsis containing pPRF2-5′ and pPRF2e-p2i constructs. A, Schematic representations of PRF2 promoter-GUS fusion constructs. The box with PRF2 pr is the promoter region of PRF2; the box with e is the first exon of PRF2; the black box with GUS is the GUS coding region; the line with p2i is the first intron of PRF2; ATG is the start codon. B to G, From pPRF2-5′; H to M, from pPRF2e-p2i; B and H, 3-d-old seedlings; C and I, roots of 3-d-old seedlings; D and J, 15-d-old plants; E and K, rosette leaves; F and L, flowers from 5-week-old plants; G and M, young siliques. GUS staining was performed for 12 h. Scale bars, 1 mm (B, D, E, H, J, and K), 0.2 mm (C, G, I, and M), and 0.5 mm (F and L).
We analyzed the expression of the PRF2 promoter-GUS fusion construct in various developmental stages of transgenic Arabidopsis. In 3-d-old seedlings containing pPRF2-5′, GUS staining was mainly observed in the vascular bundles of cotyledons. Some faint staining was detected near the vascular bundles, and there was no expression in the shoot apical meristem region (Fig. 1B). Most parts of the root showed GUS expression, except the root tip (Fig. 1C), and relatively strong staining was observed in the vascular bundles of the root (data not shown in detail). This preferential expression in vascular bundles was also maintained in 15-d-old plants. In this case, GUS expression was detected in the vascular bundles of rosette leaves and petioles (Fig. 1, D and E). Most of the roots were stained, but no GUS expression was observed in the root tips. The older expanded leaves showed stronger expression than smaller ones (Fig. 1D). In flowers, the filaments of stamens and the veins of sepals were stained. No GUS staining was observed in anthers, pollen, carpels, or petals (Fig. 1F). In young siliques, staining was detected in the receptacle (Fig. 1G). These expression patterns were largely in agreement with previous reports (Christensen et al., 1996). However, expression of the PRF2 promoter-GUS was completely changed by the inclusion of the first intron within the construct. In 3-d-old seedlings containing pPRF2e-p2i, strong GUS staining was detected throughout the plant. Most parts of the seedlings, including the root hairs, were deeply stained. Shoot apical meristems and root tips also showed GUS expression (Fig. 1, H and I). These strong and constitutive GUS expressions were observed also in 15-d-old plants. The deep blue staining was detected in the rosette leaves, petioles, roots, root hairs, and trichomes (Fig. 1, J and K). Almost every part of the floral organs, such as the petals, sepals, anthers, filaments, and stigmas, were stained (Fig. 1L). In young developing siliques, the stigma region and receptacle were stained (Fig. 1M). Although these results suggest a significant role of the intron in PRF2 gene expression, they cannot rule out the effect of coding regions on gene expression because pPRF2e-p2i has a full length of the first exon encoding 41 amino acids. Therefore, to examine the effect of the coding region, a pPRF2e construct that harbors the promoter region and the first exon fused with the GUS gene was generated, and the expression patterns of this construct were analyzed in 23 lines of T1 transgenic plants. Because of copy number variations of the transgene or positional effects, there were some variations, but most of the plants showed similar GUS expression patterns with pPRF2-5′ (data not shown), suggesting that the first exon of PRF2 has no significant effect on the gene expression pattern. Furthermore, in a transient assay, the entire open reading frame (ORF) in the pPRF2ORFGP construct showed no positive effect on GUS expression and the expression level of the pPRF2geGP that contains the entire PRF2 gene except the 3′-UTR was similar to that of the pPRF2e-p2iGP (Fig. 7B). These results support the fact that the sequences of PRF2 mRNA or amino acids fused to the GUS gene do not affect enhanced gene expression. Therefore, these data demonstrate that the first intron of PRF2 is responsible for the strong and constitutive expression of PRF2 throughout plant tissues.
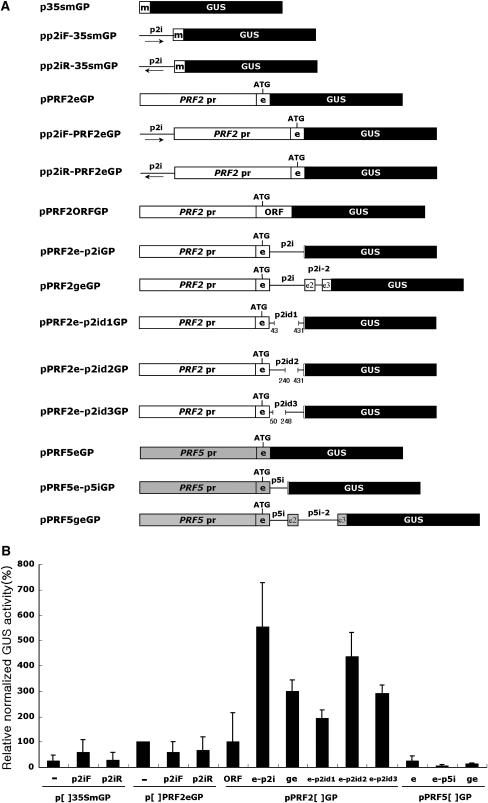
Figure 7.
Transient expression analysis of various PRF2 and PRF5 promoter-GUS fusion constructs. A, Schematic representation of construction for transient expression. GP at the end of the construct name indicates a vector for transient expression. The white box with m is a 35S minimal promoter; the black box with GUS is the GUS ORF; the thin line with p2i is the first intron of PRF2; the white box with PRF2 pr is the PRF2 promoter region; the white box with e is the first exon of PRF2; ATG is the start codon; ORF is the ORF of PRF2 cDNA; the white boxes with e2 and e3 are the second and third exons of PRF2; the line with p2i-2 is the second intron of PRF2; the gray box with PRF5 pr is the PRF5 promoter region; the gray boxes with e2 and e3 are the second and third exons of PRF5; the lines with p5i and p5i-2 are the first and second introns of PRF5. Arrows indicate the orientation of intron. The numbers in intron deletion constructs indicate the deletion points from the 5′ end of the intron. B, Relative normalized GUS activity in percentage. The full name of each construct is the addition of each insert name into the brackets of the vector name. For example, p[p2iF-]35SmGP is the pp2iF-35SmGP construct. Each transfection was performed at least three times. GUS activity was normalized with cotransfected luciferase activity and relative activity of pPRF2eGP was set to 100%. Bars are means ± sd.
The First Intron of PRF2 Increases Both mRNA and Protein Levels
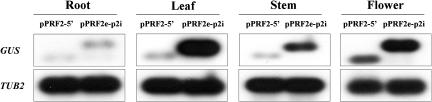
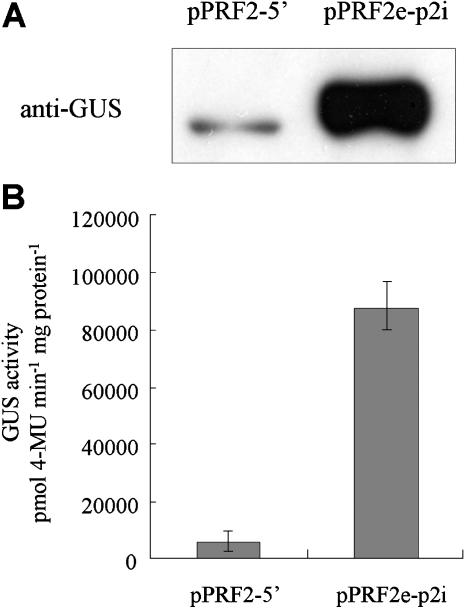
To investigate the basis for different expression of pPRF2-5′ and pPRF2e-p2i constructs, levels of mRNA and protein were analyzed. The accumulation of GUS transcripts was analyzed by reverse transcription (RT)-PCR analyses. One microgram of total RNA from roots, leaves, stems, and flowers was used for RT, and PCR was performed for 25 cycles to prevent saturation of PCR products. Sequencing of RT-PCR products of pPRF2e-p2i indicated that splicing occurred precisely (data not shown). In all tissues, the transcript level of transgenic plants carrying pPRF2e-p2i was increased when compared to pPRF2-5′ (Fig. 2), suggesting that different expression occurred at the transcription level. The increased mRNA levels were particularly apparent in the leaf and stem. To examine whether the increase in transcript level is fully reflected in the protein level, protein accumulation in the leaves was analyzed by western-blot analysis and GUS assay (Fig. 3). Twenty micrograms of total soluble proteins from the leaves of 4-week-old plants were used for immunoblot analysis with the GUS antibody. Similar to the transcript accumulation pattern, the levels of protein were much higher in pPRF2e-p2i than pPRF2-5′ plants. Results of fluorometric GUS assay indicated that pPRF2e-p2i gene expression was enhanced by about 15-fold relative to that of pPRF2-5′ (Fig. 3). Thus, the increase in transcript accumulation induced by the first intron was reflected in the GUS protein level. Therefore, strong expression of pPRF2e-p2i is correlated with increased accumulation of the GUS transcript. These results suggest that the role of the first intron of PRF2 is closely related to increasing steady-state mRNA levels.
Figure 2.
RT-PCR analysis of transcript expression of transgenic Arabidopsis containing pPRF2-5′ and pPRF2e-p2i constructs in various tissues. One microgram of total RNA from each organ was used for RT. TUBULIN2 (TUB2) amplified with TUB-F (5′-CTCAAGAGGTTCTCAGCAGTA-3′) and TUB-R (5′-CTCAAGAGGTTCTCAGCAGTT-3′) primers was used as a control. After electrophoresis of PCR products, it was transferred to the nylon membrane and hybridized with 32P-labeled GUS or TUB2 probes.
Figure 3.
Western-blot analysis and determination of GUS activity in transgenic Arabidopsis containing pPRF2-5′ and pPRF2e-p2i. A, Total soluble protein (20 μg) from mature leaves was loaded per lane and analyzed using monoclonal GUS antibody. B, GUS activity is expressed as pmol 4-methyl-umbelliferone min−1 mg−1 protein. Values are means ± sd (n = 3).
The First Intron of PRF5 Has Little Effect on PRF5 Gene Expression
We demonstrated the important role of the first intron in gene expression of PRF2, the vegetative profilin. To determine whether the role of first introns is conserved in a reproductive profilin, we examined the first intron of PRF5. About 1.5 kb of the promoter region of PRF5 and the first exon were translationally fused with the GUS reporter gene in a pPRF5e construct that tests promoter activity. To analyze the intron's role, a pPRF5e-p5i construct, which includes all parts of pPRF5e as well as the first intron of PRF5 with 24 bp of the second exon, was generated. The schematic structures of pPRF5e and pPRF5e-p5i are depicted in Figure 4A. Because we mainly focused on the effect of the intron on the PRF5 spatial expression pattern, we did not isolate the single-copy lines, and more than 10 lines of T1 plants harboring pPRF5e and pPRF5e-p5i constructs were examined for the analysis. GUS histochemical analysis of transgenic plants harboring pPRF5e showed strong expression in pollen. Staining was also observed in the stigmas and anthers. Very weak staining was detected in the upper part of filaments and petals (Table I; Fig. 4D). GUS expression was not detected in 3-d-old seedlings and 15-d-old plants in most lines that were examined (Fig. 4, B and C). Although it is not clear whether GUS staining in other floral organs is an actual expression or just diffusion from pollen, GUS expression of pPRF5e was predominant in pollen. A pollen-specific expression pattern was in agreement with a previous report (Christensen et al., 1996). pPRF5e-p5i, which includes its own first intron within the construct, showed almost identical expression with pPRF5e. GUS staining was not observed in the vegetative tissues of nearly all transgenic plants (Fig. 4, E and F). Like pPRF5e, GUS staining in flowers was mainly detected in the pollen and anthers (Table I; Fig. 4G). These results suggest that the first intron of PRF5 has no significant effect on the expression pattern of the PRF5 gene.
Figure 4.
Histochemical analysis of GUS expression in transgenic Arabidopsis containing pPRF5e and pPRF5e-p5i constructs. A, Schematic representations of PRF5 promoter-GUS fusion constructs. The gray box with PRF5 pr is the promoter region of PRF5; the gray box with e is the first exon of PRF5; the black box with GUS is the GUS coding region; the line with p5i is the first intron of PRF5; ATG is the start codon. B to D, From pPRF5e; E to G, from pPRF5e-p5i; B and E, 3-d-old seedlings; C and F, 15-d-old plants; D and G, flowers. Scale bars, 0.5 mm (B, D, E, and G), 1 mm (C and F), and 0.1 mm (anthers in small box).
Table I.
Summary of expression patterns of PRF constructs in reproductive organs
GUS staining intensity in various tissues was indicated by a minus (−), negative or weakly detectable; plus (+), light blue; ++, blue; +++, dark blue. The flower stalks were cut and incubated in GUS staining solution for 12 h. The ratio of the number of transgenic plants that show GUS staining to the total number of independent transgenic plants is indicated below the staining intensity.
| Construct | Stigma | Anther | Filament | Petal | Sepal | Cut Edge |
|---|---|---|---|---|---|---|
| pPRF2e | − | − | ++ | − | ++ | + |
| (0/23) | (0/23) | (21/23) | (0/23) | (16/23) | (16/23) | |
| pPRF2e-p5i | − | − | ++ | − | + | − |
| (0/23) | (0/23) | (14/23) | (0/23) | (7/23) | (2/23) | |
| pPRF5e | ++ | +++ | − | − | − | − |
| (12/12) | (12/12) | (2/12) | (1/12) | (0/12) | (0/12) | |
| pPRF5e-p5i | ++ | +++ | − | − | − | − |
| (10/13) | (13/13) | (1/13) | (1/13) | (0/13) | (0/13) | |
| pPRF5e-p2i | ++ | +++ | ++ | + | ++ | +++ |
| (15/24) | (21/24) | (19/24) | (8/24) | (15/24) | (23/24) |
The Role of Introns under the Control of Heterogeneous Promoters
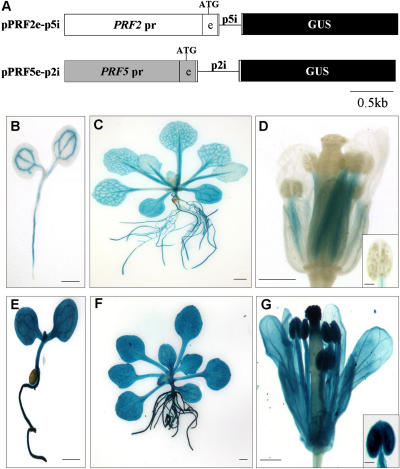
The analysis of transgenic plants carrying PRF2 and PRF5 promoter-GUS fusion constructs revealed that the first intron of PRF2 plays an important role in gene expression, while the first intron of PRF5 has little effect. However, it is unclear whether only the intron itself is responsible for these differences. Other factors, such as interaction between intron and promoter, could be involved in profilin gene expression. Thus, to investigate the roles of introns in detail, introns were exchanged between PRF2 and PRF5. For the construction of pPRF2e-p5i, the first intron of PRF5 was inserted at the 3′ end of the first exon in the pPRF2e construct. In the same way, the first intron of PRF2 was introduced into the pPRF5e construct, thus giving pPRF5e-p2i (Fig. 5A). For efficient splicing, both introns were accompanied with short flanking exons. The resulting constructs were transformed into Arabidopsis, and the expression pattern was analyzed in more than 20 independent T1 plants. In 3-d-old seedlings of pPRF2-p5i, expression was mainly observed in cotyledons and roots (Fig. 5B). This vascular bundle-specific expression was also observed in leaves, petioles, and roots of 15-d-old plants (Fig. 5C). In flowers, expression was observed in the filaments and sepals. No GUS staining was detected in pollen or in anthers (Table I; Fig. 5D). The overall expression patterns of pPRF2-p5i plants were very similar to those of pPRF2-5′ (Fig. 1, B–G). These data indicate that the first intron of PRF5 has no significant effect on the expression of the PRF2 promoter. However, unlike the first intron of PRF5, the first intron of PRF2 had significant influence on the expression of the PRF5 promoter. GUS histochemical analysis of pPRF5e-p2i showed strong expression throughout the plant. Deep blue staining was observed in nearly every part of the plant, including root hairs and tips of 3-d-old seedlings (Fig. 5E). The expression pattern was totally different from that of pPRF5e or pPRF5e-p5i (Table I; Fig. 4, B–G). This strong expression was also observed in 15-d-old plants (Fig. 5F). Most of the floral organs, including stigmas, anthers, pollen, filaments, petals, and sepals, also showed GUS expression (Table I; Fig. 5G). This constitutive expression pattern was quite similar to that of pPRF2e-p2i (Fig. 1, H–M). Thus, the properties of the two introns were maintained under heterogeneous promoters, and this confirmed the functional distinctiveness of the two introns. Particularly, the first intron of PRF2 had a significant effect on the expression pattern of both the PRF2 and PRF5 promoters, and this demonstrates that the first intron of PRF2 is responsible for the constitutive expression of pPRF2e-p2i and pPRF5e-p2i.
Figure 5.
Histochemical analysis of GUS expression in transgenic Arabidopsis containing pPRF2e-p5i and pPRF5e-p2i. A, Schematic representation of promoter-GUS fusion constructs. The white box with PRF2 pr is the promoter region of PRF2; the white box with e is the first exon of PRF2; the line with p2i is the first intron of PRF2; the gray box with PRF5 pr is the promoter region of PRF5; the gray box with e is the first exon of PRF5; the line with p5i is the first intron of PRF5; the black box with GUS is the GUS coding region; ATG is the start codon. B to D, From pPRF2e-p5i; E to G, from pPRF5e-p2i; B and E, 3-d-old seedlings; C and F, 15-d-old plants; D and G, flowers. Scale bars, 0.5 mm (B, D, E, and G), 1 mm (C and F), and 0.1 mm (anthers in small box).
The First Intron of PRF2 Alters the Spatial Expression Pattern of PRF Genes
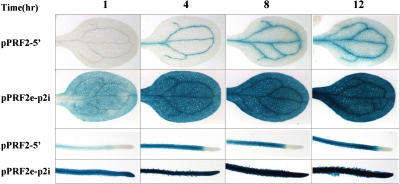
The expression of pPRF2-5′ was mainly observed in the vascular bundles of vegetative tissues. But faint GUS staining was detected in nonvascular tissues. In the pPRF2e-p2i plants, GUS staining is slightly stronger in the vascular tissues. In addition, increased expression induced by the intron was observed in the RT-PCR and western-blot analysis. Therefore, it is possible that strong expression of pPRF2e-p2i could be a result of a quantitative increase in the expression pattern of pPRF2-5′. However, the result with pPRF5-p2i suggested that the intron alters the spatial expression pattern of the PRF5 promoter. To determine the role of the intron in detail, GUS staining was performed in 3-d-old seedlings at various incubation times (Fig. 6). In pPRF2-5′, GUS staining was observed to occur weakly in the vascular bundles of cotyledons after 1-h staining. This vascular expression pattern was maintained and staining intensity was increased according to the staining time. After 12-h staining, weak staining was observed in the vascular bundles and nearby tissues. In pPRF2e-p2i plants, no vascular patterning was observed in the cotyledons after 1-h incubation. The overall parts of the cotyledons were evenly stained. This pattern was maintained after staining for 4, 8, and 12 h. In the roots, a different pattern was observed and maintained in both samples from the beginning. If the intron just increased expression, the early stage of the staining pattern of pPRF2e-p2i would be similar to that of pPRF2-5′. Therefore, these results suggest that the intron does not just increase or enhance gene expression, but instead is also involved in regulating spatial gene expression patterns.
Figure 6.
GUS staining patterns of transgenic Arabidopsis containing pPRF2-5′ and pPRF2e-p2i constructs at various incubation time points. Seedlings grown on Murashige and Skoog medium for 3 d were incubated for 1, 4, 8, and 12 h, respectively, and the reaction was stopped and the tissues cleared by incubation with 100% ethanol.
The First Intron of PRF2 Affects Gene Expression in a Position-Dependent Manner
The results above demonstrate that the first intron of PRF2 enhances gene expression and can induce ectopic expression under the control of a tissue-specific promoter. To determine whether controlling elements exist within the intron, three intron deletion constructs were generated (Fig. 7A). The resulting constructs were transiently expressed in Arabidopsis leaf mesophyll protoplasts, and normalized GUS activity with cotransfected luciferase activity was taken as a measure of promoter activity. Among the three intron deletions, the intron deletion d1 showed decreased GUS expression when compared to the full-length intron (Fig. 7B). The intron deletion analysis indicates that the enhancing elements would exist within the intron.
To examine the possibility that the first intron of PRF2 acts as an enhancer, the first intron of PRF2 was cloned upstream of the PRF2 promoter-GUS fusion construct in a forward or reverse direction (Fig. 7A). The resulting constructs were transiently expressed in Arabidopsis leaf mesophyll protoplasts. Although both pp2iF-PRF2eGP and pp2iR-PRF2eGP constructs have the intron upstream of the promoter region, their relative GUS activity was similar to that of the pPRF2eGP construct (Fig. 7B). This suggested that the first intron of PRF2 is not a classical enhancer. To further confirm this result, the intron was cloned upstream of the cauliflower mosaic virus 35S minimal promoter-GUS fusion construct (Fig. 7A). There was no detectable enhancer activity under the control of the minimal promoter (Fig. 7B). Therefore, these results demonstrate that the intron is not a classical transcriptional enhancer and it enhances gene expression in a position-dependent manner.
The Role of the First Intron in the Profilin Gene Family
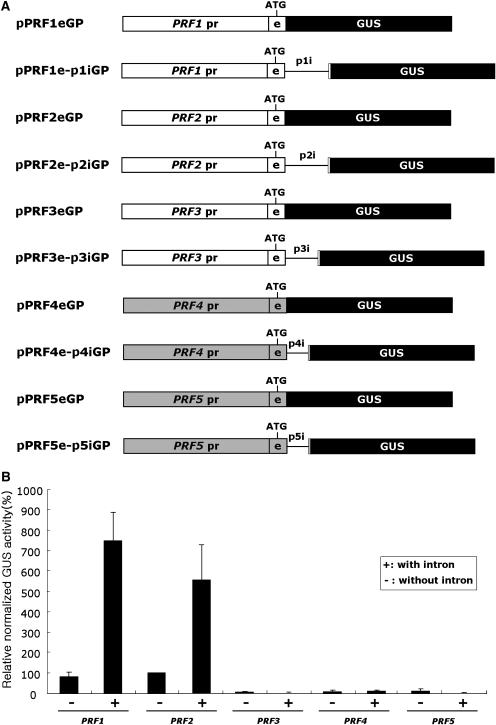
To examine whether the role of the first intron observed in PRF2 and PRF5 gene expression is also applicable to the entire Arabidopsis profilin gene family members, the promoter-GUS fusion constructs with or without their first introns were generated and their expression levels were compared in the Arabidopsis protoplast transient expression system (Fig. 8). The promoter-GUS fusion constructs of two vegetative profilins, PRF1 and PRF2, showed comparable changes in the GUS expression level depending on the presence of their own first intron. This suggests that the first introns of PRF1 and PRF2 have similar effects on gene expression. On the contrary, the promoter-GUS fusion constructs of two reproductive profilins, PRF4 and PRF5, showed a basal level GUS activity in leaf mesophyll protoplasts regardless of the presence or absence of their first intron. Because they are not expressed properly in vegetative tissues, it is uncertain whether their introns enhance gene expression or not. Thus, the role of the first introns of PRF4 and PRF5 is not clear in this system. Nonetheless, considering that the first intron of the PRF2 altered the expression pattern of the PRF5 promoter (Fig. 5, E–G), our results indicate that the first introns of reproductive profilin genes are functionally different from those of PRF1 and PRF2. Interestingly, although PRF3 encodes a vegetative-type profilin, the PRF3 promoter-GUS fusion construct was expressed at a basal level and a positive role of the first intron was not detected (Fig. 8B).
Figure 8.
Effect of the first intron on the expression of the Arabidopsis profilin gene family. A, Schematic representation of construction for transient expression. GP at the end of the construct name indicates a vector for transient expression. The white box is vegetative genes; the gray box is reproductive genes; the white or gray box with e is the first exon of each gene; the black box with GUS is GUS ORF; the thin lines with p1i, p2i, p3i, p4i, and p5i are the first introns of PRF1, PRF2, PRF3, PRF4 and PRF5, respectively; the white or gray box with gene name pr is the promoter region of each gene; ATG is the start codon. B, Relative normalized GUS activity in percentage. A plus (+) indicates the presence of an intron within the construct; a minus (−) indicates no intron. Each transfection was performed at least three times. GUS activity was normalized with cotransfected luciferase activity and relative activity of pPRF2eGP was set to 100%. Bars are means ± sd.
DISCUSSION
Profilin is a small actin-binding protein that regulates actin cytoskeleton dynamics and plays a role in cell elongation, cell shape, and flowering (Ramachandran et al., 2000). In Arabidopsis, there are five genes encoding three vegetative (PRF1, PRF2, and PRF3) and two reproductive (PRF4 and PRF5) profilins. All five members are very similar in amino acid sequences and genomic structures. All of these profilin genes are interrupted by two introns. The first intron of vegetative profilins is located between amino acids 41 and 42 (Gln and Leu) and that of reproductive profilins is located between amino acids 44 and 45 (Gln and Phe). Because PRF4 and PRF5 have three additional amino acids in the first exon, virtually all Arabidopsis profilin genes contain first introns at identical positions. In spite of conserved intron position, our results indicate that the roles of introns in gene expression are quite different between vegetative (PRF1 and PRF2) and reproductive (PRF4 and PRF5) profilin genes. GUS histochemical analyses of promoter-GUS fusions of PRF1 and PRF2 that encode major vegetative profilins indicate that their expressions are vascular bundle specific (Christensen et al., 1996; Ramachandran et al., 2000). These expression patterns were unexpected because vegetative profilin was assumed to show constitutive expression like vegetative actins. In our research, the expression of PRF2 promoter-GUS fusions was nearly identical with previous results. However, when the first intron was included in the construct, the expression pattern was greatly changed (Fig. 1). Strong and constitutive GUS staining was observed in most tissues and at diverse developmental stages. In the seedlings, expression was detected in the cotyledons, shoot apical meristems, roots, root tips, and root hairs. In 15-d-old plants, rosette leaves, roots, root hairs, and trichomes were stained. In flowers, stigmas, anthers, filaments, petals, and sepals showed GUS expression. Different expression patterns of promoter-GUS fusion constructs, with or without introns, were also evident in mRNA and protein accumulation. The presence of the intron resulted in a 15-fold increase in GUS activity in leaves (Figs. 2 and 3). This constitutive expression pattern of pPRF2-p2i is mostly in good agreement, except in ovules with in situ immunolocalization of vegetative class profilins (PRF1, PRF2, and PRF3) in Arabidopsis (Kandasamy et al., 2002). Therefore, it is evident that expression of PRF2 is not vascular bundle specific, and the first intron plays an important role in the strong and constitutive expression of PRF2.
To further investigate the role of profilin introns, we examined the first intron of the reproductive profilin PRF5. GUS staining with the pPRF5e construct was mainly detected in pollen and anthers. But, unlike the results obtained with the intron of PRF2, GUS expression patterns of the pPRF5e and pPRF5e-p5i constructs were nearly identical (Fig. 4; Table I), indicating that the first intron of PRF5 has little effect on the gene expression pattern. Although the spliceable intron was placed downstream of the first exon of PRF2 (pPRF2e-p5i), there was no significant alteration of expression pattern, confirming that the PRF5 intron had little effect on the gene expression pattern.
The result of intron exchange between PRF2 and PRF5 suggests that the first intron of PRF2 does not just enhance or increase the level of gene expression (Fig. 5). Both pPRF5e and pPRF5e-p5i showed reproductive tissue-specific expression. But, when the first intron was replaced with the intron of PRF2, strong GUS expression was observed throughout the plant body. This expression pattern of pPRF5e-p2i is quite surprising and demonstrates that the PRF2 intron completely changes tissue-specific expression of the PRF5 promoter. To determine whether this also occurred in the expression of PRF2, GUS staining patterns of plants harboring pPRF2-5′ and pPRF2e-p2i constructs were analyzed at various incubation times (Fig. 6). If the PRF2 intron just increased the expression level, the expression pattern of pPRF2e-p2i in the early stage of staining would be similar to that of pPRF2-5′. Plants of pPRF2e-p2i incubated for 1 h were stained evenly in the cotyledons and roots, and the staining was obviously different from pPRF2-5′ (Fig. 6). Therefore, these results suggest that the first intron of PRF2 is involved in regulating spatial gene expression.
Because the two first introns examined in this study maintained their own properties under the control of both the PRF2 and PRF5 promoters (Fig. 5), the different roles of these introns seem to be the properties of the introns themselves. Although sequences of introns are greatly variable in higher plants, AU richness is common in plant introns and is required for efficient splicing (Goodall and Filipowicz, 1989). Therefore, we analyzed the sequences of our two introns to determine whether there are significant differences in AU content. The AU content of the first intron was slightly lower in PRF2 (64.4%) than in PRF5 (67.7%), but both were similar to the average (67.5%) of Arabidopsis introns (Table II). Because AU content of both introns is above 60%, which is required for efficient splicing in dicot plants (Goodall and Filipowicz, 1989), it is likely that there is no significant difference in AU content. The most remarkable difference between two introns was in the length of them. The first introns of PRF2 and PRF5 are 478 and 260 bp in length, and they were 2.9 and 1.6 times longer than the average length of Arabidopsis introns (165.4 bp), respectively (Table II). Although the biological significance of long introns is not certain at present, long introns may serve as binding sites for various regulatory elements. The results of our intron deletion analysis suggest that the first intron of PRF2 may have the binding motifs or regulatory elements and these are to be specified in further research.
Table II.
Arabidopsis Genome Initiative (AGI) identification, sequence properties of introns, and number of ESTs of Arabidopsis profilin genes
The genomic distribution and structures of profilin genes were analyzed by NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/). AGI ID is the designation of the predicted genes that were given by the AGI. For example, the 1,976th annotated gene counting from the top of chromosome 2 is At2g19760. The number of ESTs was obtained from the NCBI Unigene (http://www.ncbi.nlm.nih.gov/UniGene) and TIGR (http://www.tigr.org) database.
| Gene
|
AGI ID
|
Length
|
AU Content
|
No. of ESTs
|
|||
|---|---|---|---|---|---|---|---|
| Intron I | Intron II | Intron I | Intron II | NCBI | TIGR | ||
| bp | % | ||||||
| PRF1 (PFN1) | At2g19760 | 506 | 92 | 63.2 | 67.4 | 22 | 23 |
| PRF2 (PFN2) | At4g29350 | 478 | 111 | 64.4 | 66.7 | 23 | 24 |
| PRF3 | At5g56600 | 367 | 66 | 68.7 | 69.7 | 5 | 6 |
| PRF4 (PFN3) | At4g29340 | 256 | 85 | 70.3 | 67.1 | 2 | 4 |
| PRF5 (PFN4) | At2g19770 | 260 | 575 | 67.7 | 70.2 | 0 | 2 |
Although the precise mechanism is not clear, the first intron of PRF2 increased the corresponding mRNA level, and this was reflected in the protein level as well, as shown in Figures 2 and 3. In intron-mediated enhancement, generally the enhancement is related to increased steady-state mRNA accumulation (Callis et al., 1987; Rethmeier et al., 1997; Rose, 2002). Furthermore, the first intron of PRF2 enhanced the gene expression in a position-dependent manner and could not induce the expression of the minimal promoter in a transient expression assay (Fig. 7). Therefore, it is supposed that the first intron of PRF2 increases gene expression by the cotranscriptional or posttranscriptional mechanisms as discussed in the earlier reports on intron-mediated enhancement (Jeon et al., 2000; Bourdon et al., 2001; Clancy and Hannah, 2002; Rose, 2002, 2004).
Among the five profilins of Arabidopsis, we mainly examined the role of the first introns on the expression of PRF2 and PRF5 in transgenic plants. Genes encoding vegetative (PRF1 and PRF2) and reproductive (PRF4 and PRF5) profilin are highly similar to each other in many aspects, such as DNA and amino acid sequences, expression patterns, and, especially, the length of the first intron (Table II). These high degrees of similarity seem to be closely related to the evolutionary history of the profilin gene family. In the Arabidopsis genome, genes encoding PRF1 and PRF5 are located in chromosome 2, and PRF2 and PRF4 are located in chromosome 4 where they are arranged in tandem arrays. Interestingly, both of the PRF1 and PRF2 genes encoding vegetative profilins are flanked with a gene encoding a reproductive profilin. Considering that about 60% of the Arabidopsis genome was duplicated (Arabidopsis Genome Initiative, 2000), this suggests that one set of tandem arrayed profilin genes was duplicated to give two sets. Therefore, we expected that the role of the first intron on gene expression would also be similar. As expected, only the first introns of PRF1 and PRF2 increased reporter activity, while the first introns of PRF4 and PRF5 did not. These results indicate that different regulatory mechanisms by introns control gene expression in vegetative (except PRF3) and reproductive profilins. Considering that PRF1 and PRF2 are major vegetative profilins, our data demonstrate that their first introns are required for their strong and constitutive expression in vegetative tissues. In this respect, the PRF3 gene is unique among profilin genes as shown in Table II. The PRF3 gene is not coupled with other profilin genes in the Arabidopsis genome, and the length of the first intron was smaller than other vegetative profilins, PRF1 or PRF2. The total number of PRF3 expressed sequence tags (ESTs) found in the National Center for Biotechnology Information (NCBI) and The Institute for Genomic Research (TIGR) databases was only 25% of PRF1 or PRF2, indicating that PRF3 is not expressed as much as PRF1 or PRF2. Our results indicating that PRF3 promoter-GUS fusion constructs showed basal levels of expression in the transient expression system also support the above explanation (Fig. 8). At present, we do not know whether weak expression of PRF3 is due to loss of intron-enhancing activity or significantly reduced promoter activity. Therefore, further examination of PRF3 gene expression would provide additional clues for understanding the importance of introns on expression of the profilin gene family.
Recently, the effect on gene expression of the leader intron in the Arabidopsis ACT1 gene encoding reproductive actin was reported. ACT1 is expressed most strongly in mature pollen, but it is also expressed in vegetative tissues, such as young vascular tissues and root tips. The leader intron was found to be required for the high-level expression of ACT1 in reproductive tissues. In addition, substituting its leader intron with that of ACT2 into an ACT1 promoter-GUS fusion resulted in failure of expression in pollen (Vitale et al., 2003). Although only two introns under the control of a reproductive actin promoter were tested, at least these results suggest that introns play a role in actin gene expression. Therefore, it is possible that a role of introns in gene expression may have coevolved in the cytoskeletal genes.
In this study, we have demonstrated that the first introns of PRF1 and PRF2 play an important role in constitutive expression in most vegetative tissues and are functionally different from those of PRF4 and PRF5. And we have discussed the possible importance of introns in the expression of profilin and actin genes. In future research, we will determine how the intron of the vegetative profilin genes affects gene expression and also examine the role of introns in the expression of various cytoskeletal genes.
MATERIALS AND METHODS
Plant Materials and Construction of Promoter-GUS Fusions for Transgenic Plants
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was grown in a growth chamber (23°C, 16 h day/8 h night, 300 μE m−2). For construction of PRF2 promoter fusions, the 1.6-kb promoter region, including up to a 5′-UTR and the start codon (pPRF2-5′), the entire exon 1 (pPRF2e), and the first intron (pPRF2e-p2i), was amplified using P2-F (5′-GTCGACAATGTTCCACCACCTAC-3′), P2-R (5′-GGATCCCATCTTTCTTCTTCTCC-3′), P2e-R (5′-AGGAAGAAAAACGGATCCCTGAGGGAAAG-3′), and P2i-R (5′-GGATCCTGCTATCTCTGCAGG-3′) primers, respectively. The PCR products were cloned into the pGEM-T easy vector (Promega) and confirmed by sequencing. The cloned fragments were digested with SalI/BamHI and ligated into the pBI101 vector (CLONTECH). Similar methods were used for construction of PRF5 promoter fusions, pPRF5e and pPRF5e-p5i, using PRF5-specific primers: P5-F (5′-AAGCTTCTGGTCCTGTATTTGCCTAACCAAG-3′), P5e-R (5′-GGATCCCTGAGGAAAATTAGCGCTCTGAG-3′), and P5i-R (5′-GGATCCTGTGATCTCTTGAGGTTTGAAC-3′). For intron exchange experiments, introns were amplified with P2int-F (5′-GGATCCGCTTTCCCTCAGGTTTTTCTTC-3′) and P2i-R for the first intron of PRF2 and with P5int-F (5′-GGATCCAATTTTCCTCAGGTATAATTAC-3′) and P5i-R for the intron of PRF5. Amplified introns were inserted into the BamHI site of pPRF2e or pPRF5e. The resulting constructs were inserted into Agrobacterium tumefaciens strain C58C1Rif+.
Plant Transformation and GUS Histochemical Analysis
Transgenic Arabidopsis was generated by the floral-dip method (Clough and Bent, 1998). Transformed plants were selected on 0.5× Murashige and Skoog 0.8% (w/v) agar plates containing 50 mg L−1 kanamycin and 100 mg L−1 cefotaxim. For analysis of pPRF2-5′ and pPRF2e-p2i lines, T1 transgenic plants were self-pollinated, and single-copy lines were selected by Southern-blot analysis and tested for the segregation ratio of T2 seeds on agar plates containing kanamycin. The selected single home lines were used for further analysis. Histochemical and fluorometric analysis of GUS activity was performed as described previously (Mun et al., 2002). Images were obtained using either Nikon (Tokyo) Coolpix 4500 or Stemi SV11 (Zeiss) with photomatrix Coolsnap cf digital camera (Photomatrix).
RT-PCR Analysis
To investigate the expression patterns of the GUS transcripts in transgenic Arabidopsis, RT-PCR analyses were performed. Total RNA was purified from leaves of mature plants by Tri-Reagent (Molecular Research Center). One microgram of total RNA was used for RT in a volume of 25 μL and incubated at 42°C for 1 h with avian myeloblastosis virus reverse transcriptase and oligo(dT) primer (Promega), and inactivated by incubating for 5 min at 95°C. After completion of RT, 2 μL of RT products were amplified using P2RT-F (5′-AAACAGTCTCATCTCGCCGGAGAAG-3′) and GUSRT-R (5′-AAAGACTTCGCGCTGATACCAGAC-3′) primers. PCR was performed using Biotherm DNA polymerase (Genecraft) in a GeneAmp PCR system 9700 (Perkin-Elmer). The PCR condition was 5 min at 95°C, followed by 25 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by 7 min at 72°C. The PCR products were run on agarose gel, transferred to Hybond XL membranes (Amersham), and hybridized. Probes were generated from GUS ORF from pBI101 and radiolabeled with [α-32P]dCTP using the Rediprime II random prime system (Amersham). After high stringent washing, blots were exposed to the film. To confirm that correct splicing had occurred, RT-PCR products were cloned into the pGEM-T easy vector and sequenced using the ABI 3100 sequence analyzer (Applied Biosystems).
Western-Blot Analysis
Monoclonal GUS antibody (Molecular Probes) was used at a titer of 1:5,000 for western-blot analysis. Twenty micrograms of total soluble proteins from leaves were loaded per lane. Western-blot analysis was performed as described previously (Mun et al., 2000).
Sequence Analysis of Arabidopsis Profilin Genes
The genomic sequences of five profilin genes in Arabidopsis were obtained from Plant Genome Central at the NCBI (http://www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html). The total numbers of ESTs cloned were obtained from Unigene (http://www.ncbi.nlm.nih.gov/UniGene/) and TIGR (http://www.tigr.org). The average length and AU content of the Arabidopsis intron was calculated based on whole intron sequences obtained from The Arabidopsis Information Resource (TAIR; ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/At_intron_20040301).
Transient Expression Analysis in Arabidopsis Leaf Mesophyll Protoplasts
For transient gene expression, the HindIII and EcoRI fragment from pBI101, which includes the GUS gene, was inserted into pUC19 and named as a pGP. The minimal promoter region of pBI121 was amplified with 35sm-F (5′-CTGCAGTCGACGCAAGACCCTTCCTCTATA-3′) and GUSRT-R primers and the PstI/BamHI fragment was ligated into pGP to generate p35smGP. For construction of pp2iF-35smGP and pp2iR-35smGP, the first intron of PRF2, which was amplified with P2int-sal-F (5′-GTCGACGCTTTCCCTCAGGTTTTTCTTC-3′) and P2int-sal-R (5′-GTCGACTGCTATCTCTGCAGGCTTCAA-3′) primers, was inserted into the SalI site of p35smGP. The SalI/BamHI fragments of PRF2 and PRF5 promoter-GUS fusion constructs in pBI101 were cloned into pGP (Fig. 8A). For construction of pp2iF-PRF2eGP and pp2iR-PRF2eGP, the first intron of PRF2 was inserted into the SalI site of pPRF2eGP. To make pP2ORFGP, the SalI/BamHI fragment of the pPRF2-5′ construct was ligated into pGP, thus generating pPRF2-5′GP, and the ORF of the PRF2 gene was amplified with P2orf-F (5′-GGATCCATGTCGTGGCAATCATACGTC-3′) and P2orf-R (5′-GGATCCGAGACCAGACTCGATAAGGTAATC-3′) primers using cDNA made from total RNA and inserted into the BamHI site of pPRF2-5′GP. The genomic region was amplified with P2-F and P2orf-R for pPRF2geGP and P5-F and P5orf-R (5′-GGATCCAAGACCCTGTTCGATCAAGTAATC-3′) for pPRF5geGP and ligated into pGP vector. For construction of intron deletion constructs, P2id1-R (5′-TTATTTGAGACAGAGGATCGGAAGAAAC-3′), P2id1-F (5′-AGTCGGTTGAAGCTAATTGCCTACTTTG-3′), p2id2-R (5′-ACACCTATAGACAAGATTTGACTACTG-3′), and p2id3-F (5′-TTTGCTTGGCACAGAATCTTATCTCC-3′) were used. Schematic representation of these transient vectors is depicted in Figure 7A. For construction of PRF1, PRF4, and PRF3 promoter-GUS fusions, genomic fragments were amplified with the following gene-specific primers: for PRF1, P1-F (5′-AAGCTTCATATACATACCAAAGCCATCATGGA-3′), P1e-R (5′-GGATCCCTGAGGAAATTTGGCGCTCTGAGC-3′), and P1i-R (5′-GGATCCATCGATTTCTTGAGGCTTCAAC-3′); for PRF3, P3-F (5′-AAGCTTCCCATAGAAAGCAATGTATATGCTC-3′), P3e-R (5′-GGATCCCTGAGGAAAATTGTTGCTCTGAGCC-3′), and P3i-R (5′-GGATCCCTGAATTTCCTCAGGCTTCACCTA-3′); and for PRF4, P4-F (5′-AAGCTTTTATCAGTCATACCATTTTTCTGGAC-3′), P4e-R (5′-GGATCCCTGAGGGAAGTTGGCGCTCTGAGCC-3′), and P4i-R (5′-GGATCCACTGAACTCCTGTCCCTTGAACTATATG-3′). They were inserted into the HindIII/BamHI sites of pGP (Fig. 8A). For construction of p35SLucP, the internal control for transient expression analysis, the 35S promoter region from pBI121 and the gene encoding a firefly luciferase amplified with Luc-F (5′-GGATCCATGGAAGACGCCAAAAACATAAAG-3′) and Luc-R (5′-GAGCTCTTACACGGCGATCTTTCCGCCCT-3′) primers using pGL3-promoter vector (Promega) were introduced into the HindIII/SacI sites of the pGP vector.
Arabidopsis mesophyll protoplasts from 3-week-old plants were transfected by the polyethylene glycol method (Sheen, 2002). Ten micrograms of plasmid DNA containing promoter-GUS fusion constructs were cotransfected with 10 μg of p35SLucP as an internal control. After transfection, protoplasts were incubated for 24 h at 24°C in the dark. Proteins were extracted by 1× cell culture lysis reagent buffer (Promega) and used for the GUS fluorometric and luciferase assay. Luciferase assay was performed according to the manufacturer's instructions (Promega) using the Victor2 multilabel microplate reader (Perkin-Elmer).
Acknowledgments
The authors wish to express sincere thanks to Prof. Donald J. Armstrong (Oregon State University) for helpful discussions and critical review. We would also like to thank Hyung-Seok Choi for help with analyzing sequences of the Arabidopsis whole intron.
This work was supported by the Ministry of Education and Human Resources Development (BK21 Research Fellowship to Y.-M.J.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sang-Gu Kim (kimsg@plaza.snu.ac.kr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071316.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Balŭska F, Jásik J, Edelmann HG, Salajová T, Volkmann D (2001) Latrunculin B induced plant dwarfism: plant cell elongation is F-actin dependent. Dev Biol 231: 113–124 [DOI] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S (1999) Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol 9: 364–370 [DOI] [PubMed] [Google Scholar]
- Bolle C, Herrmann RG, Oelmuller R (1996) Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J 10: 919–924 [DOI] [PubMed] [Google Scholar]
- Bourdon V, Harvey A, Lonsdale DM (2001) Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep 2: 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot D (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1: 1183–1200 [DOI] [PubMed] [Google Scholar]
- Christensen HEM, Ramachandran S, Tan CT, Surana U, Dong CH, Chua NH (1996) Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J 10: 269–279 [DOI] [PubMed] [Google Scholar]
- Clancy M, Hannah LC (2002) Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130: 918–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer element within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kim SY, Park WD (1995) A potato Sus3 sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell 7: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Staiger CJ (2000) Profilin. In CJ Staiger, F Baluška, D Volkmann, P Barlow, eds, Actin: A Dynamic Framework for Multiple Plant Cell Functions. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 45–65
- Goodall GJ, Filipowicz W (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 [DOI] [PubMed] [Google Scholar]
- Guillen G, Valdes-Lpoez V, Noguez R, Oliveares J, Rodriguez-Zapata LC, Perez H, Viali L, Villanueva MA, Sanchez F (1999) Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J 19: 497–508 [DOI] [PubMed] [Google Scholar]
- Huang S, McDowell JM, Weise MJ, Meagher RB (1996) The Arabidopsis profilin gene family: evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol 111: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G (2000) Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol 123: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2002) Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil Cytoskeleton 52: 22–32 [DOI] [PubMed] [Google Scholar]
- Kost B, Mathur J, Chua NH (1999) Cytoskeleton in plant development. Curr Opin Plant Biol 2: 462–467 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Drøbak BK, Staiger CJ (2000) Maize profilin isoforms are functionally distinct. Plant Cell 12: 583–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB (2001) Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development. Plant Cell 13: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Vitale AV (1999) The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet 15: 278–284 [DOI] [PubMed] [Google Scholar]
- Mittermann I, Swoboda I, Pierson E, Eller N, Kraft D, Valenta R, Heberle-Bors E (1995) Molecular cloning and characterization of profilin from tobacco (Nicotiana tabacum): increased profilin expression during pollen maturation. Plant Mol Biol 27: 137–146 [DOI] [PubMed] [Google Scholar]
- Mun JH, Lee SY, Yu HJ, Jeong YM, Shin MY, Kim H, Lee I, Kim SG (2002) Petunia actin depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene 292: 233–243 [DOI] [PubMed] [Google Scholar]
- Mun JH, Yu HJ, Lee HS, Kwon YM, Lee JS, Lee I, Kim SG (2000) Two closely related cDNAs encoding actin-depolymerizing factors of petunia are mainly expressed in vegetative tissues. Gene 257: 167–176 [DOI] [PubMed] [Google Scholar]
- Plesse B, Criqui MC, Durr A, Parmentier Y, Fleck J, Genschik P (2001) Effects of the polyubiquitin gene Ubi. U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol 45: 655–667 [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethmeier N, Kramer E, Van Montagu M, Cornelissen M (1997) Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J 12: 895–899 [DOI] [PubMed] [Google Scholar]
- Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8: 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40: 744–751 [DOI] [PubMed] [Google Scholar]
- Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11: 455–464 [DOI] [PubMed] [Google Scholar]
- Schobert C, Gottschalk M, Kovar DR, Staiger CJ, Yoo BC, Lucas WJ (2000) Characterization of Ricinus communis phloem profilin, RcPRO1. Plant Mol Biol 42: 719–730 [DOI] [PubMed] [Google Scholar]
- Sheen J (2002) A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/ (September 15, 2004)
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Goodbody KC, Hussey PJ, Valenta R, Drøbak BK, Lloyd CW (1993) The profilin multigene family of maize: differential expression of three isoforms. Plant J 4: 631–641 [DOI] [PubMed] [Google Scholar]
- Valenta R, Duchene M, Pettenburger K, Sillaber C, Valent P, Bettelheim P, Breitenbach M, Rumpold H, Kraft D, Scheiner O (1991) Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 253: 557–560 [DOI] [PubMed] [Google Scholar]
- Valenta R, Ferreira F, Grote M, Swoboda I, Vrtala S, Duchene M, Deviller P, Meagher RB, McKinney E, Heberle-Bors E, et al (1993) Identification of profilin as an actin-binding protein in higher plants. J Biol Chem 268: 22777–22781 [PubMed] [Google Scholar]
- Vidali L, Perez HE, Lopez VV, Noguez R, Zamudio F, Sanchez F (1995) Purification, characterization, and cDNA cloning of profilin from Phaseolus vulgaris. Plant Physiol 108: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Wu RJ, Cheng Z, Meagher RB (2003) Multiple conserved 5′ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Mol Biol 52: 1135–1151 [DOI] [PubMed] [Google Scholar]
- Yu LX, Nasrallah J, Valenta R, Parthasarathy MV (1998) Molecular cloning and mRNA localization of tomato pollen profilin. Plant Mol Biol 36: 699–707 [DOI] [PubMed] [Google Scholar]