Abstract
Plasmid-mediated gene therapy can restore dystrophin expression in skeletal muscle in the mdx mouse, a model of Duchenne muscular dystrophy. However, sufficient long-term expression and distribution of dystrophin remain a hurdle for translating this technology into a viable treatment for Duchenne muscular dystrophy. To improve plasmid-mediated gene therapy for muscle diseases, we studied the effects of targeted plasmid integration using a phage integrase (φC31) that can mediate the integration of suitably modified plasmids into the mammalian genome. Using a luciferase expression plasmid, we monitored plasmid gene expression noninvasively in living mice by bioluminescence imaging. Coinjection of an integrase plasmid resulted in 5- to 10-fold higher levels of sustained luciferase expression. Likewise, plasmid-mediated dystrophin expression in mdx muscle was enhanced by integration. Using a combination of dystrophin and luciferase plasmids, we analyzed the functional benefit of dystrophin expression in the dystrophic muscle. The expression of dystrophin slowed the loss of luciferase expression associated with muscle degeneration, and that protection was enhanced by targeted integration of the dystrophin plasmid. In the presence of integrase, dystrophin expression was distributed along a much greater length of individual fibers, and this was associated with increased protection against degenerative changes. These data demonstrate the importance of both the level and distribution of dystrophin expression to achieve therapeutic efficacy, and that the efficacy can be enhanced by targeted plasmid integration.
Keywords: bioluminescence, dystrophin, integrase
Duchenne muscular dystrophy (DMD), a severe X-linked recessive degenerative disease, is caused by mutations in the dystrophin gene that lead to an absence of dystrophin protein in skeletal and cardiac muscle. Plasmid-mediated gene delivery for DMD has shown promise in preclinical studies and offers several advantages over viral delivery systems (1), including the ability of plasmids to accommodate large genes, such as the 14-kb dystrophin cDNA. Although skeletal muscle appears to be remarkably efficient, compared with other tissues, to take up and express plasmids, direct injection of naked DNA leads to patchy expression of delivered genes among fibers in a given muscle (2, 3). Furthermore, as with viral vectors, plasmid-mediated gene therapy will become a viable approach for humans only when systemic delivery is achieved. These major hurdles for plasmid-mediated gene therapy for DMD are areas of active investigation (4, 5), and limited studies in humans are currently under way (6).
Although gene expression from plasmids persists for many months in muscles of rodents (7), the stability of extrachromosomal plasmids remains a concern for the treatment of DMD, where persistence for decades is necessary. Recent evidence suggests that there is a continual loss of plasmid DNA from muscles, and that loss is paralleled by a loss of gene expression (8). This decline is more rapid than could be explained by myonuclear turnover and is likely due to degradation of extrachromosomal DNA. Such a decline in gene expression would have profound implications for the use of plasmid-mediated gene therapy for DMD. The gradual loss of dystrophin, either uniformly or regionally, will render fibers susceptible to degeneration, thus leading to a spiral of loss of therapeutic efficacy. A fiber that expresses dystrophin only along a given segment would be susceptible to degeneration from the dystrophin-negative regions. Thus, advances in plasmid delivery that increase the amount and distribution of dystrophin are likely to be important developments for plasmid-mediated gene therapy for DMD.
Recent studies (9–13) have demonstrated that certain phage integrases can catalyze the integration of plasmids into the mammalian genome if those plasmids contain specific consensus sequences. The φC31 integrase can mediate integration of plasmids containing the attB recognition sequence into the mammalian genome at so-called “pseudo-attP” sites (10, 11). This integration has resulted in sustained expression of therapeutic proteins in animal models of liver and skin diseases (11, 13, 14). In the case of delivery of the factor IX gene to the liver, the expression was not only sustained, but the level of expression was 10-fold higher when integration was promoted by φC31 integrase (12). We have explored the possibility of using φC31 integrase to increase the level of plasmid-mediated gene expression in skeletal muscle and in particular to produce sustained, functional levels of dystrophin protein in muscles of mdx mice. We found that plasmid integration was associated with higher sustained levels of transgene expression. These results have important implications for the design and efficacy of plasmid-mediated gene therapy for DMD.
Methods
Plasmids. The φC31 integrase expression plasmid, pCSI (previously termed pCMVInt), and the control plasmid, pCS, have been described (9, 12). Details of the construction of plasmids and the composition of pCS and pCSI are included in the Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site.
Mice. C57BL/10SnJ and mdx mice offspring from breeder colonies were originally obtained from The Jackson Laboratory. Mice were housed and maintained in the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care Systems in accordance with the guidelines of the Administrative Panel on Laboratory Animal Care of Stanford University.
Intramuscular Injections. Mice were anesthetized by using either methoxyfluorane followed by i.p. ketamine/xylazine/water (1:1:2) at 0.9 μl/g or isofluorane through a nose cone. All mice were injected at the age of 12 days. Tibialis anterior muscles were surgically exposed through skin incisions, and injections were performed by using a 0.3-ml insulin syringe with a 30-gauge needle (Becton Dickinson). The volume of injection was maintained constant at 20 μl, and the amount of plasmid DNA was 25 μg per construct. A concentration of 2.5 μg/μl was therefore used for all experiments involving the injection of two plasmids, whereas a higher concentration (3.75 μg/μl) was used for experiments in which we injected three plasmids (Fig. 5).
Fig. 5.
Effect of integrase on long-term expression of dystrophin in mdx muscle. (A) Muscles of mdx mice were injected with CK6-dystrophin, CK6-luciferase, and either pCS (right legs) or pCSI (left legs). Luciferase activity was monitored noninvasively by using a bioluminescent imaging system. Shown are results from a single mouse at different time points (number of days indicated below each image) after plasmid injections. (B) Cumulative data from groups of mice as illustrated in A. The slower decline in muscles injected with pCSI demonstrates the greater protection conferred by integration of the dystrophin plasmid. The levels of luciferase activity in the pCS-injected muscles had declined to near baseline levels at the latest time point (n = 5 for each time point; *, P < 0.05).
All muscles received 20 μl of 0.5 units/μl hyaluronidase 2 h before plasmid injection (3, 15). After DNA injection, electroporation of the muscle was performed by using an ElectroSquare-Porator (BTX, Genetronics, San Diego) with a two-needle electrode array at a setting of five pulses of 50-ms duration at a voltage of 360 V/cm. These parameters were optimized for electroporating the dystrophin plasmid to obtain the maximal number of dystrophin-positive fibers. It should be noted that the only difference obtained by using lower electroporation voltages was a reduction in transduction efficiency. The relative levels of expression were the same, and integration was observed with all parameters tested. Electroporation results in transient muscle injury that correlates with the duration, frequency, and voltage of the pulses used, but recovery is complete, as has been noted by others (3).
Noninvasive Imaging for Luciferase Activity. Mice to be imaged were anesthetized with either methoxyfluorane and 0.9 μl/g i.p. ketamine/xylazine/water (1:1:2) or isofluorane through a nose cone. Luciferin substrate (33 mg/ml PBS; Biosynth, Basel) was injected i.p. (150 mg/kg body weight). Ten minutes after luciferin injection, luminescence was detected using an in vivo imaging system (IVIS, Xenogen, Alameda, CA) and associated software (living image, Xenogen) for quantitation (16). Luminescence images were acquired at exposure times ranging from 1 sec to 1 min, according to the degree of luminescence, and were superimposed onto still images of each mouse. Luminescence was quantitated within regions of interest in the units of photons/sec per cm2.
Luciferase Assay. Luciferase activity was assayed by using a Femtomaster FB 12 luminometer (Zylux, Oak Ridge, TN), as described (17).
PCR Methods. Detailed methods, including all PCR reaction conditions and primer sequences, are contained in Supporting Text.
Immunofluorescence Analysis. Dystrophin immunostaining was performed as described (18). Dystrophin expression was detected by using a monoclonal antibody against the rod domain (exons 31/32) of the dystrophin protein (MANDYS-8; 1:100) or a polyclonal antibody (P7, 1:1,000; a generous gift of Terence Partridge (Hammersmith Hospital, London), specific for the region of the dystrophin protein encoded by exon 58 (19). Quantitative determination of the number of dystrophin-positive fibers was performed on the entire cross-sectional area for each section analyzed.
Statistical Analysis. Data are presented as means and standard deviations. Comparisons among groups were done by using Student's t test assuming two-tailed distribution and unequal variances.
Results
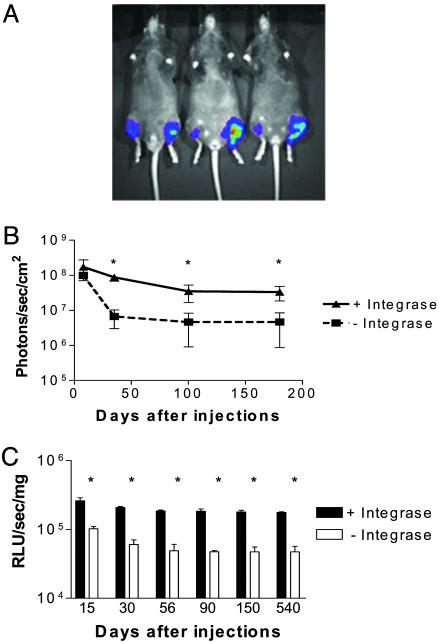
Effect of Integrase on Plasmid-Mediated Gene Expression. Muscles of wild-type mice were coinjected with an attB plasmid encoding luciferase (CK6-luciferase) and either a control plasmid (pCS) or a plasmid containing the φC31 integrase gene (pCSI). Luciferase expression was monitored noninvasively in anesthetized mice at different times after plasmid injection by using a bioluminescence imaging system (Fig. 1A). The highest levels of luciferase gene expression were achieved 1–2 weeks after injection and then began to decline (Fig. 1B). Muscles expressing integrase had higher levels of luciferase expression as early as 4 days after plasmid administration, and the expression remained higher for 6 months and beyond (Fig. 1B). The average enhancement of luciferase expression by integrase was ≈5- to 10-fold. By biochemical analysis, comparably higher levels of luciferase activity were also observed in pCSI-injected muscles (Fig. 1C).
Fig. 1.
Effects of integrase on levels of plasmid-mediated gene expression in normal muscle. (A) Muscles of C57 mice were injected with an attB luciferase reporter plasmid and either pCSI or pCS and monitored by bioluminescent imaging. This image shows three mice 4 months after plasmid injection, revealing higher levels of luciferase expression in muscles injected with pCSI (left legs) compared with muscles injected with pCS (right legs). (B) Cumulative data from studies such as that shown in A (n ≥ 5; *, P < 0.05). (C) Luciferase activity was measured on total extracts of muscles isolated at different times after injection of the attB luciferase plasmid and either pCSI or pCS (n = 3; *, P < 0.05).
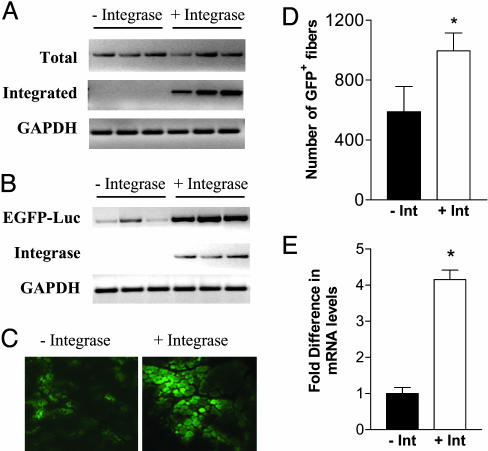
To test whether higher expression was correlated with integration, we analyzed muscles for levels of total CK6-EGFP-luciferase plasmid DNA and plasmid integrated at the mpsL1 site, a known hot spot for integration by φC31 (12). No differences in total plasmid levels were detected between pCS- and pCSI-injected muscles 5 weeks after injection, but integration was detected only in pCSI-injected muscles (Fig. 2A). RT-PCR analysis demonstrated that transcript levels were uniformly higher in pCSI-injected muscles (Fig. 2B). Furthermore, the expression of GFP was greater in pCSI-injected muscles, as reflected by the greater intensity of GFP fluorescence and the greater number of GFP-positive fibers (Fig. 2 C and D). The enhancement of expression in integrase-expressing muscles is shown more accurately by quantitative analysis of transcript levels, which were >4-fold higher in pCSI-injected muscles (Fig. 2E).
Fig. 2.
Effects of integrase on levels of plasmid and plasmid-mediated gene expression. (A) The effects of the integrase expression on plasmid levels and plasmid-mediated gene expression were studied in muscles 5 weeks after injection of CK6-EGFP-luciferase and either pCS or pCSI. Similar amounts of plasmid were present in the two groups (at the top). Plasmid integration at the mpsL1 site as determined by nested PCR, however, occurred only in pCSI-injected muscles (at the bottom). (B) Analysis at the transcript level was determined by RT-PCR on mRNA isolated from the same muscles as in A. The expression of the integrase plasmid is shown for each muscle. (C) GFP expression was examined by fluorescence microscopy in muscles injected with the CK6-GFP-luciferase plasmid and either pCS or pCSI. (D) The number of GFP-expressing fibers was determined in pCS- and pCSI-injected muscles 5 weeks after plasmid injection (n = 4; *, P < 0.05). This analysis underestimates the difference in level of expression, because it does not take into account the marked differences in fluorescence intensity as shown in C. (E) Quantitative real-time RT-PCR analysis was performed to compare the level of expression of the GFP-expressing plasmid in the presence or absence of integrase. The level of expression was >4-fold greater in pCSI-injected muscles (n = 3; *, P < 0.01).
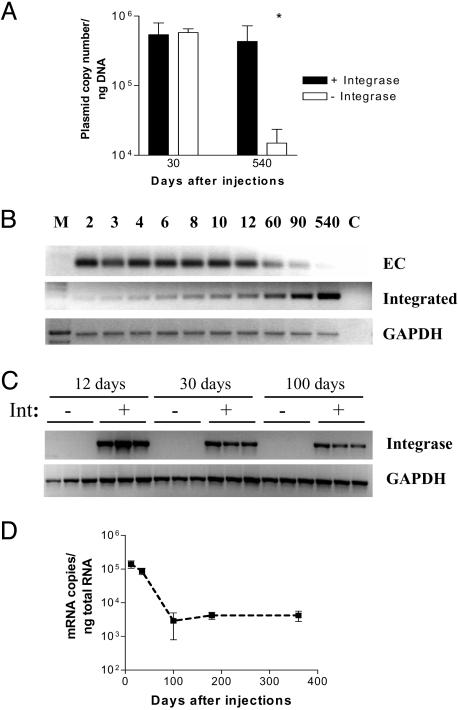
Plasmid Persistence With or Without Integration. To determine whether plasmid integration would limit loss of plasmid DNA after intramuscular injection, we quantified the levels of an attB plasmid (CK6-luciferase) at different times after coinjection with either pCS or pCSI. Real-time PCR analysis was performed on total DNA (chromosomal plus extrachromosomal; see Fig. 8, which is published as supporting information on the PNAS web site). No differences in plasmid copy number between pCS- and pCSI-injected muscles were observed 30 days after injection (Fig. 3A). Over the ensuing 18 months, the levels of plasmid stabilized and changed little in pCSI-injected muscles (Fig. 3A). By contrast, plasmid levels declined in muscles without integrase (Fig. 3A), consistent with previous reports (7, 8).
Fig. 3.
Effects of integrase on persistence of extrachromosomal and integrated plasmid DNA. (A) Genomic DNA isolated at different times after CK6-luciferase plasmid injection either with or without coinjection of pCSI. Quantitative real-time PCR was performed by using the Forw-Luc1/Rev-Luc1 primer pair to assess total plasmid levels. There was no difference between the levels at 30 days, but the difference at 540 days was statistically significant (P < 0.001). (B) Genomic DNA isolated at different times after CK6-luciferase plasmid injection with pCSI. Nested PCR (20 cycles for both rounds) was performed to assess extrachromosomal (EC) plasmid levels and the levels of plasmid that had integrated into the mpsL1 site. GAPDH was used as an internal standard to assure equal amplification, and first-round PCR reactions were stopped after 15 cycles. (C) The level of integrase expression was analyzed by real-time RT-PCR in pCS- and pCSI-injected muscles. No integrase expression was detected in muscles that received pCS. In contrast, there was prominent integrase expression in pCSI-injected muscles that declined over time but was still clearly present 100 days after injection. (D) Quantitative analysis by real-time RT-PCR of integrase transcript in pCSI-injected muscles revealed there was a gradual decline that leveled off ≈100 days after injection and remained constant thereafter up to 1 year after injection.
We examined separately the changes in levels of extrachromosomal plasmid DNA and the levels of plasmid integrated into the mpsL1 site in pCSI-injected muscles (Fig. 8). No integration was seen in pCS-injected muscles in this or any other experiment, consistent with previous reports of negligible random integration after intramuscular plasmid injections (20). For integrase-expressing muscles, we observed a continuous decline of extrachromosomal plasmid levels, whereas the level of integrated plasmid continued to increase for the duration of the study (Fig. 3B). By 18 months after injection, the amount of integrated plasmid accounted for the majority of the plasmid present in the pCSI-injected muscles. The continued integration of attB plasmid is consistent with the sustained expression of integrase from the pCSI plasmid (Fig. 3 C and D), typical of plasmid gene expression after intramuscular injection as seen in Fig. 1 and noted previously (7). The relationship among plasmid levels, either extrachromosomal or integrated, and of plasmid gene expression is considered further in Discussion.
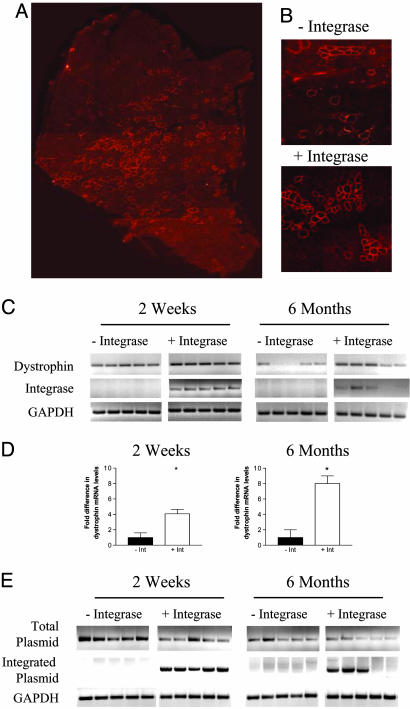
Integrase Enhancement of Plasmid-Mediated Dystrophin Expression in mdx Mice.To determine whether plasmid-mediated dystrophin expression could be similarly enhanced in muscles of mdx mice, we coinjected an attB plasmid encoding dystrophin with pCS or pCSI and analyzed the expression of dystrophin at different times thereafter. Two weeks after injection, dystrophin-positive fibers were detectable in muscles of both groups of mice (Fig. 4A), but the average number of dystrophin-positive fibers was ≈2× greater in muscles coinjected with pCSI (Table 1). As with the analysis of GFP-positive fibers (Fig. 2D), this difference was most likely due to an increase in the number of fibers expressing dystrophin at or above the level of sensitivity of detection in pCSI-injected muscles.
Fig. 4.
Integrase-enhanced dystrophin gene expression in mdx mice. (A) Muscles of mdx mice were injected with CK6-dystrophin and pCSI, followed by electroporation. Two weeks after the injections, immunostaining analysis showed broad distribution of the dystrophin expression throughout the muscle cross section. Overall, 15–20% of the fibers displayed a high level of dystrophin expression. (B) The effect of integrase was reflected in the level of dystrophin expression in pCS- and pCSI-injected muscles. Dystrophin-positive fibers were detected in both groups, but the number of dystrophin-positive fibers and the intensity of immunofluorescence were uniformly greater in pCSI-injected muscles. (C) Muscles of mdx mice were analyzed after the injection of CK6-dystrophin and either pCS or pCSI for the levels of wild-type dystrophin transcript and integrase transcript by RT-PCR at two different time points after injection. Even at early time points, higher levels of dystrophin expression were detected in muscles injected with pCSI. On average, transcript levels declined in both groups between 2 weeks and 6 months after plasmid injections but were maintained at higher levels in pCSI-injected muscles (n = 5). (D) Real-time RT-PCR analysis was performed 2 weeks and 6 months after injection of CK6-dystrophin and either pCS or pCSI to assess quantitatively the level of dystrophin transcripts. There was a 4-fold greater level of dystrophin expression 2 weeks after plasmid delivery in muscles that also expressed integrase, and this difference was even greater at 6 months, when the level of expression was 8-fold greater in pCSI-injected muscles (n = 5 for 2-week time point; n = 3 for 6-month time point; *, P < 0.05). (E) Muscles were examined for total dystrophin plasmid levels by PCR and for plasmid that had integrated into the mpsL1 site (see Fig. 3A) by nested PCR. There was a decline of total plasmid in both groups and integration of the dystrophin plasmid at the mpsL1 site in pCSI-injected muscles.
Table 1. Effect of integrase on the number of dystrophin-positive fibers.
| Number of dystrophin-positive fibers
|
||||
|---|---|---|---|---|
| Time after injection | Age at time of analysis | – Integrase | + Integrase | No. revertant fibers |
| 2 weeks | 1 month | 160 ± 40 | 359 ± 119* | 3 ± 3 |
| 5.5 months | 6 months | 55 ± 31 | 164 ± 41* | 39 ± 18 |
At indicated times after the injection of CK6-dystrophin and either pCS or pCSI, the numbers of dystrophin-positive fibers were counted in individual muscles (n = 5; *P < 0.05 compared with the – Integrase group). In separate groups of mice, the number of revertant fibers were counted (n = 5).
We tested whether integrase-enhanced dystrophin expression would result in a more sustained expression of dystrophin. Six months after the injection of CK6-dystrophin and either pCS or pCSI, the number of dystrophin-positive fibers had declined in both groups, but the number of fibers in pCSI-injected muscles remained higher (Table 1). As expected from the intensity of dystrophin staining and the number of dystrophin-positive fibers, dystrophin transcript levels were higher in pCSI-injected muscles at early and late time points (Fig. 4C). To compare these levels more quantitatively, we used real-time RT-PCR to determine the levels of dystrophin transcripts in pCS- and pCSI-injected muscles at 2 weeks and 6 months. The enhancement of expression by integrase was >4-fold at the earlier time point and increased to >8-fold at the later time point (Fig. 4D). The enhancement of dystrophin expression by integrase was again correlated with plasmid integration, which was detected only in pCSI-injected muscles (Fig. 4E). These studies of plasmid integration were done quantitatively not to compare levels of integration over time but simply to assess for the presence or absence of integrated plasmid. The overall levels of integration that had occurred at the later time point were lower than those detected in wild-type injected muscles, which was primarily due to loss of fibers associated with subtherapeutic levels of dystrophin expression (see below).
To assess the functional significance of integrase-enhanced dystrophin expression, we injected mdx muscles with CK6-dystrophin and either pCS or pCSI, but we also introduced a third plasmid encoding luciferase for noninvasive monitoring. It has been shown (21) that dystrophin expression can “protect” the expression of a coinjected reporter plasmid from loss by limiting the degenerative process. In the absence of dystrophin, luciferase activity declined to undetectable levels within a few weeks (not shown), consistent with previous reports. Concomitant dystrophin expression resulted in a much more gradual decline in luciferase activity, but muscle expressing integrase maintained higher levels of luciferase activity (Fig. 5). This demonstrates that integration results in a functionally significant enhancement of dystrophin expression.
Effect of Integrase on Dystrophin Distribution and Therapeutic Efficacy. The expression of dystrophin per se did not completely prevent the degenerative process from leading to a loss of plasmid gene expression in mdx muscle (Fig. 5B). This was also evident from the direct analysis of total dystrophin plasmid in mdx muscles over time (see Fig. 4E). Although, on average, there was a gradual continuous decline in both luciferase expression and dystrophin expression even in integrase-expressing muscles, the decline was much less pronounced in pCSI-injected muscles (Fig. 5A). To assess the characteristics of dystrophin expression in those muscles that might account for this, we examined the longitudinal distribution of dystrophin. Virtually all fibers in pCS-injected muscles were dystrophin-positive only in the midbelly region of the muscle where the plasmids had been injected. By contrast, we found that pCSI-injected muscles had groups of fibers that expressed dystrophin along their entire lengths at early time points and at 6 months after injection (Fig. 6A). This increased distribution was quantitated in both pCS- and pCSI-injected muscles at 6 months (Table 2). These results highlight the dramatic enhancement of dystrophin expression along the longitudinal axis of individual fibers associated with plasmid integration, an effect that likely accounted for the more sustained expression in pCSI-injected muscles.
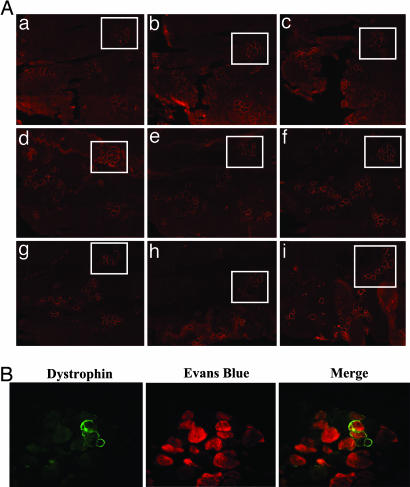
Fig. 6.
Distribution and functional significance of dystrophin expression in muscles with or without integrase. (A) Distribution of dystrophin protein expression was analyzed by immunofluorescence 5 months after delivery of CK6-dystrophin and pCSI. Serial sections of the same muscle 800 μm apart showing dystrophin expression extending the entire length of individual fibers in the tibialis anterior muscle. Boxed area shows a cluster of dystrophin-positive fibers that could be followed along the entire length of this muscle. (B) Six months after injection of CK6-dystrophin and either pCS or pCSI, mice were injected with EBD and killed 24 h later. Dystrophin expression (green) and EBD uptake (red) were assessed in each muscle. EBD uptake was seen in many dystrophin-negative (not shown) but also in dystrophin-positive fibers, as shown here. Such fibers were much more prevalent in pCS- than in pCSI-injected muscles (see Table 2).
Table 2. Characteristics of dystrophin-positive fibers in pCS- and pCSI-injected muscles.
| Longitudinal distribution of dystrophin, μm | Percent of dystrophin-positive fibers that were EBD-positive | ||
|---|---|---|---|
| – Integrase | + Integrase | – Integrase | + Integrase |
| 1,473 ± 1000 | 4,094 ± 2153* | 28.2 ± 12% | 3.5 ± 3%* |
Six months after the injection of CK6-dystrophin and either pCS or pCSI, muscles were examined for dystrophin expression and EBD uptake (injected 24 hr before death). Dystrophin-positive fibers were randomly identified in the midbelly region of each muscle, and the longitudinal distribution of dystrophin expression in that fiber was followed both rostrally and caudally in serial sections. At least 20 dystrophin-positive fibers for each muscle were analyzed (n = 4). For simultaneous analysis of dystrophin expression and EDB uptake, the total number of fibers that were both EDB and dystrophin-positive was determined in a section from the midbelly region of each muscle, and this was divided by the total number of dystrophin-positive fibers in that section (n = 3). (*, P < 0.05 compared with the – Integrase group.)
As a further test, we injected Evans blue dye (EBD), which is taken up into degenerating fibers but not intact fibers (22), into mice in which muscles had been injected with CK6-dystrophin and either pCS or pCSI 6 months previously. Examination 1 day later revealed that there were clusters of dystrophin-negative fibers that had taken up EBD in each muscle, as expected. Most of the dystrophin-positive fibers did not show evidence of EBD uptake. However, in pCS-injected muscles, we found many fibers that were both dystrophin- and EBD-positive (Fig. 6B), suggesting that the level of dystrophin was subtherapeutic in that region or in adjacent segments. The relative paucity of dystrophin-positive fibers that were also EBD-positive fibers in pCSI-injected muscles at late time points was also consistent with our findings of enhanced expression and increased longitudinal distribution of dystrophin in those muscles. Quantitative analyses of these findings are presented in Table 2 and provide further evidence that the enhanced expression and distribution of dystrophin are critical to the long-term functional efficacy of plasmid-mediated gene delivery.
Discussion
We have studied the enhancement by φC31 integrase of plasmid-mediated gene therapy for muscle diseases in general and DMD in particular. Our studies demonstrate that plasmid integration offers several advantages over standard plasmid-mediated gene therapy in which gene expression is mediated by extrachromosomal plasmids. Higher levels of gene expression were observed in muscles in which plasmid integration was promoted, and those levels remained substantially higher over time. Of note, no evidence of host immune response was observed, as determined by analysis for the presence of CD4+ and CD8+ lymphocytes (not shown), in muscle expressing integrase and/or luciferase suggesting that neither of these transgenes induces a significant cellular immune response under these experimental conditions.
The beneficial effects of the plasmid integration were observed also for dystrophin gene expression in mdx mice. When dystrophin expression was mediated by a plasmid that was targeted for integration, higher levels of dystrophin expression were detected at different times after injection. This was evident not only as an increase in the number of dystrophin positive fibers but also as an increase in the level of expression and longitudinal distribution of dystrophin in individual fibers within a muscle.
Our data suggest that the number of dystrophin-positive fibers in a muscle cross section is not a good predictor, alone, of therapeutic efficacy. It is clear that muscle fibers that express dystrophin at early time points after plasmid delivery remain susceptible to degeneration at later time points (8), and this was particularly evident in pCS-injected muscles. The uptake of EBD even in fibers that express dystrophin indicates that dystrophin expression is subtherapeutic, either because the level of expression is too low or because the distribution is inadequate to protect adjacent regions of the fiber from necrotic degeneration. The advantage of integrase-enhanced expression appears to be a higher level of dystrophin expression in general, resulting in a much higher number of fibers expressing therapeutic levels along their entire lengths.
Two interesting observations that emerged from our studies raise important questions about the stability of extrachromosomal plasmids, the transcription of genes on plasmids that have been injected and taken up into muscle fibers, and the effects of integration on these properties. One observation is that transgene expression from extrachromosomal plasmid in wild-type muscle is relatively stable over many months, even as the total plasmid levels in the muscle declines (7, 8). We observed an initial loss of plasmid during the 6–8 weeks after injection that was paralleled by a decline in transgene expression. However, over the ensuing months, there was a disproportionate loss of plasmid compared with loss of luciferase activity (Figs. 1 and 2). This clearly indicates that only a portion of the plasmid accounts for the measured gene expression. As such, studies of plasmid gene expression, with or without integration, may shed light on the mechanisms that regulate plasmid-mediated gene expression in muscle. It appears that the integrated plasmid is responsible for the higher level of expression (Fig. 2C). However, integration appears to continue for many months, such that the integrated plasmid accounts for the vast majority of plasmid remaining >1 year after injection (Fig. 2C), and yet during that time there is no increase in gene expression. These data suggest that most of the extrachromosomal plasmid is transcriptionally silent and remains so even after integration. The characteristics that determine whether an extrachromosomal plasmid will be transcriptionally active or silent remain unknown.
Plasmid silencing has been implicated a major cause of loss of transgene expression in eukaryotic cells (23–25). The cause for such loss in transgene expression has been attributed in part to the presence of highly methylated CpG sequences and bacterial backbone sequences present in the plasmid preparation (26). Recent studies have shown that depletion of CpG sequences in plasmid vectors can reduce the silencing effects and induce stable gene expression (26–29). The use of purified expression cassettes depleted of bacterial DNA and modification of the plasmid backbone structure may substantially enhance the level of plasmid expression that can be achieved in muscles (30).
Technologies to enhance plasmid-mediated gene delivery are likely to be at the forefront of nonviral gene therapy approaches to DMD in the near future. It was recently demonstrated that integrase expression can mediate the integration of attB plasmids in myoblasts in vitro (31), a finding relevant to potential ex vivo gene therapy approaches to DMD. Although there appear to be hot spots of integration, it remains to be determined how site-specific the φC31-directed integration is in the mammalian genome and the development of integrases that direct integration to few or even one site is a priority (32). It is clear from human clinical trials involving retroviral vectors that integration of exogenous elements into the human genome carries real risks (33). It will be important to identify the preferred integration sites mediated by φC31 or any other integrase that would be considered for use in humans. This remains a safety concern for any gene therapy strategy for which integration is likely. Nevertheless, the results presented here demonstrate that the possibility of enhancing plasmid-mediated gene therapy for DMD using targeted integration vectors such as φC31 integrase is very promising and warrants further study.
Supplementary Material
Acknowledgments
We thank Drs. Chris Contag and Tim Doyle of Stanford University for assistance with the bioluminescent imaging; Dr. Jeff Chamberlain of the University of Washington, Seattle, for providing the full-length dystrophin cDNA; and Dr. Terry Partridge of the Medical Research Center Clinical Sciences Centre at Hammersmith Hospital, London, for providing the polyclonal antidystrophin antibody. This work was supported by a Research Development Grant from the Muscular Dystrophy Association (to C.B.) and by a grant from the Muscular Dystrophy Association (to T.A.R.).
Author contributions: C.B, M.P.C., and T.A.R. designed research; C.B., S.J., T.M.W., Y.L., and E.C.O. performed research; C.B. and T.A.R. analyzed data; and C.B., M.P.C., and T.A.R. wrote the paper.
Conflict of interest statement: M.P.C. is a consultant and cofounder of Poetic Genetics, LLC, which has licensed the integrase technology from Stanford University.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DMD, Duchenne muscular dystrophy; EBD, Evans blue dye.
References
- 1.Herweijer, H. & Wolff, J. A. (2003) Gene Ther. 10, 453-458. [DOI] [PubMed] [Google Scholar]
- 2.Sebestyen, M. G., Ludtke, J. J., Bassik, M. C., Zhang, G., Budker, V., Lukhtanov, E. A., Hagstrom, J. E. & Wolff, J. A. (1998) Nat. Biotechnol. 16, 80-85. [DOI] [PubMed] [Google Scholar]
- 3.McMahon, J. M., Signori, E., Wells, K. E., Fazio, V. M. & Wells, D. J. (2001) Gene Ther. 8, 1264-1270. [DOI] [PubMed] [Google Scholar]
- 4.Mir, L. M., Bureau, M. F., Gehl, J., Rangara, R., Rouy, D., Caillaud, J. M., Delaere, P., Branellec, D., Schwartz, B. & Scherman, D. (1999) Proc. Natl. Acad. Sci. USA 96, 4262-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, G., Ludtke, J. J., Thioudellet, C., Kleinpeter, P., Antoniou, M., Herweijer, H., Braun, S. & Wolff, J. A. (2004) Hum. Gene Ther. 15, 770-782. [DOI] [PubMed] [Google Scholar]
- 6.Romero, N. B., Benveniste, O., Payan, C., Braun, S., Squiban, P., Herson, S. & Fardeau, M. (2002) Neuromuscul. Disord. 12, Suppl. 1, S45-S48. [DOI] [PubMed] [Google Scholar]
- 7.Wolff, J. A., Ludtke, J. J., Acsadi, G., Williams, P. & Jani, A. (1992) Hum. Mol. Genet. 1, 363-369. [DOI] [PubMed] [Google Scholar]
- 8.Molnar, M. J., Gilbert, R., Lu, Y., Liu, A. B., Guo, A., Larochelle, N., Orlopp, K., Lochmuller, H., Petrof, B. J., Nalbantoglu, J., et al. (2004) Mol. Ther. 10, 447-455. [DOI] [PubMed] [Google Scholar]
- 9.Groth, A. C., Olivares, E. C., Thyagarajan, B. & Calos, M. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thyagarajan, B., Olivares, E. C., Hollis, R. P., Ginsburg, D. S. & Calos, M. P. (2001) Mol. Cell Biol. 21, 3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olivares, E. C., Hollis, R. P. & Calos, M. P. (2001) Gene 278, 167-176. [DOI] [PubMed] [Google Scholar]
- 12.Olivares, E. C., Hollis, R. P., Chalberg, T. W., Meuse, L., Kay, M. A. & Calos, M. P. (2002) Nat. Biotechnol. 20, 1124-1128. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz-Urda, S., Thyagarajan, B., Keene, D. R., Lin, Q., Calos, M. P. & Khavari, P. A. (2003) Hum. Gene Ther. 14, 923-928. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz-Urda, S., Thyagarajan, B., Keene, D. R., Lin, Q., Fang, M., Calos, M. P. & Khavari, P. A. (2002) Nat. Med. 8, 1166-1170. [DOI] [PubMed] [Google Scholar]
- 15.Gollins, H., McMahon, J., Wells, K. E. & Wells, D. J. (2003) Gene Ther. 10, 504-512. [DOI] [PubMed] [Google Scholar]
- 16.Contag, C. H. & Bachmann, M. H. (2002) Annu. Rev. Biomed. Eng. 4, 235-260. [DOI] [PubMed] [Google Scholar]
- 17.Danko, I., Williams, P., Herweijer, H., Zhang, G., Latendresse, J. S., Bock, I. & Wolff, J. A. (1997) Hum. Mol. Genet. 6, 1435-1443. [DOI] [PubMed] [Google Scholar]
- 18.Lu, Q. L. & Partridge, T. A. (1998) J. Histochem. Cytochem. 46, 977-984. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Q. L., Mann, C. J., Lou, F., Bou-Gharios, G., Morris, G. E., Xue, S. A., Fletcher, S., Partridge, T. A. & Wilton, S. D. (2003) Nat. Med. 9, 1009-1014. [DOI] [PubMed] [Google Scholar]
- 20.Wang, Z., Troilo, P. J., Wang, X., Griffiths, T. G., Pacchione, S. J., Barnum, A. B., Harper, L. B., Pauley, C. J., Niu, Z., Denisova, L., et al. (2004) Gene Ther. 11, 711-721. [DOI] [PubMed] [Google Scholar]
- 21.Danko, I., Fritz, J. D., Latendresse, J. S., Herweijer, H., Schultz, E. & Wolff, J. A. (1993) Hum. Mol. Genet. 2, 2055-2061. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda, R., Nishikawa, A. & Tanaka, H. (1995) J. Biochem. (Tokyo) 118, 959-964. [DOI] [PubMed] [Google Scholar]
- 23.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., et al. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 24.Paillard, F. (1999) Hum. Gene Ther. 10, 2089-2090. [DOI] [PubMed] [Google Scholar]
- 25.Chen, Z. Y., He, C. Y., Meuse, L. & Kay, M. A. (2004) Gene Ther. 11, 856-864. [DOI] [PubMed] [Google Scholar]
- 26.Yew, N. S., Zhao, H., Przybylska, M., Wu, I. H., Tousignant, J. D., Scheule, R. K. & Cheng, S. H. (2002) Mol. Ther. 5, 731-738. [DOI] [PubMed] [Google Scholar]
- 27.Gill, D. R., Smyth, S. E., Goddard, C. A., Pringle, I. A., Higgins, C. F., Colledge, W. H. & Hyde, S. C. (2001) Gene Ther. 8, 1539-1546. [DOI] [PubMed] [Google Scholar]
- 28.Yew, N. S., Przybylska, M., Ziegler, R. J., Liu, D. & Cheng, S. H. (2001) Mol. Ther. 4, 75-82. [DOI] [PubMed] [Google Scholar]
- 29.Chen, Z. Y., He, C. Y., Ehrhardt, A. & Kay, M. A. (2003) Mol. Ther. 8, 495-500. [DOI] [PubMed] [Google Scholar]
- 30.Hartikka, J., Sawdey, M., Cornefert-Jensen, F., Margalith, M., Barnhart, K., Nolasco, M., Vahlsing, H. L., Meek, J., Marquet, M., Hobart, P., et al. (1996) Hum. Gene Ther. 7, 1205-1217. [DOI] [PubMed] [Google Scholar]
- 31.Quenneville, S. P., Chapdelaine, P., Rousseau, J., Beaulieu, J., Caron, N. J., Skuk, D., Mills, P., Olivares, E. C., Calos, M. P. & Tremblay, J. P. (2004) Mol. Ther. 10, 679-687. [DOI] [PubMed] [Google Scholar]
- 32.Sclimenti, C. R., Thyagarajan, B. & Calos, M. P. (2001) Nucleic Acids Res. 29, 5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi, Y., Hahm, S. H. & Lee, K. H. (2005) Curr. Gene Ther. 5, 25-35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.