Abstract
Sickle cell anemia is one of the most common genetic diseases worldwide. Patients often suffer from anemia, painful crises, infections, strokes, and cardiopulmonary complications. Although current management has improved the quality of life and survival of patients, cure can be achieved only with bone marrow transplantation when histocompatible donors are available. The ES cell technology suggests that a therapeutic cloning approach may be feasible for treatment of this disease. Using a transgenic/knockout sickle cell anemia mouse model, which harbors 240 kb of human DNA sequences containing the βS-globin gene, we prepared ES cells from blastocysts that had the sickle cells anemia genotype and carried out homologous recombination with DNA constructs that contained the βA-globin gene. We obtained ES cells in which the βS was corrected to the βA sequence. Hematopoietic cells differentiated from these ES cells produced both hemoglobin A and hemoglobin S. This approach can be applied to human ES cells to correct the sickle mutation as well as β-thalassemia mutations.
Keywords: correction of mutation, homologous recombination, sickle cell anemia, hematopoietic differentiation, β-thalassemia
Sickle cell anemia, one of the most common single-gene disorders worldwide, primarily affects people in Africa, the Mediterranean area, the Middle East, and the Indian subcontinent (1). In the United States, the gene frequency among African Americans is ≈0.08, and hence ≈1 in 600 births may be affected by sickle cell anemia (2). The severity of sickle cell anemia varies from a mild clinical course to severe anemia with frequent painful crises, infections, strokes, and cardiopulmonary and renal complications (3). Treatment consists of prevention of infection, blood transfusion for severe anemia and stroke, and treatment of complications. Although the search for antisickling agents is ongoing, no clinically effective agent has yet been found; compounds that increase fetal hemoglobin to inhibit sickling are also being sought (4, 5). The use of hydroxyurea has decreased the frequency of crises and hospitalization and increased the fetal hemoglobin level in some patients (6). At present, sickle cell anemia can be cured by bone marrow transplantation when there is a histocompatible donor (7). However, only a small fraction of patients in the United States have suitable donors (8). The use of cord blood stem cells for transplant has somewhat extended the donor pool (9). However, the management of a group of patients with severe disease is still limited to supportive treatments.
The introduction of ES cell technology suggests that a therapeutic cloning approach can be investigated for the treatment of genetic disorders such as sickle cell anemia. ES cells can be differentiated into hematopoietic cells for the treatment of blood disorders. Thus, Rideout et al. (10) demonstrated the possibility of ES cell therapy in a mouse model of immunodeficiency that was created by knockout of the Rag2 gene. They made ES cells by transferring the nuclei of skin cells cultured from the diseased mice into donor mouse oocytes. The missing Rag2 gene was reinserted by homologous recombination with a construct that contained the normal Rag2 gene. The ES cells were then differentiated into hematopoietic cells and transplanted back to the mouse to cure the immunodeficiency.
Theoretically, such an approach can be used to treat sickle cell anemia for those clinically severe patients who do not have histocompatible donors for transplantation. Skin or other nucleated cells can be cultured from patients, and the nuclei can be transferred to oocytes from donors to make ES cells. The mutation in the β-globin gene in these ES cells can then be corrected by homologous recombination, and the cells can be differentiated into hematopoietic cells for transplant into the patients.
The availability of mouse models for sickle cell anemia can provide a test for such an approach to treat this disease. There are several mouse models of sickle cell anemia, all carrying the human α-, βS-, and γ-globin transgenes and knockouts of the endogenous mouse α- and β-globin genes (11–13). Although some of the models were made by injecting truncated β-globin gene complex under the control of the locus control region (LCR), the one that we have made carries a βS-globin transgene within a 240-kb yeast artificial chromosome that contains the LCR and the ε-, Gγ-, Aγ-, δ-, and βS-globin genes in their native context. Therefore, the ES cells from this sickle cell anemia mouse are likely to have the chromatin structure at the β-globin gene region that resembles that of the human. Hence, this mouse may offer an ideal model to test homologous recombination in ES cells to convert the β-globin sequence from βS to βA. This model may also be used as a test for the ES cell approach for the treatment of β-thalassemia, because similar corrections are applicable to many of the β-thalassemia mutations. In this study, we made ES cells from the sickle cell anemia mouse, corrected the βS mutation to the normal βA sequence by homologous recombination, differentiated the ES cells to hematopoietic cells, and demonstrated that the corrected ES cells synthesized hemoglobin A as well as hemoglobin S.
Results
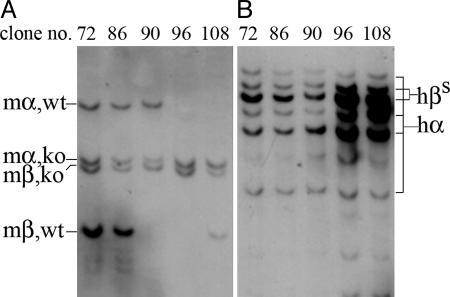
Generation of an ES Cell Line That Carries the Sickle Cell Anemia Genotype. The sickle cell anemia mouse line carrying a yeast artificial chromosome containing 240 kb of human β-globin gene cluster was used in these experiments (13). Female mice carrying homozygous or heterozygous mouse α-globin, heterozygous mouse β-globin gene knockouts, and homozygous human α- and βS-globin yeast artificial chromosome transgenes were mated with male mice with the same genotype. Blastocysts were isolated and embryonic stem cell lines were prepared according to the standard procedure. We isolated 129 blastocysts and generated 12 ES cell lines from them. The genotypes of the ES cell lines were identified by Southern blot analysis using digoxigenin-labeled mouse α- and mouse β-globin genes as well as by using human α- and human γ-globin genes as probes (Fig. 1 and Table 1). We detected the genotypes expected from the mating pairs in these 12 ES cell lines. Among the 12, 10 showed the presence of some mouse α- and/or mouse β-globin genes. Two cell lines, clone 96 and clone 106, contained complete knockouts of the mouse α- and the mouse β-globin genes and were homozygous for the human α- and βS-globin genes. Thus they had the same genotype of the sickle cell anemia mouse with which we started. We selected ES cell line 96 for subsequent targeting and in vitro differentiation experiments
Fig. 1.
Genotyping of ES cell clones by Southern blot analysis using mouse (m) α- and β-globin gene probes (A) and human (h) α- and γ-globin gene probes (B). ko, knockout; wt, wild type.
Table 1. Genotypes of ES clones isolated from total of 129 blastocysts.
| Genotype of ES cell clone
|
No. of ES clones
|
||||
|---|---|---|---|---|---|
| mα | mβ | hα | hβs | ES clone nos. | |
| -/- | +/- | +/+ | +/+ | 3 | 17, 61, 108 |
| +/- | -/- | +/+ | +/+ | 1 | 90 |
| +/+ | -/- | +/+ | +/+ | 1 | 86 |
| +/- | +/- | +/+ | +/+ | 5 | 72, 73, 84, 97, 100 |
| -/- | -/- | +/+ | +/+ | 2 | 96, 106 |
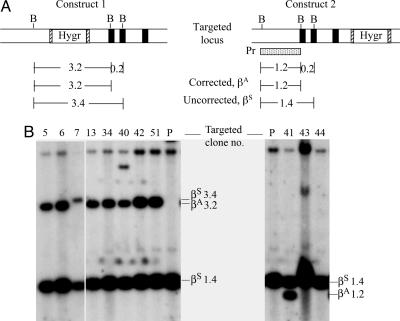
Identification of Homologous Recombinants. Gene targeting with the positive/negative selection scheme was used to facilitate homologous recombination (14). Because the sickle cell anemia mice already harbored the neomycin-resistance gene in the mouse α-globin gene knockout locus, we chose the hygromycin resistance gene as the positive selection marker. We further made use of the cre/lox system by inserting the loxP site on each side of the hygromycin-resistance gene (15). This gene was flanked with loxP sites in case it became necessary to remove it for maximum gene expression. The herpes virus thymidine kinase gene at the 3′ end of the construct was used for negative selection in homologous recombination in ES cells. We made two constructs, construct 1 and construct 2, for homologous recombination experiments (Fig. 2A). In construct 1, the hygromycin-resistance gene was inserted into the HpaI site 0.8 kb upstream from the β6 codon between 3.3 kb of the 5′ noncoding sequence and 5.9 kb containing the βA-globin gene. Because we were concerned that the hygromycin-resistance gene might interrupt some yet-unknown control elements at the upstream region, we made construct 2, in which the hygromycin-resistance gene was inserted into the AvrII site after the 3′ enhancer between 5.0 kb containing the βA-globin gene and 2.5 kb of the 3′ noncoding region.
Fig. 2.
Homologous recombination in ES cells prepared from the sickle cell anemia mice. (A) Targeting construct 1 and 2 and the βS genomic structure. The β-globin gene and flanking sequence of 5.9 kb and 5.0 kb is shown with the three exons indicated by three black boxes for the βA gene and the first exon indicated by the stippled box for the βS gene. The positive selection marker, the hygromycin-resistance gene (Hygr), is flanked by the loxP site (hatched boxes). The negative selectable marker, herpes simplex virus thymidine kinase gene (hsvTK), is at the 3′ end of the constructs. The box Pr is a 400-bp probe used for Southern blot analysis. (B) Two types of recombinants resulting from homologous recombination depending on the site of crossing-over: one correcting βS to βA, the other not correcting βS to βA. (C) Southern blot analysis of DNA of the parental line 96 (P) and the drug-resistant clones (indicated by the numbers). With construct 1, EcoRV digestion produces a 15-kb genomic βS fragment and a 9-kb recombinant fragment. With construct 2, PvuII digestion produces a 12-kb genomic βS and a 14-kb recombinant fragment. The 5-kb band A is the result of star activity of the enzyme Pvull, possibly due to low ionic strength, high pH, or high enzyme concentration in the incubation condition (22).
The ES cell clone 96 was electroporated with DNA of construct 1 or 2 and subjected to selection for hygromycin and ganciclovir. By using a genomic probe outside of the targeting construct, for construct 1, digestion of the DNA of the resistant clones with EcoRV yielded a 9-kb recombinant band as well as the 15-kb untargeted genomic band. We screened 74 ES cell clones and found 8 with the correct recombination. For construct 2, digestion of the DNA of the resistant clones with PvuII yielded a 14-kb recombinant band in addition to the 12-kb untargeted genomic band. Four of 50 clones that were screened showed this recombination. (Fig. 2 B and C and Table 2, second column).
Table 2. Comparison between construct 1 and 2 of frequencies in targeting and expression of human βA-globin gene after repair.
| Expression of hemoglobin A in targeted clones
|
||||
|---|---|---|---|---|
| Targeting construct | Targeting frequency | Crossover frequency at β6 | Yes | No |
| Construct 1 | 8/74 | 7/8 | 6, 13, 34, 42, 51 | 5, 40 |
| Construct 2 | 4/50 | 1/4 | 41 | 11, 43, 44 |
Correction of βS to βA Sequence. During homologous recombination, crossing-over could occur at any point along the whole region of the 5.9 kb or the 5.0 kb containing the β-globin gene. The conversion of the βS to the βA sequence during recombination depended on the site of crossing-over. If it occurred before the β6 codon, the βS would remain uncorrected, but if it occurred at or after the β6 codon, βS would be corrected to βA. To determine if the crossover had corrected the βS to the βA sequence, the DNAs of the targeted clones were digested with Bsu36I, which cleaves the βA but not the βS sequence at the β6 position. The corrected βA DNA would generate a 3.2-kb band instead of a 3.4-kb βS band with construct 1 because of the insertion of the hygromycin-resistance gene between these two Bsu36 I sites. Of the eight recombinant clones targeted with construct 1, seven gave the 3.2-kb βA in addition to the 1.4-kb βS band and were therefore corrected from βS to βA in one allele (Fig. 3). With construct 2, the corrected βA DNA would generate a 1.2-kb βA band instead of the 1.4-kb βS band. One of the four recombinant clones gave the 1.2-kb in addition to the 1.4-kb βS band and therefore was corrected from βS to βA. The higher frequency of correction by construct 1 than by construct 2 appears to agree with the notion that the frequency of crossing-over at the β6 position increases when the selectable marker is placed closer to the mutation site in construct 1 (0.8 kb) than in construct 2 (2.4 kb).
Fig. 3.
Identification of corrected or uncorrected β6 sequences in the targeted genes by Southern blot analysis. Genomic DNAs from the parental cell line (P) and the homologous recombinant clones were digested with Bsu36I (B) and hybridized with a 32P-labeled 1.2-kb Bsu36I fragment 5′ to the β6 position as shown under construct 2 (Pr). With construct 1, because of the position of insertion of the hygromycin gene, the corrected genome, βA, generates a 3.2-kb fragment, whereas the uncorrected βS generates a 3.4-kb fragment. The 1.4-kb fragment is from the unrecombined allele. With construct 2, the corrected and uncorrected genomes produce a 1.2-kb and 1.4-kb fragment, respectively. (A) Restriction map. (B) Southern blot analysis.
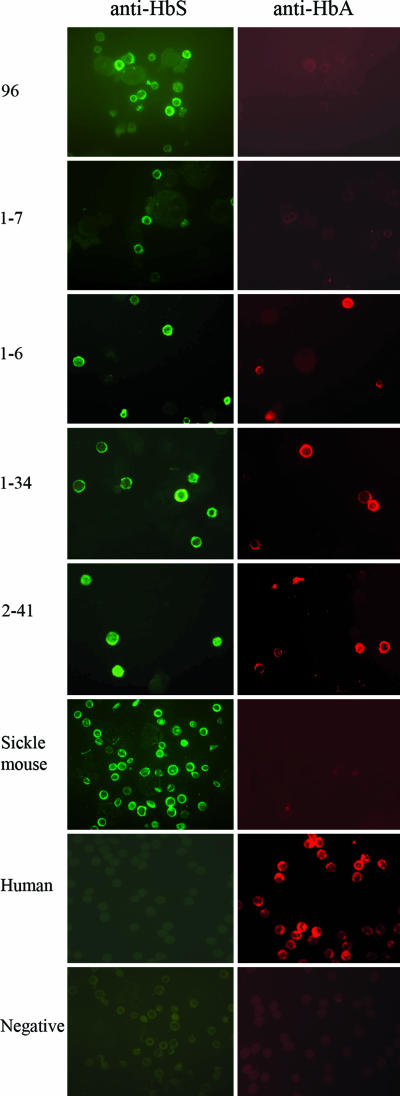
Hemoglobin A and S Synthesis in Hematopoietic Cells Differentiated from the Correctly Targeted ES Cells. The parental ES cell line 96 and the recombinant clones were cultured in vitro and differentiated into hematopoietic cells by using the two-step differentiation protocol (16, 17). After 14-day culture in the secondary differentiation medium, colonies of erythroid, lymphoid, or mixed cells were seen. Cells in the colonies were displayed on two slides and stained with either the anti-hemoglobin A or the anti-hemoglobin S monoclonal antibody (Fig. 4). The parental cell line 96 was stained positive for hemoglobin S and negative for hemoglobin A, as were the red cells from the peripheral blood of a sickle cell anemia mouse. Of the seven clones corrected with construct 1, five expressed hemoglobin A and S, whereas two expressed only hemoglobin S. Presumably, some rearrangements of the gene at crossing-over might have inhibited β-globin gene expression in these two clones. With construct 2, clone 41, the only one with the corrected sequence, expressed both hemoglobin A and hemoglobin S (Table 2). Hence, the majority of the hematopoietic cells differentiated from the corrected recombinant ES cells produced hemoglobin A and hemoglobin S.
Fig. 4.
Identification of hemoglobin A and S with specific monoclonal antibodies in the hematopoietic cells differentiated from the ES cell clones. Cells on the first slide were stained with anti-hemoglobin S antibody and then incubated with secondary anti-mouse antibody conjugated with Alexa Fluor 488 (green), and on a second slide, the cells were stained with anti-A antibody and then incubated with secondary anti-mouse antibody conjugated with Alex-fluor 555 (red). The numbers indicate hematopoietic cells differentiated from ES cells. 96, parental ES cells; 1-7, uncorrected recombinant clone not producing hemoglobin A; 1-6, 1-34, and 2-41, corrected clones producing both hemoglobin A and hemoglobin S. Blood from the sickle cell anemia mouse serves as control for hemoglobin S, and blood from a human serves as a control for hemoglobin A. Negative indicates no primary anti-hemoglobin A or anti-hemoglobin S antibody was used.
Discussion
In this study, we used a mouse model to demonstrate the feasibility of using the ES cell approach to correct the β-globin gene mutation in sickle cell anemia. We prepared ES cells from the blastocysts of sickle cell anemia mice, performed homologous recombination to correct the βS mutation to βA, and differentiated the ES cell into hematopoietic cells. The cells could synthesize hemoglobin A and hemoglobin S and are therefore heterozygous for the sickle mutation. From the experience of human bone marrow transplantation, sickle cell carriers are suitable donors for patients with sickle cell anemia.
These studies show that it is advantageous to use a selectable marker close to the sequence that is to be corrected. With construct 1 in which the hygromycin-resistance gene is 0.8 kb from the β6 position, seven of eight cells showed the correction from βS to βA. In contrast, with construct 2, in which the selectable marker is 2.4 kb from the βS mutation, the β6 sequence was corrected in only one of four clones. Because the upstream region may contain an as-yet-undefined control region, the incorporation of the loxP site would allow the removal of the selectable marker. Although removal of the hygromycin-resistance gene appears not to be necessary in these experiments, because most of the corrected cells could express hemoglobin A as well as hemoglobin S, it may be desirable to incorporate the loxP site to remove foreign genes in some cases because their gene products may cause immune reactions.
In our experiments, we prepared ES cells directly from the sickle cell anemia mouse. It would be possible to use the hematopoietic cells differentiated from these cells to transplant into recipient sickle cell anemia mice because they have the same genotype. If this technique is to be applied to human diseases, it would be necessary to avoid immune rejection by nuclear transfer. Fibroblasts or other cells could be derived from the patients and their nuclei could be transferred into enucleated oocytes from normal donors. ES cells would then be prepared from them. If ES cells could be prepared from human oocytes after nuclear transfer, homologous recombination with the constructs we describe here could be used to correct the sickle mutation. The possibility of differentiation of ES cells to hematopoietic cells for transplantation to cure the Rag2-deficient mouse also makes it likely that this approach may well be successful in treating sickle cell anemia (10).
The sickle cell anemia mouse that we used had 240 kb of human sequences in the β-globin gene region integrated into the mouse chromosome. Hence, the chromosome conformation in this region is likely to resemble that in human ES cells. Also, the two targeting constructs that we have made can be used to correct mutations of the β-globin gene in human ES cells in diseases other than sickle cell anemia; i.e., they can be applied to correct mutations in β-thalassemia as well. Construct 1 will favor correction of mutations in the promoter region, the first exon, and the first intron, and construct 2 will favor correction of mutations in the third exon and the second intron. Both constructs could be tested with mutations in between.
In summary, we have shown that it is possible to correct the mutation in sickle cell anemia in ES cells that contains 240 kb of human DNA sequence. This approach can potentially be applied to human ES cells to correct mutations in the β-globin gene in sickle cell anemia and β-thalassemia.
Materials and Methods
Generation of ES Cell Line. Mice homozygous for human α- and βS-globin transgenes, heterozygous for mouse α-globin, and heterozygous for mouse β-globin gene knockouts were mated. ES cell lines were isolated according to the standard protocol (18, 19). Briefly, blastocysts were harvested from 3.5-day-old embryos and placed individually on mitomycin-treated primary mouse embryonic fibroblast feeder layers in a 96-well gelatinized plate in ES cell medium as previously described in ref. 20. After 4–5 days of culture in vitro, clumps of ES cells were formed in some wells. The cell clumps were dissociated with trypsin and replated on fresh feeder layer, and ES cell colonies became visible in 3–5 days. These ES cell colonies were expanded into a six-well plate to confluency and frozen as passage zero stocks.
Construction of Targeting Vectors. The targeting vector we used is a modification of the vector pPNT in which the neomycin was replaced by a hygromycin-resistance gene (21). To correct the β6 GAG to GTG mutation in sickle cell disease, we made two targeting constructs, construct 1 and construct 2, that carried, respectively, a 5.9-kb or a 5.0-kb sequence containing the human β-globin gene. To maximize the frequency of correcting the βS to βA sequence during crossing-over, the hygromycin-resistance gene was placed as close as possible to the β6 position without interfering with globin gene expression. Because the sequences 5′ to the β-globin gene contain elements that control gene expression, we inserted the hygromycin resistance gene in construct 1 at the HpaI site 0.8 kb 5′ to the β6 position, flanked 5′ by 3.3 kb of homologous noncoding sequences and 3′ by 5.9 kb containing the β-globin gene. Because this location of the hygromycin resistance gene might still disrupt some as-yet-undefined control elements in the upstream region, we made construct 2, in which the hygromycin-resistance gene was inserted after the 3′ enhancer of the β-globin gene, at the AvrII site 2.4 kb 3′ to the β6 codon. The two arms were 5.0 kb containing the β-globin 5′ and 2.5 kb of homologous noncoding sequence 3′ to the hygromycin-resistance gene, which was also flanked by loxP sites (Fig. 2 A).
Gene Targeting and Identification of Homologous Recombinants. Gene targeting in the ES cells carrying the sickle cell genotype was carried out as described in ref. 13. Briefly, 20 μg of construct was linearized at the NotI site and electroporated into 3 × 106 ES cells in 0.8 ml of Hepes-buffered saline in a 0.4-cm gap cuvette with a single pulse of 240 V and 125 μF in a Gene Pulser (Bio-Rad). Hygromycin (150 μg/ml) and ganciclovir (2 μM final concentration) were added 24 h after electroporation. Resistant colonies were picked after 2 weeks of culture in the selective medium and expanded, and DNA was extracted from them. Targeted clones were identified by Southern blot analysis using a 5′ probe upstream from the sequence present in the targeting constructs.
Identification of the ES Cell Clones Corrected from βS to βA. To identify whether the crossover had corrected the βS to the βA sequence, the DNAs of the targeted clones were digested with Bsu36I, which cleaves the βA but not the βS sequence at the β6 position. The corrected βA DNA would generate a 3.2-kb band instead of a 3.4-kb βS band in construct 1 and a 1.2-kb βA band instead of a 1.4-kb βS band in construct 2 (Fig. 3).
In Vitro Hematopoietic Differentiation of ES Cells. ES cell lines carrying the sickle cell anemia or corrected βA genotype were differentiated into hematopoietic cells in vitro by using the two-step differentiation procedure in semisolid methylcellulose-based medium according to the manufacturer's recommendation (StemCell Technologies, Vancouver). In the first step, single ES cells were suspended in methylcellulose-based medium for 10 days to promote their primary differentiation into embryoid bodies (EBs). In the second step, EBs were disrupted by collagenase and single cells were replated in methycellulose-based medium containing erythropoietin, mouse IL-3, mouse IL-6, and stem cell factor. After 10–14 days of incubation, colonies were counted, harvested, washed with PBS, and immunostained.
Immunostaining of Hemoglobins. Hematopoietic colonies from in vitro differentiation were collected in PBS and cytospun onto glass slides, followed by fixation with 100% methanol for 20 min at room temperature. The cells were rehydrated in PBS and sequentially incubated in blocking solution of PBS containing 5% skim milk, PBS containing 10% goat serum, and PBS containing 0.5% human γ-globulin and 1 mM EDTA. The cells were then stained with the primary monoclonal antibodies, anti-hemoglobin A, or anti-hemoglobin S (PerkinElmer) for 40 min at 37°C, washed three times with PBS, and incubated with Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary anti-mouse antibodies (Molecular Probes) in PBS for 30 min at 37°C. The cells were then washed three times in PBS, mounted with mounting medium with DAPI (Vector Laboratories), and observed under fluorescence microscopy.
Acknowledgments
This work was supported in part by National Institutes of Health Grants AM16666, HL053762, and HL070583.
Conflict of interest statement: No conflicts declared.
References
- 1.Serjeant, G. R. & Serjeant, B. E. (2001) Sickle Cell Disease (Oxford Univ. Press, Oxford), 3rd Ed.
- 2.Beutler, E. (2001) in Williams Hematology, eds. Beutler, E., Lichtman, M. A., Coller, B. S., Kipps, T. J. & Seligsohn, U. (McGraw–Hill, New York), 6th Ed., pp. 584–605.
- 3.Sauntharajah, Y., Vichinsky, E. P. & Embury, S. H. (2005) in Hematology: Basic Principle and Practice, eds. Hoffman, R., Benz, E. J., Shattil, S. J., Furie, B., Cohen, H., Silberstein, L. E. & McGlave, P. (Elsevier, Philadephia), pp. 605–644.
- 4.Al-Khatti, A., Papayannopoulou, T., Knitter, G., Fritsch, E. F. & Stamatoyannopoulos, G. (1988) Blood 72, 817–819. [PubMed] [Google Scholar]
- 5.Veith, R., Galanello, R., Papayannopoulou, T. & Stamatoyannopoulos, G. (1985) N. Engl. J. Med. 313, 1571–1575. [DOI] [PubMed] [Google Scholar]
- 6.Charache, S., Terrin, M. L., Moore, R. D., Dover, G. J., McMahon, R. P., Barton, F. B., Waclawiw, M. & Eckert, S. V. (1995) Control Clin. Trials 16, 432–446. [DOI] [PubMed] [Google Scholar]
- 7.Claster, S. & Vichinsky, E. P. (2003) BMJ 327, 1151–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballas, S. K. (1992) Lancet 340, 1226 (lett.). [DOI] [PubMed] [Google Scholar]
- 9.Locatelli, F., Rocha, V., Reed, W., Bernaudin, F., Ertem, M., Grafakos, S., Brichard, B., Li, X., Nagler, A., Giorgiani, G., et al. (2003) Blood 101, 2137–2143. [DOI] [PubMed] [Google Scholar]
- 10.Rideout, W. M., III, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109, 17–27. [DOI] [PubMed] [Google Scholar]
- 11.Paszty, C., Brion, C. M., Manci, E., Witkowska, H. E., Stevens, M. E., Mohandas, N. & Rubin, E. M. (1997) Science 278, 876–878. [DOI] [PubMed] [Google Scholar]
- 12.Ryan, T. M., Ciavatta, D. J. & Townes, T. M. (1997) Science 278, 873–876. [DOI] [PubMed] [Google Scholar]
- 13.Chang, J. C., Lu, R., Lin, C., Xu, S. M., Kan, Y. W., Porcu, S., Carlson, E., Kitamura, M., Yang, S., Flebbe-Rehwaldt, L. & Gaensler, K. M. (1998) Proc. Natl. Acad. Sci. USA 95, 14886–14890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansour, S. L., Thomas, K. R. & Capecchi, M. R. (1988) Nature 336, 348–352. [DOI] [PubMed] [Google Scholar]
- 15.Fukushige, S. & Sauer, B. (1992) Proc. Natl. Acad. Sci. USA 89, 7905–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, G., Kennedy, M., Papayannopoulou, T. & Wiles, M. V. (1993) Mol. Cell. Biol. 13, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller, G. M. (1995) Curr. Opin. Cell Biol. 7, 862–869. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman, M. H., Robertson, E. J., Handyside, A. H. & Evans, M. J. (1983) J. Embryol. Exp. Morphol. 73, 249–261. [PubMed] [Google Scholar]
- 19.Robertson, E. J. (1987) Embryo-Derived Stem Cell Lines (IRL, Oxford).
- 20.Chang, J., Lu, R. H., Xu, S. M., Meneses, J., Chan, K., Pedersen, R. & Kan, Y. W. (1996) Blood 88, 1846–1851. [PubMed] [Google Scholar]
- 21.Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T. & Mulligan, R. C. (1991) Cell 65, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 22.Nasri, M. & Thomas, D. (1987) Nucleic Acids Res. 19, 7677–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]