Abstract
The molecular diversity of the luminal endothelial cell surface arising in vivo from local variations in genetic expression and tissue microenvironment may create opportunities for achieving targeted molecular imaging and therapies. Here, we describe a strategy to identify probes and their cognate antigens for targeting vascular endothelia of specific organs in vivo. We differentially screen phage libraries to select organ-targeting antibodies by using luminal endothelial cell plasma membranes isolated directly from tissue and highly enriched in natively expressed proteins exposed to the bloodstream. To obviate liver uptake of intravenously injected phage, we convert the phage-displayed antibodies into scFv-Fc fusion proteins, which then are able to rapidly target select organ(s) in vivo as visualized directly by γ-scintigraphic whole-body imaging. Mass spectrometry helps identify the antigen targets. This comprehensive strategy provides new promise for harnessing the power of phage display for mapping vascular endothelia natively in tissue and for achieving vascular targeting of specific tissues in vivo.
Keywords: antibody, vascular targeting, endothelial cell, protein expression
Recent advances in biomedical research have generated a wide array of potential diagnostic and therapeutic opportunities in many areas of medicine (1-5). Unfortunately, the ability to overcome in vivo barriers to effectively deliver imaging agents, drugs, or genes to specific tissues in sufficient effective quantities remains an elusive goal (1, 2, 6-10). Targeting the vasculature, the main conduit through which active molecules are distributed to target cells, is receiving increased attention as a powerful means to achieve more efficient delivery to specific organs and tissues that may improve the efficacy of many promising therapeutics (8, 11-25).
The vascular targeting strategy intends to exploit the inherent accessibility of the vascular endothelium through direct contact with the circulating blood for directing pharmacodelivery to select normal or diseased tissues (15-18). Endothelial cell morphology can vary significantly in normal organs and with disease. Tissue-restricted variations at the luminal endothelial cell surface of blood vessels involve numerous proteins, many of which are modulated in vivo and lack expression ex vivo (19, 20). For example, ≈40% of endothelial cell surface proteins expressed in rat lungs in vivo are not detected in isolated rat lung endothelial cells grown in cell culture (20). But few validated targets and targeting probes have been discovered, in part, because of technical constraints. The rather extensive and quite rapid loss of normal morphology and protein expression by endothelial cells ex vivo can drastically limit their utility in target-discovery efforts. Thus, a major challenge for the vascular targeting field becomes mapping, screening, and validating potential targets and their probes under native conditions found in vivo. Although proteomic and antibody-based mapping of the vascular endothelium by using mass spectrometry and classic hybridoma technology, respectively, offer two promising alternatives (19-21), the inherent advantages in using the immense diversity of complex combinatorial libraries compels the development of robust strategies to screen such libraries under the most physiologically relevant conditions in vivo. In the last decade, the screening of phage libraries after i.v. injection to search for tissue-homing peptides has been reported (22-26), but, so far, the yield of promising tissue-specific targets has been rather modest (19). To obviate many past limitations, we endeavor to use the power of antibody diversity displayed in phage libraries by creating a screen directly against isolated endothelial cell membranes and their proteins at the tissue-blood interface in vivo.
Materials and Methods
Additional methods and vector construction are shown in Supporting Text, which is published as supporting information on the PNAS web site.
Library Construction. BALB/c mice were immunized, each with a total of 500 μg of rat lung endothelial cell plasma membrane proteins in Freund's adjuvant. The day before killing, 100 μg of endothelial plasma membranes in PBS were injected intrasplenically. Antibody-variable regions VH and VL were amplified by PCR from spleen cell cDNA by using a custom set of mouse primers. Amplified fragments were gel-purified and mixed in equal amounts for library construction or analyzed individually during the cloning of single antibody. For the assembly PCR, 50 ng of VH and 50 ng of VL DNA were mixed together with 14 ng of linker primer 5′-GGTGGTCTTAATATAACTTCGTATAATGTATACTATACGAAGTTATTAGGTGGTGGTGGT. After 10 cycles (94°C for 1 min, 55°C for 1 min, and 68°C for 1 min) in the presence of Pfx (Stratagene), a pair of biotinylated terminal primers, biotin-TACTCGCGGCCCAGCCGGCCA and biotin-CTAGGTTGGCCTCCCGGGCCA, was added, and the assembly product was then amplified for 25 cycles. The final assembly product was cleaned over anion-exchange resin (Qiagen, Valencia, CA), digested by SfiI, incubated 10 min at 22°C with 10 μl of BioMag Streptavidin beads (Polysciences) to remove the free ends, and purified by phenol-chloroform extraction and ethanol precipitation before ligation. The library was built by using an early version of our cloning vector that contained only a truncated version of the pIII coat protein-encoding DNA. Therefore, the virions lacked a full-length pIII protein necessary to infect bacteria. To build the first sublibrary, we amplified the DNA of the eluted virions by PCR and recloned them into the pSK12 vector. A total of 2.1 million full-length antibody clones were thus generated.
Library Screening. For the first round, 1 × 1013 virions from the lung library were preadsorbed three times with 1 mg of rat liver endothelial cell plasma membranes and incubated for 4 h at 22°C in Tris-buffered saline, pH 7.5, 0.1% Tween 20, 5% milk, and 0.1% polyacrylic acid over 100 μg of lung endothelial cell plasma membrane proteins dried on a polystyrene well. After five washes with Tris-buffered saline and Tween 20, bound phages were eluted with 0.1 M glycine/HCl, pH 2.0, with 1 mg/ml BSA and then neutralized with 1 M Tris, pH 8.0. Subsequent rounds used 20 μg of lung endothelial cell plasma membrane proteins and 2 × 1011 virions as input.
ScFv-Fc Cloning and Expression. The pDisplay vector (Invitrogen) was mutated with the oligonucleotide 5′-PGAGATCTGGCCGGCTGGGCCGCTAAGAGCAGCAGTACCCATAG (27) to introduce a SfiI site into the κ leader sequence. The murine IgG1 Fc fragment, including hinge, CH2, and CH3, was amplified by PCR from murine hybridoma cDNA by using the primers 5′-CACATCCTCCTAGATCTACTAGTGGCCCGGGAGGCCCCAGGGATTGTGGTTGTAAG and 5′-CCATATGTCGACTCGAGTTATCATTTACCAGGAGAGTGGGAGAG. The Fc fragment was cloned into the mutated pDisplay cut with BglII and XhoI. The vector mSK1 was then generated by ligating two complementary 21-mer stuffer primers in the double SfiI cloning site. Final constructs containing scFv inserts were transfected into Chinese hamster ovary (CHO) cells by using Lipofectamine (Invitrogen). The transfected cells were grown in serum-free medium (CHO protein-free medium; Sigma) containing 10 mM sodium butyrate for 4 days. Pure recombinant proteins were obtained after chromatography on protein A in high-salt conditions.
Results
Phage Display of Lung-Targeting Aminopeptidase P Antibody. To develop and optimize screening methods for selecting phages, we chose the antibody TX3.833, which rapidly and specifically targets the rat lung endothelium when injected intravenously (19). The variable regions of TX3.833 were cloned in a single chain Fv format (scFv) into the phagemid vector pSK12, and the predicted amino acid sequences were confirmed by mass-spectrometric analysis of the parent antibody (data not shown). Phages displaying TX3.833 scFv recognized the membrane-bound form of aminopeptidase P (APP) by Western blot analysis as a single 90-kDa band from endothelial cell plasma membranes isolated from rat lungs by using the colloidal silica coating technique (28, 29) (see Fig. 6, which is published as supporting information on the PNAS web site). This binding was specifically inhibited by preincubation with TX3.833. Confocal fluorescence microscopy confirmed cell-surface binding of TX3.833 phage to APP by showing a cell-surface punctate signal on intact MA104 cells only when transfected to express APP (see Fig. 6). Finally, this phage recognized purified recombinant soluble APP (rAPP, extracellular portion) by Western blot analysis and ELISA (data not shown).
Next, we optimized conditions for specific binding of TX3.833 phages to luminal endothelial cell plasma membranes isolated from lung. Based on numerous antibodies and functional assays, these membranes appear to preserve native protein expression, conformation, and function, making them an ideal substrate for phage selection and screening of large antibody libraries (19-21, 28-31). These endothelial membranes were dried onto a polystyrene well and blocked by using 0.1% buffered polyacrylate, which reduced the otherwise intense background binding of phage to negligible levels, thereby rendering specific binding detectable by ELISA (see Fig. 6).
Optimizing Antibody Library Screening. We prepared a library containing 300 million in-frame antibody clones from the splenocyte cDNA of a mouse immunized with the luminal endothelial cell plasma membranes isolated directly from rat lung tissue. We chose to package the library using the defective VCSM13d3 helper phage (32), because it resulted in higher levels of display compared with VCSM13. An aliquot of the lung library underwent negative selection on luminal endothelial membranes isolated from rat liver to remove unwanted panendothelial phage specificities, followed by a round of panning with the lung endothelial cell plasma membranes.
We first tested for the presence of APP-binding clones. After two additional rounds of selection by the recombinant soluble APP, the phage yield showed a 75-fold increase (see Table 2, which is published as supporting information on the PNAS web site). Accordingly, 20 of 24 clones picked randomly in the last round recognize rAPP. TX3.833 preincubation inhibited the binding of all clones, indicating overlapping, if not identical, epitopes (data not shown). Sequence analysis revealed six unique sequences with striking homologies to TX3.833 (see sequence data, which is published as supporting information on the PNAS web site). Western blot analysis showed that each phage bound a specific band at 90 kDa that was also inhibited by TX3.833 (data not shown). Now, with the knowledge that the first sublibrary contained several anti-APP clones, we wished to optimize screening directly to the membranes by following APP antibody selection.
The first sublibrary was panned twice more on isolated rat lung luminal endothelial cell plasma membranes, resulting in a 180-fold increase in phage yield. ELISA was able to detect APP binders in the second round at a frequency of between 1:250 and 1:500. In the third round sublibrary, 96 clones were analyzed, of which 5 tested positive for APP binding (PH2 group in Table 1). All five clones recognized the 90-kDa band and four bound MA104/APP cells. Phage from the second round failed to show any detectable signal by Western blot analysis, suggesting that APP binders of progressively higher affinity were enriched during selection. Nucleotide sequence analysis showed an average homology to TX3.833 sequences of 71% for the light chain and 84% for the heavy chain compared with 92% and 96.5%, respectively, observed when antibodies were directly selected by APP. These assay conditions appear appropriate for specific phage binding and, thus, for screening the antibody library to identify potential targeting probes for lung endothelium.
Table 1. Properties of selected phage antibodies.
| Clone
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P3 | P8 | P9 | P7 | P26 | P27 | P97 | P35 | PE3 | PA4 | PF12 | PH2 | |
| Frequency | ||||||||||||
| Round II (216)* | 7 | 3 | 1 | 4 | 1 | 16 | 1 | 28 | ||||
| Round III (96) | 6 | 4 | 5 | 1 | 57 | 1 | 3 | 1 | 5 | |||
| Variant no.† | 3 | 4 | 1 | 2 | 1 | 2 | 2 | 11 | 1 | 1 | 1 | 5 |
| Cited variants | PM8 | P20 | P34 | |||||||||
| Phage ELISA | ||||||||||||
| Reactivity on Lung P | + | + | + | + | + | + | + | + | + | + | + | + |
| Reactivity on Liver P | + | + | + | – | – | – | + | – | – | – | + | – |
| Phage Western blots | ||||||||||||
| Size (kDa) | 55/70 | 60 | 100 | 120‡ | 200 | 24 | 250 | 100 | 100 | 120 | 50 | 90 |
| P enriched | + | + | + | + | + | + | – | + | + | + | + | + |
| Reducing conditions‡ | + | – | + | + | + | + | ND | – | – | + | – | – |
| Phage immunohistochemistry on rat lung fresh frozen sections | ||||||||||||
| Large blood vessels§ | – | ++ | +++ | + | + | ++ | ND | +++ | +++ | – | – | – |
| Microvessels | ? | ? | ++ | ++ | ++ | ++ | ND | ++ | ++ | – | – | – |
| Alveolear staining | +++ | ++¶ | ++ | + | + | ++ | ND | + | + | + | – | – |
?, unknown; alveolar staining, endothelial and/or epithelial staining at the alveoli level. ND, not done.
Total number of clones analyzed is shown in parentheses
Based on BSTN1 digestion and binding intensity
Negative clones react only in nonreduced conditions; P7 gives a band at 120 kDa in nonreducing and 60 kDa in reducing conditions
Staining intensity is given on a scale from 1 to 3 crosses
Also reacts with the ciliated respiratory epithelium
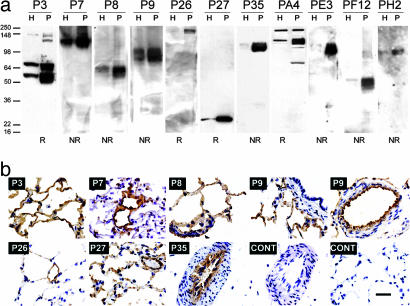
Screening for Antibodies to Endothelial Cell Surface Proteins. Phage clones from the second and third rounds of membrane panning described above were tested randomly for binding to both lung and liver endothelial cell plasma membrane proteins. Of 216 clones analyzed from the second round, 118 (54%) showed reactivity to lung endothelial membranes by ELISA. Only 15 (7%) also reacted with liver membranes. Of the 96 clones from the third round, 90 (94%) bound to lung endothelial membranes, and 12 (13%) also bound liver endothelial membranes. Individual clones were tested by Western blot analysis in both reduced and nonreduced conditions; about one-half of the binders from the second sublibrary and 85% of all of the binders from the third round produced distinct bands. We found large numbers of somatic variants by either BSTN1 restriction analysis or inhibition using scFv-Fc fusion molecules (see below) and finally identified 12 binding patterns (Fig. 1a and Table 1), with 11 showing a significantly greater binding to endothelial membranes than whole-tissue homogenates. Thus, 11 of 12 selected antibodies appear to recognize determinants associated with the endothelial cell surface in the lung, and 7 of 12 react with lungs but not liver.
Fig. 1.
Phage reactivity to rat lung luminal endothelial cell plasma membranes. (a) Western blot analysis of lung endothelial plasma membranes (P, 10 μg) and homogenate (H, 10 μg) using the indicated phage. NR and R indicate nonreducing and reducing conditions, respectively. (b) Phages were incubated on fresh frozen lung sections, washed, and reacted with an anti-PVIII antibody conjugated to horseradish peroxidase, followed by diaminobenzidine staining. Arrowhead on P8 display points to staining of bronchial epithelium. (Scale bar, 20 μm.)
Binding to vascular determinants was analyzed further by immunohistochemistry. The use of a horseradish peroxidase immunoconjugate directed to the main phage coat protein pVIII acted as a natural amplification system and, in many cases, provided clear reactivity on fresh frozen sections incubated with phages (Fig. 1b). All but clones PF12 and PH2 exhibited binding on lung sections. Table 1 summarizes the different patterns of reactivity with respect to localization in alveoli, microvessels, and large blood vessels. Clear staining of the endothelium of large blood vessels and microvessels provided evidence that enrichment in the isolated luminal endothelial cell membranes was endothelial specific. Nevertheless, binding to epithelium determinants could not be excluded in the alveoli, in particular for clones P3 and PA4. Thus, screening on endothelial plasma membranes led to the isolation of multiple phage clones that exhibited endothelial cell reactivity in the lung but with different localization patterns in capillaries, microvessels, and large blood vessels (Table 1).
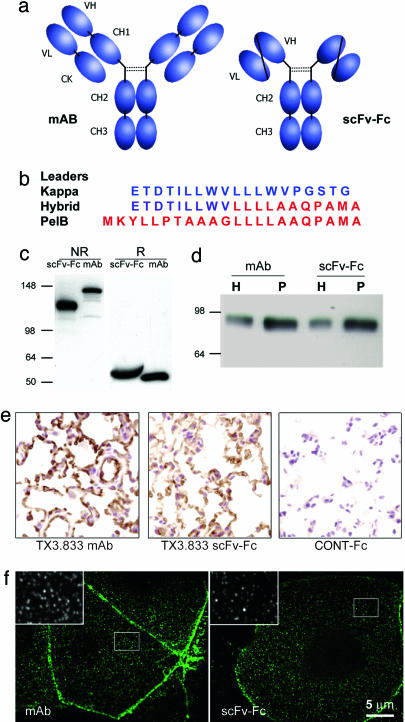
In Vivo Validation of Immunotargeting by Using Molecular Imaging. We attempted to assess possible lung-specific phage targeting objectively and visually using planar γ-scintigraphic imaging (19). All of the phages injected intravenously, including TX3.833 derivatives and nonspecific controls, showed rapid and substantial liver and spleen targeting by standard planar γ-scintigraphy imaging (Fig. 2a). We also performed dynamic live imaging in the first minute immediately after i.v. injection and found significant liver uptake with little to no lung targeting. More than 90% of the injected phage accumulated in the liver and spleen within 60 sec (Fig. 2c). To mitigate this problem, we designed an antibody-like probe by fusing TX3.833 scFv to the mouse Fc γ1 through the hinge region (Fig. 3a). To allow the transfer of scFvs of unknown sequence from our phagemid vector, we engineered the κ leader peptide to accommodate a SfiI site compatible with the bacterial PelB leader sequence. The composite sequence is a hybrid between the two leaders (Fig. 3b) and is functional in CHO cells. The resulting construct produced a scFv-Fc fusion molecule of 50 kDa that was secreted as dimers of 100 kDa (Fig. 3c). As expected, TX3.833 scFv-Fc showed reactivity to APP that appeared very similar in specificity to the parent antibody by Western blot analysis, immunohistochemistry, and confocal microscopy (Fig. 3 d-f). Tail-vein injection of the TX3.833 scFv-Fc revealed specific lung targeting by γ-scintigraphic planar imaging similar to that of the parent antibody even as early as 10-min postinjection (Figs. 2b and 5). One hour after injection, the amount of targeted scFv-Fc was 35% of the injected dose per gram of tissue (ID per g) compared with 77% ID per g for the whole antibody (see Table 3, which is published as supporting information on the PNAS web site). The significant amounts of radioactivity still circulating in the blood accounted for the difference, indicating that only part of the scFv-Fc had reached its target at that time. Imaging taken after longer time periods did not improve the overall lung targeting, because dehalogenation and body clearance became a competitive process. These observations were in accord with the apparently lower affinity of TX3.833 scFv to APP, although we did not exclude that a fraction of the recombinant scFv-Fc was unable to bind APP, possibly because of protein misfolding.
Fig. 2.
Comparative in vivo targeting by imaging biodistribution of TX3.833 phage versus scFv-Fc. Planar γ-scintigraphic images of rats at the indicated time after injection of TX3.833 phages (a) or TX3.833 scFv-Fc (b). The number in the bottom left corner of each image indicates dynamic intensity scale range of the given image starting at 0 to 100% as per color scale shown; for example, with a setting of 500, a yellow/white signal indicative of 100% will be 5-fold greater than the same color achieved at a setting of 100. S, spleen; Lv, liver; L, lungs; HC, heart cavity. (c) Live dynamic imaging of rat injected with 125I-TX3.833 phage. Representative static frames captured at the indicated times are shown.
Fig. 3.
TX3.833 scFv-Fc fusion. Schematic representation (a) and details of the composite peptide leader to facilitate transfer of the scFv insert between the phage and the scFv-Fc vector (b). TX3.833 scFv-Fc is secreted as a 100-kDa dimer (c) and has the same reactivity as the parent mAb by Western blot on lung endothelium plasma membranes (d), by histochemistry analysis on fresh-frozen rat lung sections (e), and by confocal microscopy on MA104/APP cells (f). H, tissue homogenate; P, silica-coated luminal endothelial cell plasma membranes; NR, nonreducing SDS/PAGE; R, reducing SDS/PAGE.
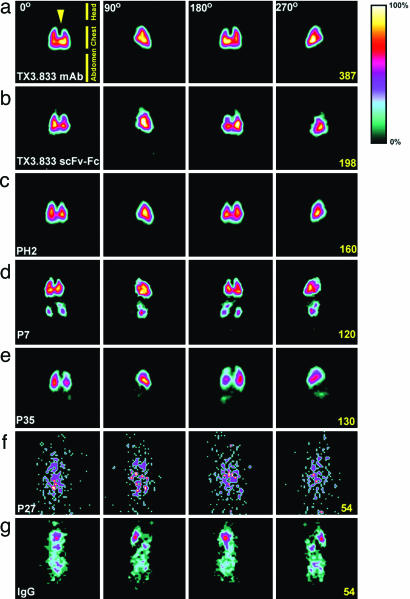
Fig. 5.
In vivo planar γ-scintigraphic imaging of scFv-Fcs in rat. (a-g) Planar images were collected by using a single-photon-emission computed tomography imaging system after tail-vein injection of the indicated 125I-scFv-Fcs. Degrees of rotation of the animal along the body axis are indicated on top; scale values are indicated on the right side of each row. Frames were collected 1 h (a-c, f, and g) or 2 h (d and e) after injection. The arrow (Upper Left) indicates the position of the heart cavity.
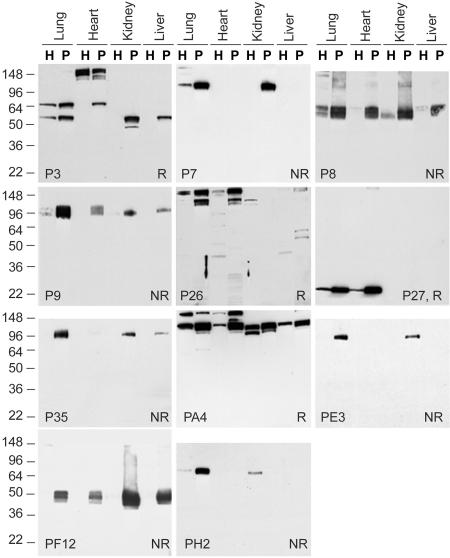
ScFv-Fc Fusions Uncover Varied Tissue-Specificity and Target Distinct Organs in Vivo. To test further the specificity of reactive antibodies from the phagemid library, we generated and characterized recombinant scFv-Fcs for each of the phages previously isolated. Fig. 4 shows the Western blot analysis of endothelial cell plasma membranes isolated from major rat organs. Most scFv-Fcs gave very strong and specific binding compared with the phage. All reproduced the original binding, except for a few variants giving initially weak signals. A few patterns were restricted to distinct organs: P7 reacted strongly in both lung and kidney endothelial membranes; P35 and PE3 showed the same pattern with higher reactivity for lung than kidney; PF12 bound most strongly to kidney endothelial cell plasma membranes, although it also reacted with all tissues; P27, P26, and PA4 were all expressed at a higher level in both heart and lung endothelial membranes.
Fig. 4.
Tissue distribution of scFv-Fc reactivity. Western blot analyses using the indicated scFv-Fc fusions were conducted on organ homogenates (H, 10 μg) or endothelial cell plasma membrane fraction (P, 10 μg) from lung, heart, brain, kidney, and liver under reducing conditions (R) or nonreducing conditions (NR).
To analyze antibody targeting to organs in vivo, radioiodinated scFv-Fcs injected into rat tail veins were imaged for 1-2 h by planar γ-scintigraphy, followed by organ excision for biodistribution analysis (see Table 3). Organ-targeting and specific imaging are highlighted in Fig. 5, where several views taken at different rotation angles along the body axis are shown (see also 3-D single-photon-emission computed tomography Movies 1-3, which are published as supporting information on the PNAS web site). ScFv-Fc P7 was rapidly taken up in lungs, kidneys, and spleen, with 31.9%, 8.5%, and 5.6% ID per g, respectively. Tissue targeting indexes (TTI; antibody in tissue per g of tissue per antibody in blood per g of blood) of 213, 56, and 37 in lung, kidney, and spleen, respectively, and tissue-selectivity indexes (TSI; TTI for targeting antibody per TTI for control antibody) of 253, 112, and 88, respectively in the same organs, confirmed selective organ-targeting.
Upon injection, P35 scFv-Fc accumulated rapidly in the lungs but not in kidneys, with 39.8% ID per g after 2 h (Fig. 5e). The high values of TTI and TSI for P35 in the lungs, 508 and 604 respectively, reflected the low level of circulating scFv-Fc at the 2-h time point. In fact, all studied organs but brain showed a TTI >1 with a mean between 20 and 27, indicating possible uptake by the blood vessels of large and medium caliber. Accordingly, immunohistochemistry analysis on lungs (Fig. 1) and heart (data not shown) exhibited intense staining of large blood vessels. Spleen accumulation was reproducible, with nearly 2% ID per g and was visible on the 3-D single-photon-emission computed tomography movies (see Movies 1-3). Lastly, PH2, which was isolated by screening lung endothelial cell membranes and shown to recognize APP (see above), targeted the lung vasculature similarly to TX3.833 scFv-Fc to give a very striking and specific image of the lungs (Figs. 2b and 5b), with an uptake of 34.9% ID per g, almost identical to TX3.833 scFv-Fc lung uptake. We also analyzed scFv-Fcs PF12, PE3, and P27 (Fig. 5f), but none showed specific lung or other organ uptake. The reactive epitopes may be inside the endothelial cell and, thus, may not be accessible for these targets.
Identification of Antigens and Vascular Targets. To date, we have identified several antigens to our antibody-like constructs using primarily mass spectrometry after immunoprecipitation. P7 antigen is plasmalemma vesicle protein 1 (PV-1), with 21% peptide sequence coverage (see Fig. 7a, which is published as supporting information on the PNAS web site). PV-1 has been associated with the stomatal diaphragms of rat lung endothelial caveolae and the diaphragms of fenestrated capillaries (33, 34). PH2 antigen is the membrane-bound APP (see above). P27 specifically recognizes caveolin-1 (see Fig. 7 b-c). Caveolin-1 embeds in the lipid bilayer with both N and C termini inside the cell, explaining the lack of tissue uptake after i.v. injection (Fig. 5f). Experiments to uncover the nature of P35 antigen have not yet succeeded.
Discussion
Over the last decade, we have developed and optimized numerous subcellular tissue-fractionation techniques to isolate luminal endothelial cell plasma membranes and their caveolae directly from tissue to map the molecular landscape of the endothelial cell surface in vivo (19-21, 28-30, 35-37). We recently described in vitro and in vivo mapping of the rat lung endothelial cell surface proteome, with ≈450 proteins identified (20). The lack of in vitro expression for ≈40% of these proteins may reveal the degree to which the unique microenvironment in each tissue can regulate endothelial cell phenotype at the molecular level. Subtractive analysis of these in vitro vs. in vivo expression differences led to the discovery of APP as a sufficiently lung-restricted endothelial cell surface protein to permit lung-specific immunotargeting in vivo (19, 20). Complementary efforts using classic hybridoma-based antibody generation identified TX3.833 as an mAb that is capable of rapid and specific lung vascular targeting (21) and recognizes APP. One challenging aspect of the discovery of vascular targets like APP is the phenotypic shift that occurs in endothelial cells once they are removed from the native tissue environment (20). We find that tissue-induced expression of endothelial proteins such as APP is rapidly lost ex vivo and yet to be regained in cell culture [refs. 19 and 20 and our unpublished observation (J.E.S.)].
Here, we report a screening approach based on the concept of assaying naturally expressed endothelial cell proteins that permits the rapid identification of not only antibodies showing organ-restricted and even organ-specific vascular targeting but also of the target endothelial antigens. This approach takes advantage of the power of phage display, especially under standard, well controlled screening conditions and, in the end, may help eliminate some of the obvious disadvantages inherent in screening phage libraries in vivo by i.v. injection. We have generated an antibody phagemid library from a mouse immunized with luminal endothelial cell surface membranes isolated from rat lung and then demonstrated that the screening of this library on these membranes identifies numerous phage clones that are reactive to endothelial cell surface determinants in lung. We have overcome key problems in validating phages by creating a cassette-type system for rapidly converting phage-displayed antibodies into scFv-Fc probes, thereby avoiding phage scavenging by the liver in vivo and enabling assessment of probe effectiveness after i.v. injection. A crucial step during the selection of phage libraries is a rapid validation process that is done under in vivo conditions closely mimicking those expected to occur with therapeutics and imaging agents. Although intravenously injected TX3.833 antibody targets the lung rather specifically, TX3.833 phage fails to show any lung targeting, with nearly all viral particles being rapidly trapped by the spleen and the liver. To counter this trapping, multivalent recombinant antibody fragments may be used and offer much improved functional affinity over free scFvs (including reduced plasma clearance in vivo) (38, 39). Therefore, we chose to use scFv-Fc fusions because they benefit from all of the purification and detection tools already available for antibodies and have long serum half-lives (40). Together with a single cloning step between phagemid and scFv-Fc format, they greatly accelerate the validation of our phage lead candidates. Using this format, we have identified three antibodies, PH2, P7, and P35, that target the lung with an uptake between 30% and 40% ID per g. Concomitant blood levels are low, resulting in very high targeting indices and precise lung imaging by planar γ-scintigraphy. Using mass spectrometry as well as antibody precipitation and blotting, we identify many of the target antigens.
The technology presented here allows the rapid screening of large libraries of probes on luminal endothelial cell plasma membranes isolated directly from tissue to preserve native protein expression in vivo. This screening translates rapidly into successful “hits” validated through in vivo imaging. Because the screen is conducted ex vivo, the panning can be performed under conditions customary to phage-display technology and has shown classically to work well over the last two decades since its discovery (41-44). All aspects of the phage-display technology can be applied, apparently without limitation. In particular, several problems encountered during in vivo panning are no longer relevant including (i) the additional complexity introduced by substantial convective forces from blood flow on phage binding and retention at the luminal endothelial cell surface in vivo, (ii) the inability to block apriori high nonspecific binding of phage (“stickiness”), and (iii) the unwarranted randomness in the assay from the short contact time and incomplete exposure of the phage-display diversity to the endothelial cell surface of each organ, because nearly all of the i.v. injected phages are scavenged within minutes or less by the liver and spleen. As a rather large structure, the phage will experience significant hydrodynamic drag and may, in fact, be streamlined to a central coaxial profile, tending to be distributed away from the vessel wall where it can bind.
Whole-body imaging appears to be an important complement to ascertain targeting and, coupled with biodistribution analysis, provides a reasonably complete validation of targeting in vivo. The data so far provide an initial proof of principle for this approach, which, ultimately, may complement other robust technologies, including genomic and proteomic approaches (19, 20, 45), to explore more comprehensively endothelial molecular diversity in vivo and to realize more fully the prospects of the vascular targeting strategy to deliver molecular-imaging agents as well as drugs and gene vectors for enhancing noninvasive diagnosis and therapy of many important diseases.
Supplementary Material
Acknowledgments
We thank Traci Smith, Michelle Bourne, and Lisa Pang for technical assistance with the preparation of endothelial cell plasma membranes; Alex Wempren and Melinda Schnitzer for performing immunohistochemical analysis; Nancy L. Andon for mass-spectrometric analysis; and Tim Stinchcombe (Tera Biotech, Inc., San Diego) for generously providing the pSurfScrip vector. This research was supported by National Institutes of Health Grants HL58216, CA95893, CA97528, and CA104898 (to J.E.S.).
Author contributions: P.V. and J.E.S. designed research; P.V., J.D.G., J.E.T., M.B., P.O., and J.E.S. performed research; and P.V. and J.E.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APP, aminopeptidase P; ID per g, injected dose per gram of tissue; TTI, tissue targeting indexes.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. DQ321336-DQ321369).
References
- 1.Massoud, T. F. & Gambhir, S. S. (2003) Genes Dev. 17, 545-580. [DOI] [PubMed] [Google Scholar]
- 2.Herschman, H. R. (2003) Science 302, 605-608. [DOI] [PubMed] [Google Scholar]
- 3.Drews, J. (2000) Science 287, 1960-1964. [DOI] [PubMed] [Google Scholar]
- 4.Lindsay, M. A. (2003) Nat. Rev. Drug Discov. 2, 831-838. [DOI] [PubMed] [Google Scholar]
- 5.Cavenee, W. K. (2002) Carcinogenesis 23, 683-686. [DOI] [PubMed] [Google Scholar]
- 6.Rudin, M. & Weissleder, R. (2003) Nat. Rev. Drug Discov. 2, 123-131. [DOI] [PubMed] [Google Scholar]
- 7.Ranney, D. F. (1986) Biochem. Pharmacol. 35, 1063-1069. [DOI] [PubMed] [Google Scholar]
- 8.Tomlinson, E. (1987) Adv. Drug Del. Rev. 1, 87-198. [Google Scholar]
- 9.Dvorak, H. F., Nagy, J. A. & Dvorak, A. M. (1991) Cancer Cells 3, 77-85. [PubMed] [Google Scholar]
- 10.Jain, R. K. (1998) Nat. Med. 4, 655-657. [DOI] [PubMed] [Google Scholar]
- 11.von Mehren, M., Adams, G. P. & Weiner, L. M. (2003) Annu. Rev. Med. 54, 343-369. [DOI] [PubMed] [Google Scholar]
- 12.Farah, R. A., Clinchy, B., Herrera, L. & Vitetta, E. S. (1998) Crit. Rev. Eukaryotic Gene Expression 8, 321-356. [DOI] [PubMed] [Google Scholar]
- 13.Carver, L. A. & Schnitzer, J. E. (2003) Nat. Rev. Cancer 3, 571-581. [DOI] [PubMed] [Google Scholar]
- 14.Schnitzer, J. E. (1998) N. Engl. J. Med. 339, 472-474. [DOI] [PubMed] [Google Scholar]
- 15.Denekamp, J. (1984) Progr. Appl. Microcirc. 4, 28-38. [Google Scholar]
- 16.Burrows, F. J. & Thorpe, P. E. (1994) Pharmacol. Ther. 64, 155-174. [DOI] [PubMed] [Google Scholar]
- 17.Schnitzer, J. E. (2001) Adv. Drug Deliv. Rev. 49, 265-280. [DOI] [PubMed] [Google Scholar]
- 18.Carver, L. A. & Schnitzer, J. E. (2002) in Biomedical Aspects of Drug Targeting, eds. Muzykantov, V. & Torchilin, B. (Kluwer, Boston), pp. 107-128.
- 19.Oh, P., Li, Y., Yu, J., Krasinska, K., Carver, L. A., Teta, J. E. & Schnitzer, J. E. (2004) Nature 429, 629-635. [DOI] [PubMed] [Google Scholar]
- 20.Durr, E., Yu, J., Krasinska, K. M., Carver, L. A., Yates, J. R. I., Testa, J. E., Oh, P. & Schnitzer, J. E. (2004) Nat. Biotechnol. 22, 985-992. [DOI] [PubMed] [Google Scholar]
- 21.McIntosh, D. P., Tan, X.-Y., Oh, P. & Schnitzer, J. E. (2002) Proc. Natl. Acad. Sci. USA 99, 1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualini, R. & Ruoslahti, E. (1996) Nature 380, 364-366. [DOI] [PubMed] [Google Scholar]
- 23.Rajotte, D., Arap, W., Hagedorn, M., Koivunen, E., Pasqualini, R. & Ruoslahti, E. (1998) J. Clin. Invest. 102, 430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arap, W., Haedicke, W., Bernasconi, M., Kain, R., Rajotte, D., Krajewski, S., Ellerby, H. M., Bredesen, D. E., Pasqualini, R. & Ruoslahti, E. (2002) Proc. Natl. Acad. Sci. USA 99, 1527-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arap, W., Kolonin, M. G., Trepel, M., Lahdenranta, J., Cardo-Vila, M., Giordano, R. J., Mintz, P. J., Ardelt, P. U., Yao, V. J., Vidal, C. I., et al. (2002) Nat. Med. 8, 121-127. [DOI] [PubMed] [Google Scholar]
- 26.Pasqualini, R., Koivunen, E., Kain, R., Lahdenranta, J., Sakamoto, M., Stryhn, A., Ashmun, R. A., Shapiro, L. H., Arap, W. & Ruoslahti, E. (2000) Cancer Res. 60, 722-727. [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, T. A., Roberts, J. D. & Zakour, R. A. (1987) Methods Enzymol. 154, 367-382. [DOI] [PubMed] [Google Scholar]
- 28.Oh, P. & Schnitzer, J. E. (1998) in Cell Biology: A Laboratory Handbook, ed. Celis, J. (Academic, Orlando, FL), Vol. 2, pp. 34-45. [Google Scholar]
- 29.Schnitzer, J. E., McIntosh, D. P., Dvorak, A. M., Liu, J. & Oh, P. (1995) Science 269, 1435-1439. [DOI] [PubMed] [Google Scholar]
- 30.Schnitzer, J. E., Oh, P. & McIntosh, D. P. (1996) Science 274, 239-242. [DOI] [PubMed] [Google Scholar]
- 31.Oh, P., McIntosh, D. P. & Schnitzer, J. E. (1998) J. Cell Biol. 141, 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rondot, S., Koch, J., Breitling, F. & Dubel, S. (2001) Nat. Biotechnol. 19, 75-78. [DOI] [PubMed] [Google Scholar]
- 33.Stan, R. V., Ghitescu, L., Jacobson, B. S. & Palade, G. E. (1999) J. Cell Biol. 145, 1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stan, R. V., Kubitza, M. & Palade, G. E. (1999) Proc. Natl. Acad. Sci. USA 96, 13203-13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnitzer, J. E., Sung, A., Horvat, R. & Bravo, J. (1992) J. Biol. Chem. 264, 24544-24553. [PubMed] [Google Scholar]
- 36.Schnitzer, J. E. & Oh, P. (1994) J. Biol. Chem. 269, 6072-6082. [PubMed] [Google Scholar]
- 37.Schnitzer, J. E., Liu, J. & Oh, P. (1995) J. Biol. Chem. 270, 14399-14404. [DOI] [PubMed] [Google Scholar]
- 38.Pluckthun, A. & Pack, P. (1997) Immunotechnology 3, 83-105. [DOI] [PubMed] [Google Scholar]
- 39.Adams, G. P. & Schier, R. (1999) J. Immunol. Methods 231, 249-260. [DOI] [PubMed] [Google Scholar]
- 40.Powers, D. B., Amersdorfer, P., Poul, M., Nielsen, U. B., Shalaby, M. R., Adams, G. P., Weiner, L. M. & Marks, J. D. (2001) J. Immunol. Methods 251, 123-135. [DOI] [PubMed] [Google Scholar]
- 41.Smith, G. P. (1985) Science 228, 1315-1317. [DOI] [PubMed] [Google Scholar]
- 42.Winter, G., Griffiths, A. D., Hawkins, R. E. & Hoogenboom, H. R. (1994) Annu. Rev. Immunol. 12, 433-455. [DOI] [PubMed] [Google Scholar]
- 43.Hoogenboom, H. R., de Bruine, A. P., Hufton, S. E., Hoet, R. M., Arends, J. W. & Roovers, R. C. (1998) Immunotechnology 4, 1-20. [DOI] [PubMed] [Google Scholar]
- 44.Azzazy, H. M. & Highsmith, W. E., Jr. (2002) Clin. Biochem. 35, 425-445. [DOI] [PubMed] [Google Scholar]
- 45.St. Croix, B., Rago, C., Velculescu, V., Traverso, G., Romans, K. E., Montgomery, E., Lal, A., Riggins, G. J., Lengauer, C., Vogelstein, B. & Kinzler, K. W. (2000) Science 289, 1197-1202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.