Abstract
The classic role of SUCROSE NONFERMENTING-1 (Snf1)-like kinases in eukaryotes is to adapt metabolism to environmental conditions such as nutrition, energy, and stress. During pea (Pisum sativum) seed maturation, developmental programs of growing embryos are adjusted to changing physiological and metabolic conditions. To understand regulation of the switch from cell proliferation to differentiation, SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE (SnRK1) was antisense repressed in pea seeds. Transgenic seeds show maturation defects, reduced conversion of sucrose into storage products, lower globulin content, frequently altered cotyledon surface, shape, and symmetry, as well as occasional precocious germination. Gene expression analysis of embryos using macroarrays of 5,548 seed-specific genes revealed 183 differentially expressed genes in two clusters, either delayed down-regulated or delayed up-regulated during transition. Delayed down-regulated genes are related to mitotic activity, gibberellic acid/brassinosteroid synthesis, stress response, and Ca2+ signal transduction. This specifies a developmentally younger status and conditional stress. Higher gene expression related to respiration/gluconeogenesis/fermentation is consistent with a role of SnRK1 in repressing energy-consuming processes in maturing cotyledons under low oxygen/energy availability. Delayed up-regulated genes are mainly related to storage protein synthesis and stress tolerance. Most of the phenotype resembles abscisic acid (ABA) insensitivity and may be explained by reduced Abi-3 expression. This may cause a reduction in ABA functions and/or a disconnection between metabolic and ABA signals, suggesting that SnRK1 is a mediator of ABA functions during pea seed maturation. SnRK1 repression also impairs gene expression associated with differentiation, independent from ABA functions, like regulation and signaling of developmental events, chromatin reorganization, cell wall synthesis, biosynthetic activity of plastids, and regulated proteolysis.
Maturing seeds differentiate from meristem-like tissues into highly specialized storage organs. In legumes, this occurs sequentially, involving a decrease in mitotic activity, Suc uptake, cell expansion, and accumulation of storage products (Borisjuk et al., 1995). Maturation is evident by morphology, changes in gene expression, and metabolite profiles (Weber et al., 2005). In Arabidopsis (Arabidopsis thaliana) and barley (Hordeum vulgare) seeds, fundamental transcriptional reprogramming accompanies maturation, evident by up-regulated genes related to storage metabolism (Ruuska et al., 2002; Sreenivasulu et al., 2004). On the metabolite level, specific sugars correspond to development in fava bean (Vicia faba) embryos. Glc in nondifferentiated premature regions correlates with mitotic activity (Borisjuk et al., 1998), whereas high Suc is present in maturing, actively elongating, and starch-accumulating cells (Borisjuk et al., 2002). Local ATP concentrations (energy state) depend on differentiation grade. Low ATP levels are present before maturation, whereas the highest values are found in differentiated, storage-active regions (Borisjuk et al., 2003). Thus, maturing embryos adapt to specific environmental and nutrient conditions, which leads to adjustment of metabolic fluxes and energy state (Rolletschek et al., 2002; Borisjuk et al., 2003).
Controlling the switch from cell division to maturation during the transition phase is particularly interesting for crop seeds because of the initiation of storage metabolism. Work on legume and Arabidopsis seeds suggests regulation mediated by sugars and hormones and their interaction (Wobus and Weber, 1999; Gibson, 2004). Suc also has signaling functions and triggers storage-associated processes (Koch, 1996). Increasing concentrations in fava bean cotyledons at maturation are mediated by Suc transporter activity within abaxial epidermal transfer cells (Weber et al., 1997). Suc feeding disrupts the meristematic state, induces cell expansion and endopolyploidization in young cotyledons (Weber et al., 1996), promotes storage activity (Ambrose et al., 1987; Corke et al., 1990), and up-regulates enzymes of the Suc-to-starch pathway at the transcriptional level (Heim et al., 1993; Weber et al., 1998). Fava bean seeds (Weber et al., 1998) and potato (Solanum tuberosum) tubers (Trethewey et al., 1998) with decreased Suc levels have impaired storage metabolism.
The hormone abscisic acid (ABA) regulates a wide range of developmental events and mediates responses to stress. In seeds, ABA is necessary to proceed through maturation, acquire desiccation tolerance, and prevent precocious germination (Phillips et al., 1997; Finkelstein et al., 2002). ABA increases during early maturation and induces expression of a cyclin-dependent kinase inhibitor that down-regulates mitotic activity (Riou-Khamlichi et al., 2000). In Arabidopsis seed mutants, the loss of ABA synthesis (aba) or sensitivity (abi) strongly affects storage protein accumulation and desiccation tolerance. Sugar responses are linked to that of ABA. Several known mutants affected in ABA synthesis (aba1, aba2, aba3) and ABA sensitivity (abi3, abi4, abi5, abi8) are also sugar-sensing mutants, indicating that sugar signaling requires an intact ABA transduction chain (Finkelstein et al., 2002; Brocard-Gifford et al., 2003; Léon and Sheen, 2003; Gibson, 2004). It is possible that Suc increases ABA sensitivity or levels. During Arabidopsis germination, exogenous sugars can modulate internal ABA concentration, increasing its synthesis or inhibiting degradation (Price et al., 2003). Alternatively, ABA could enhance the ability to respond to sugars (Smeekens, 2000; Rook et al., 2001). Whereas this knowledge comes mainly from seedling analysis, little information is available for seeds on ABA and sugar interactions and the upstream-sensing and downstream-modulating events of the ABA/sugar signal. Protein phosphorylation is associated with cotyledon maturation in fava bean. Phosphoenolpyruvate carboxylase, which specifically channels carbon into amino acid biosynthesis via the anaplerotic pathway, is activated at maturation as seen by decreased sensitivity against malate inhibition (Golombek et al., 1999). The switch from hexose to Suc sugars during transition accompanies an increased energy state and decreased AMP levels (Borisjuk et al., 2003). Phosphorylation is possibly triggered by metabolic signals and/or energy state, and thus enzyme activities and metabolic fluxes are adapted to changing environmental and/or nutritional conditions.
SUCROSE NONFERMENTING-1 (Snf1)-RELATED PROTEIN KINASES (SnRK1s) are metabolic regulators in yeast, animals, and plants. In yeast, Snf1 is expressed in response to nutrient depletion and mediates usage of alternative substrates. It works in concert with histone acetylation to regulate transcription (Lo et al., 2005). In mammals, AMP-activated protein kinase (AMPK) is activated upon stress like heat, hypoxia, ATP depletion, or reactive oxygen and modulates phosphorylation of metabolic enzymes, leading to inactivation of ATP-consuming processes and energy preservation (Hardie, 1999; Halford and Paul, 2003).
Plants contain several isoforms of the three components common to the yeast and mammalian Snf1/AMPK heterotrimeric complex: the kinase α- or Snf1 subunit, the regulatory subunit γ or Snf4, and the β- or GAL83/SIP subunit with adapter function (Buitink et al., 2003). In tomato (Lycopersicon esculentum) seeds, the α-subunit is expressed constitutively, the β-subunit is induced during maturation, and the regulatory γ-subunit (LeSnf4) is responsive to GA, ABA, and stress (Bradford et al., 2003). In germinating Medicago seeds, β- and γ-subunits are up-regulated by starvation, whereas a direct effect of sugars has not been shown (Buitink et al., 2003).
Reduction of SnRK1 in transgenic plants mainly affects cleavage and use of Suc in potato (Purcell et al., 1998; Tiessen et al., 2003), whereas in barley it impairs pollen development, possibly due to the failure to incorporate Suc into starch (Zhang et al., 2001). SnRK1 has a range of other functions. In Physcomitrella, Snf1-like kinase is required for growing in normal day-night light cycles (Thelander et al., 2004), possibly by adaptation of the metabolism to changing nutrient/energy conditions. Repression of GAL83 (β-subunit) affects development of roots and tubers and sensitivity to salt stress (Lovas et al., 2003).
To understand the role of SnRK1 for the switch from cell proliferation to differentiation during transition stages, SnRK1 was repressed by an antisense approach in transgenic pea (Pisum sativum) seeds. This caused pleiotropic defects of maturation similar to an ABA-insensitive phenotype in Arabidopsis. Array-based gene expression analysis of transgenic embryos indicates a developmentally younger status and an inability to cope with stress and to adjust metabolism to environmental conditions. It is concluded that SnRK1s are regulators of embryo maturation in legumes and interact with ABA-dependent and -independent pathways.
RESULTS
Characterization of SnRK1 in Fava Bean and Pea Seeds
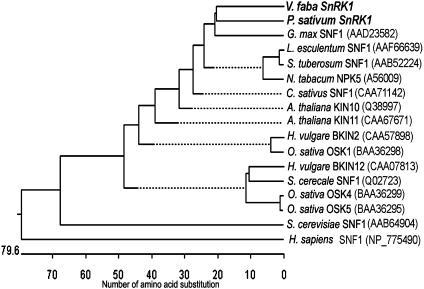
A 200-bp SnRK1 cDNA fragment was cloned from a cDNA library of fava bean cotyledons using reverse transcription-PCR. A library screen revealed 12 clones representing a single isoform, designated VfSnRK1 (α-subunit), with a calculated protein of 509 amino acids and a molecular mass of 57.9 kD. A corresponding full-length pea isoform (PsSnRK1) with 98.8% identity to the VfSnRK1 protein was found in the expressed sequence tag (EST) collection (see “Materials and Methods”). Homologies between α-subunit isoforms are shown in Figure 1.
Figure 1.
Phylogenetic tree of SnRK1 sequences from higher plants. Amino acid sequences were aligned according to GenBank, EMBL, and DDBJ accession numbers. The tree was constructed by the neighbor-joining method using ClustalW software (DNA-STAR).
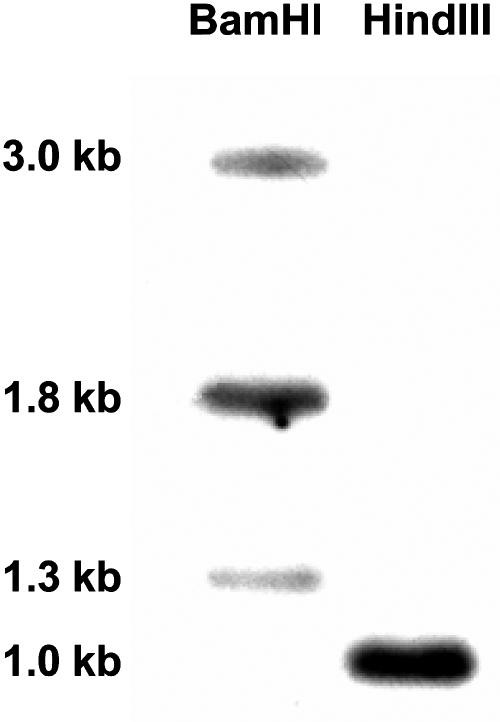
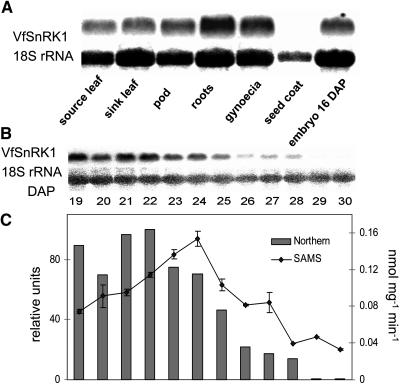
DNA gel-blot hybridization of fava bean at high stringency conditions after BamHI digestion yielded three bands, larger than expected from cDNA restriction, suggesting that VfSnRK1 contains introns. HindIII digestion revealed a single band. Because the cDNA does not have HindIII restrictions sites, additional sites may exist within introns (Fig. 2). The restriction pattern indicates that VfSnRK1 is present as a single- or low-copy gene. Tissue-specific RNA gel-blot analysis revealed expression in all tissues with decreasing transcript abundance in roots, gynoecia, embryos, pods, and sink and source leaves (Fig. 3A). In growing embryos, transcript levels were highest between 19 and 22 d after fertilization (DAF), corresponding to the transition stage. Levels continuously decreased from 23 to 30 DAF (Fig. 3B).
Figure 2.
DNA gel-blot analysis of VfSnRK1. Ten micrograms of total DNA were restricted, gel separated, and hybridized with a 200-bp probe of [32P]-labeled VfSnRK1.
Figure 3.
VfSnRK1 gene expression and kinase activity. A, RNA gel-blot analysis of tissue-specific mRNA levels of VfSnRK1. B, RNA gel-blot analysis of VfSnRK1 in growing fava bean embryos. C, Comparison of kinase activity and normalized signal intensity of SnRK1 in growing fava bean embryos as determined by the SAMS peptide kinase assay The line represents kinase activity calculated as nanomoles of phosphate incorporated per minute per milligram of plant material (±sd, n = 3). Bars represent the normalized signal intensity of mRNA levels.
VfSnRK1 kinase activity was measured in protein extracts from developing embryos using a SAMS peptide (HMRSAMSGLHLVKRR) kinase assay. Because the concentration of storage proteins strongly increased at later stages, kinase activity was calculated as nanomoles of phosphate incorporated per minute per milligram of plant material and not on a protein basis. The temporal profile of SAMS peptide kinase activity was similar to that of VfSnRK1 transcripts, but the peak of activity occurred 2 d later (Fig. 3C).
Transgenic Pea Seeds Expressing VfSnRK1 in Antisense Orientation
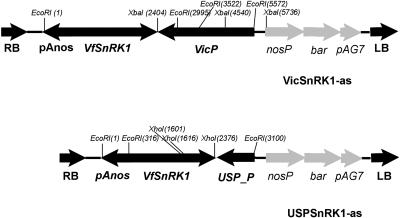
VfSnRK1 was fused in antisense orientation to the seed-specific vicilin and unknown seed protein (USP) promoters and cloned into the binary vector pGPTV-bar. Vic-SnRK1 antisense (pVicilin-SnRK1-as) and USP-SnRK1 antisense (pUSP-SnRK1-as; Fig. 4) were used for pea transformation. Nine independent transformants containing Vic-SnRK1-as and three transformants with USP-SnRK1-as were regenerated. To obtain stable lines, the plants were allowed to self-pollinate and PCR-positive plants were further propagated.
Figure 4.
Vectors used for pea transformation. Vector constructs were vicilin promoter/VfSnRK1/pGPTV-bar and USP promoter/VfSnRK1/pGPTV-bar for transformation of pea.
Approximately 30% of the antisense plants were partially or completely sterile. Because promoters of storage protein genes are active in pollen (Zakharov et al., 2004), strong inhibition of SnRK1 activity in pollen may cause abortion and subsequent sterility. Cytological analysis revealed high percentages of smaller and wrinkled pollen in these plants (data not shown). Similar effects of pollen abortion have been described in transgenic barley plants expressing the SnRK1 antisense construct under the control of the wheat (Triticum aestivum) HMG subunit promoter (Zhang et al., 2001). Nonfertile pea plants showed extensive branching, probably caused by an altered sink-source relationship due to the failure of seed development. Because we aimed to study the role of SnRK1 for seed development, transgenic lines were chosen without obvious reduction in fertility.
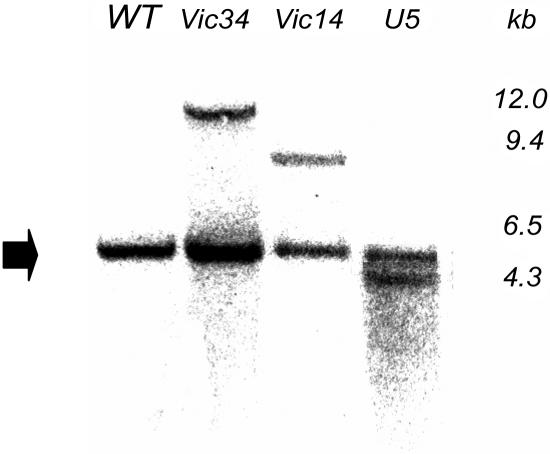
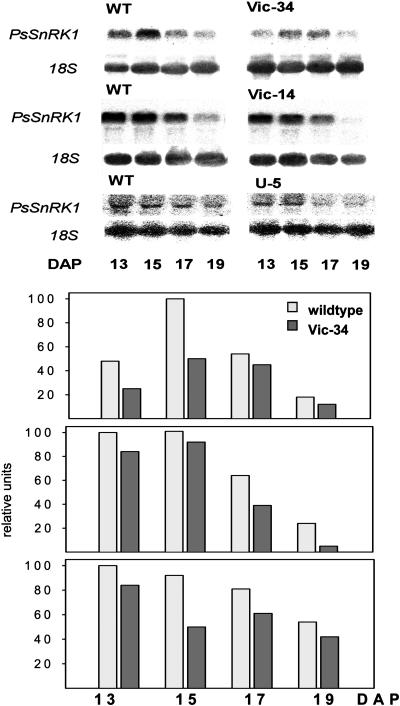
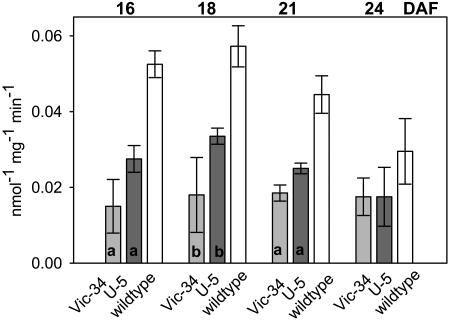
Seeds from three independent transgenic lines (Vic-34 and Vic-14 transformed with Vic-SnRK1-as, and U-5 transformed with USP-SnRK1-as) of the F4 to F6 generation were further analyzed. DNA gel-blot analysis revealed one band indicating single inserts in each line. The 5-kb hybridizing band corresponds to the endogenous PsSnRK1 (Fig. 5). The plants have no obvious growth phenotype. RNA gel-blot analysis revealed that PsSnRK1 mRNA levels were decreased in seeds during the transition stage of lines Vic 14, Vic-34, and U-5 (Fig. 6). SnRK1 activity was estimated using a SAMS peptide assay in protein extracts of embryos of lines Vic-34, U-5, and the wild type at 16, 18, 21, and 24 DAF. Activity was significantly reduced by approximately 50% to 70% at 16, 18, and 21 DAF, the period that corresponds to the transition stage. At later stages (24 DAF), activities were not significantly different (Fig. 7).
Figure 5.
Integration of transgenes in pea. DNA gel-blot analysis of transgenic pea lines Vic-34, Vic-14, and U-5. Ten micrograms of genomic pea DNA were digested with HindIII, gel separated, and hybridized using a [32P]-labeled 800-bp fragment of PsSnRK1. The 5-kb hybridizing band in each line represents the endogenous PsSnRK1.
Figure 6.
Transgene expression in growing pea embryos. RNA gel-blot analysis of growing embryos of the lines Vic-34, Vic-14, and U-5 and wild type. Signal intensities have been quantified after normalization using an 18S rDNA probe; Vic-34 (top); Vic-14 (middle); and U-5 (bottom).
Figure 7.
SnRK1 kinase activity in SnRK1 antisense and wild-type embryos. Activity was estimated using the SAMS peptide kinase assay in protein extracts of embryos between at 16, 18, 21, and 24 DAF. Bars are means (±sd), n = 3. Significant differences according to t test: P < 0.05 (a); P < 0.01 (b).
Taken together, the antisense expression of VfSnRK1 causes a decrease of 50% to 70% of both PsSnRK1 mRNA and SAMS peptide activity in embryos during the transition stage.
Seed Phenotype and Composition
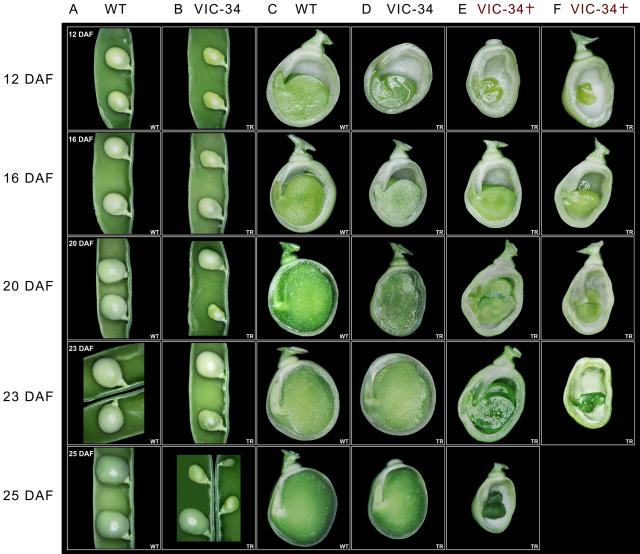
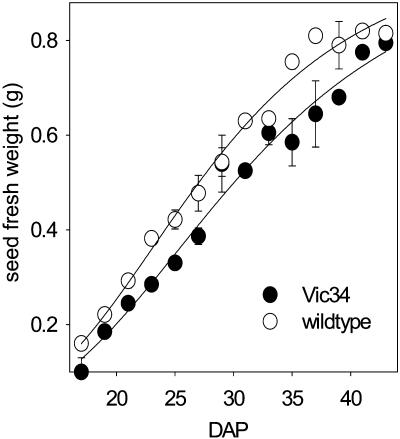
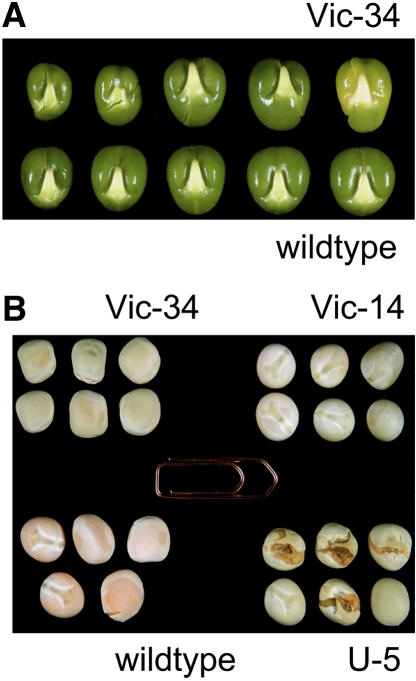
Detailed analysis of growing seeds was done for line Vic-34. Compared to the wild type (Fig. 8, A and C), the Vic-34 seeds (Fig. 8, B and D–F) are growth retarded and could be classified into two categories. A smaller fraction of seeds was more strongly growth inhibited, remained smaller, and frequently aborted before having filled the seed coat (Fig. 8, E and F). Abortion occurred at various stages from the transition stage onward. The fraction of aborted seeds was approximately 10%, but changed slightly from one generation to the other, although we grew the plants under constant conditions. Probably small differences of environment might play a role. The majority of seeds were fully developed and able to germinate (Fig. 8D). However, they were somewhat delayed in growth as shown by lower fresh-weight accumulation rates (Fig. 9). Transgenic seeds frequently showed alterations in cotyledon shape, surface, and symmetry (Figs. 8, D–F, and 10A). Dry, mature seeds of all three lines have a remarkable greenish phenotype instead of the yellow color of wild-type seeds (Fig. 10B). Occasionally, seeds germinate precociously, as shown, for example, in the U-5 line (Fig. 10B). After desiccation, the viviparous seeds are not able to germinate.
Figure 8.
Phenotype of wild-type SnRK1 antisense seeds and embryos. A, Wild-type pea seeds in pods. B, Corresponding SnRK1 antisense seeds. C, Cut-open seeds of the wild type. D, Corresponding Vic-34 SnRK1 antisense seeds. E and F, Cut-open transgenic seeds of Vic-34 embryos that do not develop enough sink strength and are going to abort. Notice that embryos do not fill the seed coat at 23 to 24 DAF.
Figure 9.
Seed growth analysis of SnRK1 antisense seeds. Fresh-weight accumulation curve of freshly harvested seeds of line Vic-34 and the wild type. Symbols are means (±sd), n = 3.
Figure 10.
Phenotype of SnRK1 antisense embryos and seeds. A, Cotyledons of line Vic-34 (top), compared to the wild type (bottom). Notice the altered symmetry of Vic-34 cotyledons. B, Phenotype of dry, mature seeds of lines Vic-34, Vic-14, U-5, and the wild type. Notice the greenish appearance and precocious germination of the U-5 seeds.
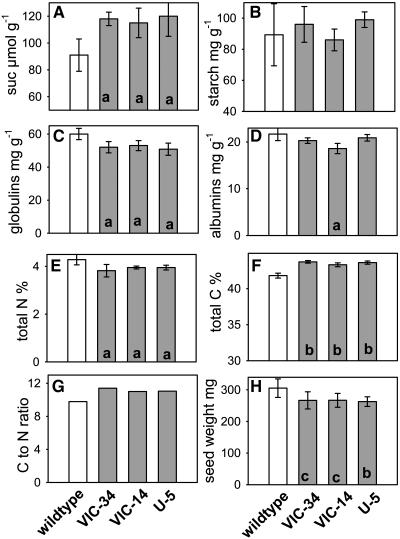
Suc and starch were analyzed in cotyledons of lines Vic-34, Vic-14, U-5, and the wild type during the early storage phase (21–24 DAF). Suc levels at that stage were clearly higher in all transgenic cotyledons (Fig. 11A), whereas starch was not different (Fig. 11B). The higher Suc-to-starch levels correspond to the observed growth retardation and indicate that conversion of Suc into storage compounds is affected. Seed storage compounds were then measured in mature, dry seeds. Starch levels were not different (data not shown). However, the globulin content was significantly lower by approximately 10% to 20% in seeds of all transgenic lines (Fig. 11C), whereas albumins were less affected and significantly lower only in Vic-14 seeds (Fig. 11D). Correspondingly lower total nitrogen was measured for all lines (Fig. 11E), whereas total carbon content was slightly, but significantly, increased (Fig. 11G). Thus, the SnRK1 antisense seeds have an increased carbon-to-nitrogen ratio (Fig. 11G). Individual seed weight was decreased by 10% to 20% in all three lines (Fig. 11H).
Figure 11.
Characterization of SnRK1 antisense transgenic pea embryos. Embryos from the lines Vic-34, Vic-14, U-5, and the wild type have been analyzed. A, Suc concentration in growing embryos at 21 to 24 DAF. B, Starch content in dry, mature embryos. C, Globulin content in dry, mature embryos. D, Albumin content in dry, mature embryos. E, Percentage of total nitrogen in dry, mature embryos. F, Percentage of total carbon in dry, mature embryos. G, Carbon-to-nitrogen ratio, calculated from E and F. H, Seed weight of dry, mature seeds. The data are presented as means ± sd of four to six individual embryos; n = 50 in H; seeds per line for G, n = 50. Significant differences according to t test: P < 0.05 (a); P < 0.01 (b); and P < 0.001 (c).
These results indicate that SnRK1 antisense seeds have general defects in maturation indicated by growth retardation, impaired globulin accumulation, lower final dry weight, and occasional vivipary.
Differences in Gene Expression Pattern between Wild-Type and Vic-34 Embryos
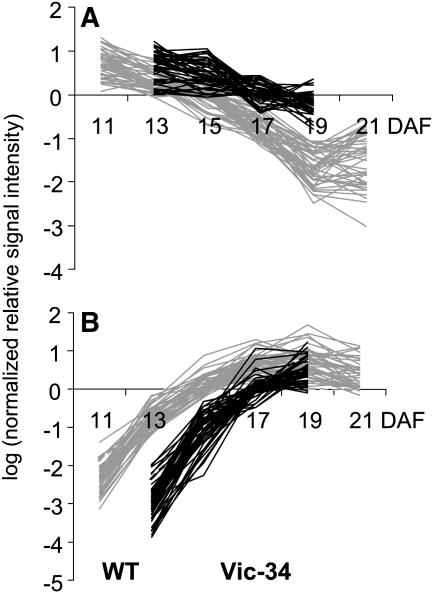
Differentially expressed sequences were arranged in two clusters. Cluster A gene transcripts are highly abundant in wild-type seeds during the prestorage phase (13–15 DAF) with a continuous decrease toward maturation. In Vic-34 embryos, this group showed higher mRNA levels between DAF 17 to 19 (Fig. 12A) and was designated as delayed down-regulated. Cluster A contains 67 genes with annotated and 36 with unknown function. Cluster B gene transcripts are low abundant in wild-type embryos at prestorage, with a continuous increase toward maturation (Fig. 12B). In Vic-34 embryos, this group showed lower values at 13 to 19 DAF and was designated as delayed up-regulated. Cluster B contains 59 genes with annotated and 22 genes with unknown function.
Figure 12.
Clusters of down- and up-regulated genes in SnRK1 antisense and wild-type embryos of pea. Temporal patterns of gene expression have been analyzed using macroarrays in growing wild-type embryos at 11, 13, 15, 17, 19, and 21 DAF (gray lines) and in SnRK1 antisense embryos, line Vic-34, at 13, 15, 17, and 19 DAF (black lines). A, Cluster A represent genes that are delayed down-regulated during the transition phase compared to wild-type seeds. B, Cluster B represents genes that are delayed up-regulated during the transition phase and compared to the wild type. Note logarithmic scale in y axis.
Delayed Down-Regulated Functions
Functional classification was based on homology (BLAST2) and literature searches. Therefore, all statements on gene identity and function have to be considered as putative. Transcript abundance does not necessarily reflect transcriptional activity, protein content, or enzyme activity. However, for simplicity reasons, higher or lower transcript levels were referred to as down- or up-regulated gene expression.
Delayed down-regulated ESTs from cluster A were divided into seven functional groups (Table I; Supplemental Fig. 1). The largest group (21 genes) is related to cell cycle and mitotic activity. Fourteen sequences encode histones H1, H1.41, H2A, H2B, H4, and H3C. mRNAs of core histones are synthesized in a cell cycle-dependent pattern at the beginning of the S-phase to allow nucleosome formation on duplicated DNA (Koning et al., 1991). Two ESTs represent annexins, two tubulins, and one actin-like protein. These proteins are associated with the microtubular system and frequently present during cell division of Medicago embryos (Gallardo et al., 2003). One EST represents a NAP/PIR-related protein with a function in actin polymerization (Li et al., 2004) and an ADP ribosylation factor-like protein, which controls vein patterning and vesicle trafficking and is related to Arabidopsis titan, controlling chromosome dynamics and cell division during seed development (McElver et al., 2000).
Table I.
Delayed down-regulated genes (cluster A)
| Group | Activity | Species |
|---|---|---|
| Group 1 | Cell Cycle and Mitotic Activity (21) | |
| PSS15F24 | Histone H1 | Pea |
| PSC35H05 | Histone H1.41 | Pea |
| PSS23F19, PSC24F23 | Histone H2A (2) | Chickpea |
| PSS06A20, PSC32K09 | Histone H2B (2) | Arabidopsis |
| PSS07N07 | Histone H3 | Pea |
| PSC32E09 PSC24G06, PSC24J03 PSC23H18, PSC26O10 PSS12A07 PSC26I07 | Histone H4 (7) | Triticum |
| PSS13P07 | Actin | Soybean |
| PSC33N04 | Tubulin β-chain | Pea |
| PSS13M16 | Tubulin α-chain | Pea |
| PSS05G23 | ADP ribosylation factor-like protein | Arabidopsis |
| PSC31D15, PSS20O14 | Annexin (2) | Medicago |
| PSS21O14 | NAP, PIR protein | Arabidopsis |
| Group 2 | Transcriptional and Translational Activity (12) | |
| PSC31I22 | 40S ribosomal protein S15 | Arabidopsis |
| PSC24P15 | 40S ribosomal protein S6 | Chickpea |
| PSS11N05 | 40S ribosomal protein S17 | Tomato |
| PSC26O24, PSS14C23 | 60S ribosomal protein L9 (2) | Pea, rice |
| PSS19C01 | 60S ribosomal protein L37e | Soybean |
| PSS21A14 | 60S ribosomal protein L4 | Arabidopsis |
| PSC30F01, PSC32G17 | 60S ribosomal protein L4-B (2) | Arabidopsis |
| PSS15B03 | Ribosomal protein L28-like | Rice |
| PSS20B07 | Poly(A)-binding protein | Cucumber |
| PSS21K24 | RNA-binding protein 45 | Tobacco |
| Group 3 | Primary and Secondary Metabolism (10) | |
| PSS06N03, PSS21L10, | Short-chain alcohol dehydrogenase (2) | Cowpea |
| PSC24G03 | ent-Kaurenoic acid oxidase | Pea |
| PSC22K07 | BR biosynthetic protein LKB | Pea |
| PSC29O14 | Isocitrate NADP-dehydrogenase, plastidial | Medicago |
| PSS08G11, PSC29E24 | Fru-bis-P aldolase (2) | Pea |
| PSS07G24 | UDP-Glc pyrophosphorylase | Pea |
| PSS15A16 | Aspartate aminotransferase | Soybean |
| PSS21I10 | Reversibly glycosylated polypeptide-2 | Arabidopsis |
| Group 4 | Regulatory and Signaling Functions (8) | |
| PSC32C05 | Calreticulin | Castor bean |
| PSS12F15 | Calmodulin | Medicago |
| PSC29E14 | Acyl-CoA-binding protein | Rape |
| PSC22A22 | Polygalacturonase inhibitor | Medicago |
| PSS11B24 | Protein kinase, SERK-like | Rice |
| PSS12F15 | HSP-70 | Arabidopsis |
| PSC33J14 | LEC-1 transcription factor | Arabidopsis |
| PSS20E21 | Peptidyl-prolyl cis-trans-isomerase | Fava bean |
| Group 5 | Mitochondrial Functions (6) | |
| PSS06I18 | Succinate dehydrogenase α-subunit | Arabidopsis |
| PSS18J18 | Cytochrome c oxidase | Arabidopsis |
| PSC26D12 | ADP/ATP carrier protein CANT1 | Lupin |
| PSC31H07 | Dicarboxylate/tricarboxylate carrier | Tobacco |
| PSC24H08 | F1-ATP synthase β-subunit | Arabidopsis |
| PSS05I05 | Mitochondrial matrix protein import related | Arabidopsis |
| Group 6 | Transport Related (6) | |
| PSC32K17 | Sugar transport protein | Arabidopsis |
| PSS16N05 | Aquaporin PIP | Medicago |
| PSC27B07 | Proton pump interactor | Arabidopsis |
| PSS21F16, PSS21E13 | Vacuolar ATP synthase catalytic subunit A (2) | Maize |
| PSS20N15 | Vacuolar H+-ATPase proteolipid | Arabidopsis |
| Group 7 | Photosynthesis (4) | |
| PSS21J10 | Plastocyanin | Pea |
| PSS22N10 | Chlorophyll a/b-binding protein | Pea |
| PSS12B15 | Transketolase | Capsicum |
| PSC31O09 | FAD | Clover |
The second group of 12 ESTs is related to transcriptional and translational activity with 10 ribosomal proteins of different classes, one poly(A)-binding, and another RNA-binding protein.
The third group represents 10 sequences related to primary and secondary metabolism. Four have possible roles in hormone synthesis: two ABA-2-like short-chain dehydrogenases involved in steroid hormone synthesis, which are Suc and GA inducible in watermelon (Citrullis vulgaris) seeds (Kim et al., 2003); one pea ent-kaurenoic acid oxidase (PsKAO2), a seed specifically expressed key enzyme of GA synthesis (Davidson et al., 2003); and one pea brassinosteroid (BR) biosynthetic protein LKB, which controls the 24-methylenecholesterol-to-campesterol step in BR biosynthesis (Nomura et al., 1999). Other sequences encode NADP-isocitrate dehydrogenase and UDP-Glc pyrophosphorylase, both of which are stress up-regulated. Two ESTs encode cytoplasmic Fru-bis-P aldolase, which is hypoxia up-regulated and related to gluconeogenesis (Konishi et al., 2004). Others encode Asp transaminase with a putative role in fermentation and reversibly glycosylated polypeptide, with UDP-Glc:protein transglucosylase activity involved in the synthesis of protein-bound α-glucans and in cell expansion.
The fourth group contains eight ESTs with different regulatory and signaling functions. Four are putatively stress induced. Three are associated with Ca2+-signaling pathways: calreticulin, calmodulin, and acyl-CoA-binding protein. The latter has Ca2+-binding and second messenger activity. Others encode polygalacturonase inhibitor, a cell wall protein belonging to the Leu-rich repeat family with functions in development and defense (Di Matteo et al., 2003), and Ser/Thr protein kinase, putatively GA induced with a role in morphogenesis/embryogenesis (Khan et al., 2005). One EST encodes a Lec1-like transcription factor, which in Arabidopsis determines embryogenic identity and in Medicago is auxin up-regulated and associated with embryogenesis and morphogenesis (Kwong et al., 2003). Other ESTs represent heat shock protein (HSP)-70 and heat shock-inducible chloroplast-targeted peptidyl-prolyl cis-trans-isomerase (Luan et al., 1994).
The fifth group of six sequences is related to mitochondrial functions. Three are associated with the mitochondrial respiration chain: succinate dehydrogenase (complex I), cytochrome c oxidase (complex IV), and ATP synthase β-subunit (complex V). Another sequence encodes a mitochondrial ADP/ATP carrier (CANT1), a mitochondrial dicarboxylate/tricarboxylate carrier, and an import-related protein associated with the mitochondrial matrix.
The sixth group contains six transport-related ESTs encoding a putative sugar transporter of unknown function and an aquaporin-like protein, which are GA and stress up-regulated (Jang et al., 2004). Four sequences are related to vacuolar H+ transport: two vacuolar ATPase catalytic subunits A, a vacuolar ATPase proteolipid, and an H+ pump interactor protein, which stimulates and binds ATPase (Morandini et al., 2002).
The seventh group of four sequences is associated with photosynthesis with plastocyanin, chlorophyll a/b-binding protein, transketolase, and Leu zipper fatty acid desaturase (FAD); the latter is light up-regulated in tobacco (Nicotiana tabacum) and required for chlorophyll synthesis (Liu et al., 2004).
Delayed Up-Regulated Functions
Delayed up-regulated genes due to SnRK1 repression are divided into eight functional groups (Table II; Supplemental Fig. 2). The largest group is related to storage activity and contains 19 ESTs. Seven encode 11S storage proteins, six vicilins, and one legumin. Another six encode seed storage or late embryogenesis-related proteins, four USPs, three lectins, one pea albumin, a Pro-rich, cold-inducible lipid transfer protein, and a secreted embryo-specific lipoprotein. One EST represents a biotin carboxyl carrier protein subunit involved in storage lipid synthesis. From the 19 genes, at least 15 are known to be ABA inducible and at least seven are related to drought and desiccation tolerance.
Table II.
Delayed up-regulated genes (cluster B)
| Group | Activity | Species |
|---|---|---|
| Group 1 | Storage Related (19) | |
| PSC21C18, PSC24I14, PSC26D22, PSC33I08, PSC21D24, PSC33J21 | Vicilin (6) | Fava bean, pea |
| PSC33E02, PSC34D04, PSC26F02, PSC24O16 | Embryo-abundant proteins, USP (4) | Fava bean |
| PSC24L17, PSC25H03, PSC25L10 | Lectins (3) | Pea, Arabidopsis |
| PSC22D16 | Legumin B | Fava bean |
| PSS05J22 | Pro-rich protein, cold-inducible protein | Medicago |
| PSC32I12 | Biotin carboxyl carrier | Soybean |
| PSC22H10 | Albumin 1 | Pea |
| PSC22D16 | Legumin B | Fava bean |
| PSC31K12 | Embryo-specific protein | Rice |
| Group 2 | Transcriptional and Translational Activity (11) | |
| PSC27P13 | 40S ribosomal protein S11 | Soybean |
| PSC31E12 | 60S ribosomal protein L30 | Lupin |
| PSC23H04 | 60S ribosomal protein L10A | Arabidopsis |
| PSC25L18 | Ribosomal protein L32 | Medicago |
| PSC30H24 | Histone H3A | Medicago |
| PSC25K01 | Translationally controlled tumor protein | Pea |
| PSC32E08 | DEAD-box RNA helicase | Pea |
| PSC29N17 | Methionyl tRNA synthetase | Arabidopsis |
| PSC27H08 | Lysyl tRNA synthetase | Tobacco |
| PSC27N04 | Heterogeneous nuclear ribonucleoprotein | Arabidopsis |
| PSC27J23 | Small nuclear ribonucleoprotein D2 | Arabidopsis |
| Group 3 | Stress Tolerance (8) | |
| PSC22H11, PSS09H10, PSS07H15 | Alcohol dehydrogenase-1 (3) | Pea |
| PSC23A09 | Mismatched repair protein | Maize |
| PSC31K03 | NIMA protein kinase | Poplar |
| PSC30P22 | Snakin-1, GA regulated | Potato |
| PSC26L10 | UDP-Glc:flavonol 3-O-glucosyltransferase | Tobacco |
| PSC23A24 | Mitochondrial pore protein | Arabidopsis |
| Group 4 | Regulatory and Signaling Functions (6) | |
| PSC30D03 | Abi-3 | Pea |
| PSC24B23 | Small G protein (ROP3) | Medicago |
| PSC22P01 | Pirin | Arabidopsis |
| PSC23G17 | Protein phosphatase 2C (Abi-1) | Arabidopsis |
| PSC32P08 | Transcription factor homeobox-Leu zipper family | Arabidopsis |
| PSC25E19 | Ago-1 | Arabidopsis |
| Group 5 | Primary and Secondary Metabolism (5) | |
| PSC30K22 | Ceramide glucosyltransferase | Gossypium |
| PSS07D09 | NAD-dependent malate dehydrogenase, cytosolic | Soybean |
| PSC30A07 | Phosphoribulokinase/uridine kinase | Rice |
| PSC25P16 | Acetohydroxy acid isomeroreductase | Pea |
| PSC31I09 | Riboflavin synthase α-chain | Arabidopsis |
| Group 6 | Cell Wall Synthesis and Metabolism (3) | |
| PSC30L06 | Cellulose synthase-4 | Rice |
| PSC27D17 | α-1,3 glycosyltransferase | Arabidopsis |
| PSC25P03 | Pectinesterase | Pea |
| Group 7 | Regulated Protein Degradation (3) | |
| PSC25B20 | 26S proteasome, regular subunit (RPN11) | Arabidopsis |
| PSC23E03 | 20S proteasome β-subunit D | Arabidopsis |
| PSC21P17 | Subtilisin protease | Chickpea |
| Group 8 | Plastidial Metabolism (3) | |
| PSC35H04 | Oxoglutarate/malate translocator (OMT) | Arabidopsis |
| PSC22H09 | EPSP2 | Tobacco |
| PSC31O08 | ADP/ATP translocator | Arabidopsis |
The second group consists of 11 sequences related to transcriptional and/or translational activity. Four encode 40S and 60S ribosomal proteins: two methionyl and lysyl tRNA synthetases, one a translationally controlled tumor protein involved in protein elongation, and one a small nuclear ribonucleoprotein involved in rRNA modification, ribosomal function, polyploidy, and chromosome rearrangement (Brown et al., 2003). One sequence encodes a pea DEAD-box RNA helicase, a multifunctional protein involved in protein synthesis (Pham and Tuteja, 2000), and histone H3A, which in yeast is the target of Snf1 phosphorylation, which enhances transcription of genes activated in response to cell-signaling events by changes in chromatin structure (Nowak and Corces, 2004).
The third group consists of eight ESTs related to stress tolerance. Three encode alcohol dehyrogenase-1, which in Arabidopsis is ABA inducible and related to drought tolerance (DeBruxelles et al., 1996). Another two are related to DNA repair and genotoxic stress tolerance, involving mismatched repair protein and noninherited maternal antigen (NIMA)-related protein kinase; the latter is activated by DNA-damaging agents (Noguchi et al., 2002). Other sequences encode antimicrobial Snakin-1-like peptide, which in potato is ABA inducible and involved in biotic stress tolerance (Berrocal-Lobo et al., 2002), and UDP-Glc:flavonol 3-O-glucosyltransferase, homologous to bronze-1 in maize (Zea mays), a key enzyme of anthocyanin biosynthesis, a substance with protective functions. One is similar to the mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family, which is important to cope with stress (Lister et al., 2004).
The fourth group of six sequences is related to regulatory and signaling functions. Four are directly involved in ABA signaling: Abi-3 transcription factor, which is a key regulator of seed maturation, confers ABA sensitivity and activates vicilin promoters (Zeng et al., 2003) and cellular differentiation in response to ABA and sugars (Rohde et al., 2002); small G protein (ROP3), which is ABA stimulated and accumulates at dormancy (Zheng et al., 2002); pirin-like protein interacting with CCAAT-box binding transcription factors (Abi-3, Lec-1) and G proteins, which is required for ABA-imposed delay of germination (Lapik and Kaufman, 2003); and protein phosphatase 2C (Abi-1), which is ABA and stress induced (Schweighofer et al., 2004). Other ESTs encode a transcription factor of the homeobox Leu zipper family, which is putatively inducible by BRs and has a role in vascular development (Baima et al., 2001), and argonaute 1 (ago-1)-like protein, with possible roles in specific RNA degradation, determination of organ polarity, and developmental timing (Bowman, 2004).
The fifth group encodes five ESTs related to primary and secondary metabolism: ceramide glucosyltransferase possibly involved in glycolipid synthesis, cytosolic NAD-malate dehydrogenase possibly involved in the production of malate from oxalacetate for amino acid production (Scheibe, 2004), and acetohydroxy acid isomeroreductase involved in Val biosynthesis. Others encode riboflavin synthase, which controls FAD and flavin mononucleotide synthesis, cofactors for enzymes of storage-lipid production and phosphoribulokinase/uridine kinase-like protein, which is light/stress/redox regulated and could regulate sugar flow through the pentose phosphate cycle (Chen et al., 2004).
A sixth group contains three ESTs involved in cell wall synthesis and modification, cellulose synthase, α-1,3 glycosyltransferase, and pectinesterase involved in cell wall weakening and elongation (Ren and Kermode, 2000).
The seventh group contains three sequences related to protein degradation, the 20S proteasome β-subunit, the 26S proteasome regulatory subunit, and the subtilisin-like protease. The latter is phosphate deficiency inducible and required for epidermal surface formation in Arabidopsis embryos (Tanaka et al., 2001).
The eighth group contains three ESTs involved in storage activity of plastids: an oxalacetate/malate translocator, with a possible role in import of carbon skeleton into plastids for amino acid synthesis; an ADP/ATP translocator, which is rate limiting for the import of ATP into plastids for biosynthesis; and 5-enolpyruvylshikimate-3-P synthase 2 (EPSP2 synthase), an enzyme of the shikimate pathway involved in defense-related aromatic biosynthesis (Forlani et al., 1994).
DISCUSSION
In plants, abiotic stress and/or environmental conditions regulate gene expression as well as development. Accordingly, internal signals in seed development are nutrient status, drought/osmotic conditions, oxygen, and/or energy. During maturation, the developmental program has to be adjusted to the changing physiological and metabolic status. Snf1-like kinases are ubiquitously present in eukaryotes. Their classic role is to adapt metabolism to changing environmental conditions, such as nutrition, energy, and stress (Hardie, 1999). We show here that in legume seeds SnRK1 kinase interacts with ABA signal transduction and is a key regulator controlling developmental programming associated with the switch from prestorage to maturation.
SnRK1 Repression Affects Seed Maturation
VfSnRK1 is expressed in various tissues, preferentially sink organs (Fig. 3). In growing embryos, highest expression as well as kinase activity occurred during the transition stage. To analyze SnRK1 functions in seed development, we used antisense expression in pea seeds, which reduced both SnRK1 mRNA and SAMS peptide kinase activity by 50% to 70%. Phenotypic analysis reveals seed abortion of 10% to 20% at various stages of early maturation. Obviously, these seeds cannot develop enough sink strength to grow sufficiently and therefore do not reach maturity. The majority of seeds, although growth retarded, reach maturity. Such a phenotype can arise when SnRK1 repression is near the threshold level of the minimal activity required to proceed through maturation. The fact that a 50% to 70% reduction of SnRK1 transcripts and activities caused partial seed abortion indicates that the residual SnRK1 activity is around a critical level, which represents a kind of threshold for initiation of seed maturation. A similar pleiotropic and heterogeneous phenotype has also been observed for seeds of the lh-2 mutant (ent-kaurene oxidase), which, although possibly different from the SnRK1 action, also affects seed sink strength (Swain et al., 1995). These results suggest that, if seed sink strength is below the critical threshold level, abortion will occur later on. The heterogeneous phenotype therefore may arise from slight variations of seed sink strength of individual seeds coming from microenvironments, location of pods at different nodes, and location of seeds within the pods.
Viable seeds clearly show maturation defects. Lower rates of fresh-weight accumulation and higher Suc levels indicate that utilization and/or conversion of Suc into storage products is affected. Seed composition analysis shows that storage defects concern globulin synthesis rather than starch. Lower mature seed weight, greenish appearance of seeds, and occasional precocious germination indicate that the seeds have problems reaching final maturation and full desiccation tolerance. Such a phenotype is reminiscent of Arabidopsis mutants with defects in ABA synthesis or sensitivity (Finkelstein et al., 2002) or tobacco seeds expressing anti-ABA antibodies (Phillips et al., 1997). More than 50% of SnRK1-repressed seeds show altered cotyledon surface, shape (Fig. 7), or symmetry (Fig. 9A), which indicates a control for SnRK1 in cotyledon growth and for coordination between cell division and expansion.
SnRK1 Repression Causes a Prolonged Prestorage Phase
Comparison of gene expression between wild-type and Vic-34 embryos reveals 183 differentially expressed genes in two clusters (Fig. 11). Cluster A genes are highly abundant during prestorage, decrease toward maturation, and are delayed down-regulated in Vic-34 embryos (Table I; Supplemental Fig. 1). Cluster B genes are stage specifically induced in wild-type embryos at maturation and are delayed up-regulated in Vic-34 embryos (Table II; Supplemental Fig. 2).
Gene expression related to GA and/or BR synthesis is increased. GAs work additively with BRs and are antagonistic to ABA. In young Arabidopsis seeds, the GA-to-ABA ratio can regulate developmental fate (Gazzarrini et al., 2004). GA is especially important for early seed growth of pea. The lh-2 mutation, which decreases GA levels in seeds, causes partial abortion (Swain et al., 1995), probably by defects in cell proliferation. Accordingly, genes involved in cell expansion are activated in Vic-34 (reversibly glycosylated polypeptide 2 and aquaporin); the latter is GA inducible (Jang et al., 2004). GAs also can stimulate mitotic activity (Fabian et al., 2000). From 68 delayed down-regulated genes with annotated functions, 21 are related to cell cycle/mitotic activity and 12 to transcriptional/translational activity (Table I). This indicates prolonged cell division and cell proliferation. We conclude that up-regulated gene expression related to GA/benzoic acid (BA) synthesis specifies a developmentally younger stage of Vic-34 embryos, which is consistent with increased gene expression of cell cycle genes. Altered balance between GA/BA and ABA could have also consequences for coordinated development and could explain changes in cotyledon shape and symmetry (Fig. 10A).
SnRK1 Repression Decouples Metabolic and Hormonal Signals
A switch from mitotic to cell expansion growth, during which sugar signals are involved, is characteristic for transition-stage embryos. Hexoses are correlated to mitotic activity, whereas Suc feeding disrupts the meristematic state and induces cell expansion (Weber et al., 1996). However, in Vic-34 embryos, increased cell cycle-related gene expression is associated with even higher Suc (Fig. 6), indicating that Suc alone is not sufficient to induce differentiation in embryos. In Arabidopsis, ABA is necessary to proceed through seed maturation and to acquire desiccation tolerance (Finkelstein et al., 2002), which are both affected in the Vic-34 embryos. ABA can inhibit cyclin-dependent kinases, which down-regulates mitotic activity in Arabidopsis seeds (Wang et al., 1998). Accordingly, ABA-insensitive seed mutants have prolonged cell division activities (Raz et al., 2001). We conclude therefore that in Vic-34 embryos repression of SnRK1 may cause either loss of ABA function and/or disconnection between metabolic signals and ABA, which could finally be the reason for prolonged expression of genes related to cell proliferation.
SnRK1 Repression Affects Metabolic Adaptation to Environmental Conditions
Vacuolar H+ transport (four genes) is up-regulated, which is an ATP-consuming process essential for vacuole enlargement, cell growth, and expansion (Shiratake et al., 1997). In rice (Oryza sativa) seedling leaves, GA3 and BA stimulate vacuolar H+ pumping (Yang et al., 2003), which is ABA inhibited in guard cells (Zhang et al., 2004). Higher gene expression related to respiration and mitochondrial assimilate transport indicates up-regulation of mitochondrial functions in Vic-34 embryos. Other up-regulated genes are related to responses to low oxygen, fermentation, and/or gluconeogenesis (Fru-bis-P aldolase, Asp-aminotransferase, mitochondrial dicarboxylate/tricarboxylate carrier, succinate dehydrogenase). Oxygen concentrations and energy state are low in young fava bean seeds. However, fermentation is only observed in young premature embryos, but disappears during transition and maturation (Rolletschek et al., 2002). We hypothesized that transition-stage embryos become adapted to oxygen/energy availability, as shown by decreasing respiration and increasing energy states (Borisjuk et al., 2003), a process that possibly involves SnRK1. The fact that gene expression related to H+ pumping remains high in Vic-34 embryos is consistent with SnRK1 kinase functions to repress these oxygen- and energy-consuming processes under low availability. In yeast, Snf1 kinases are required for the cellular response to stress conditions, such as nutrient limitation, salt stress, and heat shock (Sanz, 2003). Up-regulated stress genes (HSP-70, Fru-bis-P aldolase, peptidyl-prolyl cis-trans-isomerase) and that of Ca2+ signal transduction (calmodulin, calreticulin, acyl-CoA-binding protein) indicate that Vic-34 embryos cannot properly cope with stress conditions.
SnRK1 Repression Exhibits a Phenotype Similar to ABA Insensitivity
From 57 annotated and delayed up-regulated genes, 17 are directly related to protein storage and 10 to mRNA translation. This is consistent with decreased storage protein synthesis (Fig. 11) and indicates that its gene expression requires SnRK1. The majority of these storage protein genes are also up-regulated by ABA. Remarkably, expression of Abi-3 is strongly repressed in Vic-34. Abi-3 is a major regulator of seed maturation and Arabidopsis abi-3 mutants display an ABA-insensitive phenotype with severe maturation defects like failure to synthesize storage proteins, Suc accumulation, and loss of desiccation tolerance (Finkelstein et al., 2002). It is possible that, due to Abi-3 down-regulation, Vic-34 embryos display a similar ABA-insensitive phenotype. Interestingly, Lec-1, an upstream regulator of Abi-3 (Kagaya et al., 2005), is even up-regulated in Vic-34 embryos. The same is true for Fus-3 (data not shown). Possibly, SnRK1 directly activates Abi-3 on the transcriptional level but does not affect Lec-1/Fus-3. However, we cannot rule out that SnRK1 can also affect other signaling pathways. Three repressed genes have regulatory and signaling functions involved in ABA signal processing and amplification (small G protein, Abi-1, pirin) and are putatively ABA and/or stress inducible. The impaired ABA signal transduction could explain their repression in the Vic-34 embryos.
A larger number of repressed genes are related to stress tolerance, involving drought/desiccation (vicilins, USP, alcohol dehydrogenase), genotoxic stress (mismatched repair protein, NIMA-related kinase), and biotic stress (snakin-1). Other stress tolerance genes are UDP-Glc:flavonol 3-O-glucosyltransferase, a key enzyme of anthocyanin synthesis (Winkel-Shirley, 2002), and TIM17/22/23-like mitochondrial import translocase. Protein import is important for mitochondrial stress adaptation like respiratory inhibitors and drought (Lister et al., 2004). ABA mediates environmental stress signals and stimulates protein synthesis, which protects cellular structures and amplifies and processes the signal (Ingram and Bartels, 1996). Repression of storage protein genes and stress tolerance could therefore also be explained by the ABA-insensitive phenotype due to down-regulation of Abi-3. We conclude that SnRK1 is a modulator of ABA functions in seeds linking nutrient and/or energy state to ABA-regulated responses.
SnRK1 Repression Down-Regulates Maturation-Related Gene Expression Independent of ABA
From the histones especially, isoform H3A is repressed in Vic-34 embryos. Histone H3 is the target of Snf1 phosphorylation in yeast, which results in gene activation probably through chromatin modification (Lo et al., 2005). The latter is important, especially during phase transitions, and activates gene transcription in response to cell-signaling events (Nowak and Corces, 2004). This suggests that SnRK1 in legume seeds is involved in transcriptional programming during the transition phase through chromatin modification.
Cell wall synthesis and modification, a cellular function related to maturation, is repressed in Vic-34 embryos. Massive cell enlargement during legume seed maturation requires de novo synthesis and/or reconstruction of cell wall constituents. Another repressed function is related to the biosynthetic activity of plastids. Together with the effect on mitochondrial function, this shows that during embryo maturation a phase-dependent switch occurs from mitochondrial function to biosynthetic activities of plastids. In Vic-34 embryos, this process is affected, suggesting that SnRK1 could play a role.
Other repressed genes encode enzymes directly or indirectly involved in storage activity like amino acid biosynthesis (malate dehydrogenase, acetohydroxy acid isomeroreductase), glycolipid synthesis (ceramide glucosyltransferase), and aromatic biosynthesis (riboflavin synthase, EPSP2). Two genes have regulatory and signaling functions related to development: a transcription factor of the class III homeodomain Leu zipper family, which in Arabidopsis is an early marker of procambial cells and may promote vascular differentiation (Baima et al., 2001), and ago-1 specifying organ polarity and developmental timing (Bowman, 2004). Arabidopsis ago-1 mutants are embryo lethal (Lynn et al., 1999). Some alleles affect organ polarity in leaves with a loss of adaxial regions (Kidner and Martienssen, 2005). Such a role is consistent with the observed phenotype of changed cotyledon shape and symmetry in the Vic-34 embryos.
Three repressed genes are associated with regulated proteolysis (20S and 26S proteasomal subunits and subtilisin protease). Phase transitions in plant development involve altered patterns of protein expression. Regulated proteolysis by the proteasome complex is a key regulatory component for this process. In Arabidopsis, ubiquitin-specific proteolysis is essential for early embryo development (Doelling et al., 2001). Arabidopsis SnRKs interact with the 20S proteasome complex (Farras et al., 2001). This suggests a role of SnRK1 in the developmental events mediated by regulated proteolysis in transition-phase seeds.
MATERIALS AND METHODS
Plant Material
Pea (Pisum sativum cv Erbi) plants were grown in 2-L pots in growth chambers under a light-to-dark regime of 16 h light (20°C) and 8 h dark (18°C). Plants were fertilized once a week with nitrate and ammonium in order to keep nonlimiting nitrogen conditions. For the isolation of embryos, pods were tagged according to DAFs, collected in the middle of the light phase, and processed further. For biochemical analysis, seeds were harvested, and embryos were immediately isolated and snap frozen in liquid nitrogen.
Plant Transformation
The fava bean (Vicia faba) SnRK1 full-length cDNA was cloned under the control of the vicilin promoter or the USP promoter (Fiedler et al., 1993) into pGPTV-bar (Becker et al., 1992), yielding Vic-SnRK1-as (Fig. 4A) and USP-SnRK1-as (Fig. 4B). Pea transformation was performed after Schroeder et al. (1993), and modified as described by Giersberg et al. (2004). Because it is critical to compare datasets from different plant batches, comparisons between wild-type and antisense seeds have been performed only within a batch where plants have been grown at the same time together in the same growth chamber.
SAMS Peptide Kinase Activity Assay
Embryo material (100 mg) was ground in liquid nitrogen with 0.009 g Polyclar AT and suspended in 500 μL extraction buffer (0.25 m mannitol, 50 mm HEPES, 50 μm sodium fluoride, 1 mm EDTA, 1 mm EGTA, 1 mm benzamidine, 1 mm dithiothreitol [DTT], 0.1 mm phenylmethylsulfonyl fluoride [PMSF], pH 8.2). After centrifugation, ammonium sulfate was slowly added to the supernatant to 40% saturation while stirring for 20 min at 4°C. Precipitated protein was suspended in 50 μL fractionation buffer (50 mm Tris-HCl, 50 mm sodium fluoride, 1 mm EDTA, 1 mm EGTA, 1 mm benzamidine, 1 mm DTT, 0.1 mm PMSF, 0.02% [v/v] Brij-35, 10% [v/v] glycerol, pH 8.2) and concentration was determined according to the Bradford method. SAMS peptide kinase activity was performed as described by Davies et al. (1989). Protein extract (5 μL) was mixed with 5 μL kinase buffer (50 mm HEPES, 50 mm sodium fluoride, 1 mm DTT, pH 7.0), 5 μL sterile water, 5 μL SAMS peptide stock solution (peptide sequence HMRSAMSGLHLVKRR; 200 μm), and 5 μL labeled ATP stock solution (1 mm [γ-32P]ATP, 1 mm unlabeled ATP, 25 mm magnesium chloride). The samples were incubated for 30 min at 30°C and 10-μL aliquots were spotted onto 1 cm2 of phosphocellulose P81 paper. The pieces were washed twice in 1% (v/v) phosphoric acid for 4 min and once in acetone, dried, and transferred to scintillation counting. Activity was expressed as nanomoles of phosphate incorporated into peptide per minute and per milligram of plant tissue.
DNA and RNA Procedures and Gel-Blot Analysis
To obtain full-length clones of VfSnRK, the PCR-amplified DNA bands of approximately 200 bp were used to screen a λZAPII (Stratagene) cDNA library from fava bean cotyledons (Heim et al., 1993). Nucleic acids (DNA and RNA) were isolated and gel-blot analysis was performed as described by Heim et al. (1993). Full-length cDNA fragments were used as probes after labeling with [32P]dCTP.
EST Generation and Characterization, and Macroarray Hybridization
Two cDNA libraries were constructed from growing pea embryos and seed coats from four different stages covering transition to midmaturation stages using the pBluescript II XR cDNA library construction kit (Stratagene). Equal amounts of RNA from different stages were mixed and used for library construction. Average insert size of the libraries was estimated between 1,000 and 1,600 bp; 5,538 cDNA clones from embryo (PSC, Pisum sativum cotyledon) and 7,680 cDNAs from seed coat (PSS, Pisum sativum seed coat) libraries were sequenced from 3′ ends with an average length of 620 nucleotides. A total of 8,414 ESTs (4,958 from embryo and 3,756 ESTs from seed coat) were used for clustering using StackPack software (www.egenetics.com), resulting in 1,082 clusters (1,465 contigs) of 6,061 sequences and 2,353 singletons. Chimeric clones were removed manually. Contigs with a high number encode vicilins (136 ESTs), histone H3 (132 ESTs), vicilin precursors (119 ESTs), embryonic-abundant protein (101 ESTs), and chlorophyll a/b-binding proteins (97). The EST set was annotated using BLASTX2 (www.ncbi.nlm.nih.gov). The set of 8,414 ESTs was annotated with reference to gene function using BLASTX2 comparisons with the NRPEP protein database (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz). EST sequence information is available at http://pgrc.ipk-gatersleben.de/est/index.php. A unigene set of 4,548 clones was PCR amplified and spotted on nylon filters. Macroarrays were used to investigate mRNA abundances of wild-type and Vic-34 embryos at 11, 13, 15, 17, 19, and 21 DAF and 13, 15, 17, and 19 DAF, respectively, from the end of the prestorage to the midmaturation phase covering the transition stage.
Macroarray Data Evaluation
Array hybridization signals were detected by phosphor imager (Fuji BAS 2000; Fuji Photo Film) and analyzed using ArrayVision (Imaging Research), J-EXPRESS (Dysvik and Jonassen, 2001), as well as software for normalization of array data developed in Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK) and based on free software R (Ihaka and Gentleman, 1996; Bolstad et al., 2003). Hybridization experiments were performed twice with independent biological replicates. The ArrayVision signal was the basis for all subsequent analyses. Intensities of individual spots and of corresponding local backgrounds were determined. Local background was subtracted from spot intensities and the signal intensities of duplicated spots for each cDNA fragment were averaged. Experiment versus replica scatter plots was generated to confirm a linear distribution after normalization.
To allow comparison of signal intensities between the two experiments, the median of the logarithmically scaled (log2) intensity distribution for each experiment was set to zero (median centering of arrays; Eisen et al., 1998). The logarithmically scaled values for each gene were centered by its median across all experiments, which emphasizes differential expression regardless of absolute signal intensities. The log2-normalized signals were subjected to k-mean clustering analysis performed by J-EXPRESS software.
Because median centering does not yield information about signal intensity, we used the data after the first round of median centering of arrays to calculate nonlogarithmic, normalized signal intensities. To exclude fragments with signals close to background, the normalized nonlogarithmic signal intensities have to exceed three arbitrary units for at least one experiment.
The complete dataset was reduced to cDNA fragments with differential expression. To identify significant regulated genes, the signals showing 3-fold and higher differences in temporal expression profiles were selected. To group together genes with similar properties, cluster analysis was done using the program J-EXPRESS (Dysvik and Jonassen, 2001). The raw dataset of the two experiments and all EST sequence data along with additional information is available at http://pgrc.ipk-gatersleben.de/est/index.php (Kunne et al., 2005).
Extraction and Determination of Starch, Protein, Total Carbon, and Nitrogen
After ethanol extraction, the starch-containing insoluble material was solubilized in 1 n KOH for 1 h at 95°C and neutralized with 5 n HCl. Starch was hydrolyzed with amyloglucosidase and determined enzymatically. To determine albumin and globulin fractions of extractable proteins, powdered samples were extracted in acetate buffer (50 mm acetate, 1 mm KCl, 10% [v/v] DMSO, 0.5% [v/v] butanol, pH 4.5) and, subsequently, in phosphate buffer (100 mm KH2PO4, 100 mm Na2HPO4, 500 mm KCl, pH 7). Protein was measured with bovine serum albumin as standard. Relative content of total carbon and nitrogen in dried, powdered samples of cotyledons was measured using an elemental analyzer (Vario EL; Elementaranalysensysteme). Statistical analysis was done using Student's t test with Sigma Stat software (Jandel Scientific).
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers AJ971809 (VfSnRK1) and AJ971810 (PsSnRK1).
Supplementary Material
Acknowledgments
We are grateful to Katrin Blaschek, Elsa Fessel, and Angela Schwarz for excellent technical assistance. We thank Isolde Saalbach for help with pea transformation and Ulrich Wobus for discussions and continuous support. We also thank Uwe Scholz, Thomas Rutkowski, and Matthias Lange for support with bioinformatics; Lothar Altschmied for help in EST annotation, clustering analysis, and array preparation; and Ursula Tiemann and Karin Lipfert for help with preparation of the artwork.
This work was supported by the Deutsche Forschungsgemeinschaft and by Institut für Pflanzengenetik und Kulturpflanzenforschung (cDNA sequencing).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with journal policy described in the Instructions for Authors (http://www.plantphysiol.org) is: Hans Weber (weber@ipk-gatersleben.de).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071167.
References
- Ambrose MJ, Wang TL, Cook SK, Hedley CL (1987) An analysis of seed development in Pisum sativum L. IV. Cotyledon cell population in vitro and in vivo. J Exp Bot 38: 1909–1920 [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Segura A, Moreno M, Lopez G, Garcia-Olmedo F, Molina A (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H, Walenta S, Panitz P, Wobus U, Weber H (2003) Energy status and its control on embryogenesis of legumes: ATP distribution within Vicia faba embryos is developmentally regulated and correlated with photosynthetic capacity. Plant J 36: 318–329 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Walenta S, Rolletschek H, Mueller-Klieser W, Wobus U, Weber H (2002) Spatial analysis of plant development: sucrose imaging within Vicia faba cotyledons reveals specific developmental patterns. Plant J 29: 521–530 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Walenta S, Weber H, Mueller-Klieser W, Wobus U (1998) High resolution histographical mapping of glucose concentrations in developing cotyledons of V. faba in relation to mitotic activity and starch accumulation: glucose as a possible developmental trigger. Plant J 15: 583–591 [Google Scholar]
- Borisjuk L, Weber H, Panitz R, Manteuffel R, Wobus U (1995) Embryogenesis of Vicia faba: histodifferentiation in relation to starch and storage protein synthesis. J Plant Physiol 147: 203–218 [Google Scholar]
- Bowman JL (2004) Class III HD-Zip gene regulation, the golden fleece of ARGONAUTE activity? Bioessays 26: 938–942 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Downie AB, Gee OH, Alvarado V, Yang H, Dahal P (2003) Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Echeverria M, Qu LH (2003) Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci 8: 42–49 [DOI] [PubMed] [Google Scholar]
- Buitink J, Thoma M, Gissot L, Leprince O (2003) Starvation, osmotic stress and desiccation tolerance lead to expression of different genes of the regulatory γ and β subunits of the SnRK1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ 27: 55–67 [Google Scholar]
- Chen X, Yu T, Xiong J, Zhang Y, Hua Y, Li Y, Zhu Y (2004) Molecular cloning and expression analysis of rice phosphoribulokinase gene that is regulated by environmental stresses. Mol Biol Rep 31: 249–255 [DOI] [PubMed] [Google Scholar]
- Corke FMK, Hedley CL, Wang TL (1990) An analysis of seed development in Pisum sativum. XI. Cellular development and the position of storage protein in immature embryos grown in vivo and in vitro. Protoplasma 155: 127–135 [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG (1989) Tissue distribution of the AMP-activated protein kinase and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem 186: 123–128 [DOI] [PubMed] [Google Scholar]
- De Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R (1996) Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 111: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F (2003) The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100: 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD (2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I (2001) J-Express: exploring gene expression data using Java. Bioinformatics 17: 369–370 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian T, Lorbiecke R, Umeda M, Sauter M (2000) The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta 211: 376–383 [DOI] [PubMed] [Google Scholar]
- Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20: 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler U, Filistein R, Wobus U, Baumlein H (1993) A complex ensemble of cis-regulatory elements controls the expression of a Vicia faba non-storage seed protein gene. Plant Mol Biol 22: 669–679 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani G, Parisi B, Nielsen E (1994) 5-enol-Pyruvyl-shikimate-3-phosphate synthase from Zea mays cultured cells. Plant Physiol 105: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133: 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Golombek S, Heim U, Horstmann C, Wobus U, Weber H (1999) PEP-carboxylase in developing seeds of Vicia faba. Gene expression and metabolic regulation. Planta 208: 66–72 [DOI] [PubMed] [Google Scholar]
- Gibson SI (2004) Sugar and phytohormone response pathway: navigating a signalling network. J Exp Bot 55: 253–264 [DOI] [PubMed] [Google Scholar]
- Giersberg M, Saalbach I, Bäumlein H (2004) Gene farming in pea under field conditions. In R Fischer, S Schillberg, eds, Molecular Farming. Wiley-Interscience, New York, pp 1–8
- Halford NG, Paul MJ (2003) Carbon metabolite signalling. Plant Biotechnol J 1: 381–398 [DOI] [PubMed] [Google Scholar]
- Hardie DG (1999) Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp 64: 13–27 [PubMed] [Google Scholar]
- Heim U, Weber H, Bäumlein H, Wobus U (1993) A sucrose-synthase gene of V. faba L.: expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta 191: 394–401 [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman RR (1996) A language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Jang JY, Kim DG, Kim YO, Kim JS, Kang H (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol 54: 713–725 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Khan MMK, Yang S, Iwasaki Y, Fujisawa Y, Fukuda H, Komatsu S (2005) A gibberellin-regulated protein phosphorylated by a putative Ca2+-dependent protein kinase is G-protein mediated in rice root. Plant Cell Environ 28: 679–687 [Google Scholar]
- Kidner CA, Martienssen RA (2005) The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol 280: 504–517 [DOI] [PubMed] [Google Scholar]
- Kim J, Kang HG, Jun SH, Lee J, Yim J, An G (2003) CvADH1, a member of short-chain alcohol dehydrogenase family, is inducible by gibberellin and sucrose in developing watermelon seeds. Plant Cell Physiol 44: 85–92 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Koning AJ, Tanimoto EY, Kiehne K, Rost T, Comai L (1991) Cell-specific expression of plant histone H2A genes. Plant Cell 3: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Yamane H, Maeshima M, Komatsu S (2004) Characterization of fructose-bisphosphate aldolase regulated by gibberellin in roots of rice seedling. Plant Mol Biol 56: 839–848 [DOI] [PubMed] [Google Scholar]
- Kunne C, Lange M, Funke T, Miene H, Thiel T, Grosse I, Scholz U (2005) CR-EST: a resource for crop ESTs. Nucleic Acids Res 33: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15: 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon P, Sheen J (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Li Y, Sorefan K, Hemmann G, Bevan MW (2004) Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol 136: 3616–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Yang YT, Liu HH, Yang GD, Zhang NH, Zheng CC (2004) NTZIP antisense plants show reduced chlorophyll levels. Plant Physiol Biochem 42: 321–327 [DOI] [PubMed] [Google Scholar]
- Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL (2005) Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J 24: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas A, Sos-Hegedus A, Bimbo A, Banfalvi Z (2003) Functional diversity of potato SNF1-related kinases tested in Saccharomyces cerevisiae. Gene 321: 123–129 [DOI] [PubMed] [Google Scholar]
- Luan S, Lane WS, Schreiber SL (1994) pCyP B: a chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell 6: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D (2000) The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12: 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini P, Valera M, Albumi C, Bonza MC, Giacometti S, Ravera G, Murgia I, Soave C, De Michelis MI (2002) A novel interaction partner for the C-terminus of Arabidopsis thaliana plasma membrane H+-ATPase (AHA1 isoform): site and mechanism of action on H+-ATPase activity differ from those of 14-3-3 proteins. Plant J 31: 487–497 [DOI] [PubMed] [Google Scholar]
- Noguchi K, Fukazawa H, Murakami Y, Uehara Y (2002) Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses. J Biol Chem 277: 39655–39665 [DOI] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T (1999) Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol 119: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG (2004) Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet 20: 214–220 [DOI] [PubMed] [Google Scholar]
- Pham XH, Tuteja N (2000) Potent inhibition of DNA unwinding and ATPase activities of pea DNA helicase 45 by DNA-binding agents. Biochem Biophys Res Commun 294: 334–339 [DOI] [PubMed] [Google Scholar]
- Phillips J, Artsaenko O, Fiedler U, Horstmann C, Mock HP, Muntz K, Conrad U (1997) Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J 16: 4489–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P, Li TC, Kang SG, Na JK, Jang J-C (2003) Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132: 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford MG (1998) Antisense expression of a sucrose non-fermenting 1 related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Raz V, Bergervoet J, Koorneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Ren C, Kermode A (2000) An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol 124: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murra JA (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 13: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W (2002) PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14: 1885–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Koschorreck M, Wobus U, Weber H (2002) Legume embryos develop in a hypoxic environment. J Exp Bot 53: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapunctal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P (2003) Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem Soc Trans 31: 178–181 [DOI] [PubMed] [Google Scholar]
- Scheibe R (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Schroeder HE, Schotz AH, Wardley-Richardson T, Spencer D, Higgins TJV (1993) Transformation and regeneration of two cultivars of pea. Plant Physiol 101: 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Shiratake K, Kanayama Y, Maeshima M, Yamaki S (1997) Changes in H(+)-pumps and a tonoplast intrinsic protein of vacuolar membranes during the development of pear fruit. Plant Cell Physiol 38: 1039–1045 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Altschmied L, Radchuk V, Gubatz S, Wobus U, Weschke W (2004) Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J 37: 539–553 [DOI] [PubMed] [Google Scholar]
- Swain SM, Ross JJ, Reid JB, Kamiya Y (1995) Gibberellins and pea seed development. Expression of the lhi, ls and le5839 mutations. Planta 195: 426–433 [Google Scholar]
- Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, Machida Y (2001) A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128: 4681–4689 [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H (2004) Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J 23: 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibban R, Halford N, Geigenberger P (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating posttranslational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500 [DOI] [PubMed] [Google Scholar]
- Trethewey RN, Geigenberger P, Hajirezaei M, Sonnewald U, Stitt M, Riesmeier J, Willmitzer L (1998) Combined expression of glukokinase and invertase in potato tubers leads to a dramatic reduction in starch accumulation and a stimulation of glycolysis. Plant J 15: 109–118 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U (1997) A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell 9: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10: 823–834 [Google Scholar]
- Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Weber H, Heim U, Golombek S, Borisjuk L, Manteuffel R, Wobus U (1998) Expression of a yeast-derived invertase in developing cotyledons of Vicia narbonensis alters the carbohydrate state and affects storage functions. Plant J 16: 163–172 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H (1999) Seed maturation: genetic programmes and control signals. Curr Opin Plant Biol 2: 33–38 [DOI] [PubMed] [Google Scholar]
- Yang S, Maeshima M, Tanaka Y, Komatsu S (2003) Modulation of vacuolar H+ -pumps and aquaporin by phytohormones in rice seedling leaf sheaths. Biol Pharm Bull 26: 88–89 [DOI] [PubMed] [Google Scholar]
- Zakharov A, Giersberg M, Hosein F, Melzer M, Muntz K, Saalbach I (2004) Seed-specific promoters direct gene expression in non-seed tissue. J Exp Bot 55: 1463–1471 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Raimondi N, Kermode AR (2003) Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol Biol 51: 39–49 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang H, Takemiya A, Song CP, Kinoshita T, Shimazaki K (2004) Inhibition of blue light-dependent H+ pumping by abscisic acid through hydrogen peroxide-induced dephosphorylation of the plasma membrane H+-ATPase in guard cell protoplasts. Plant Physiol 136: 4150–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28: 431–442 [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.