Abstract
Bacterial cell-surface attachment of macromolecular complexes maintains the microorganism in close proximity to extracellular substrates and allows for optimal uptake of hydrolytic byproducts. The cellulosome is a large multienzyme complex used by many anaerobic bacteria for the efficient degradation of plant cell-wall polysaccharides. The mechanism of cellulosome retention to the bacterial cell surface involves a calcium-mediated protein-protein interaction between the dockerin (Doc) module from the cellulosomal scaffold and a cohesin (Coh) module of cell-surface proteins located within the proteoglycan layer. Here, we report the structure of an ultra-high-affinity (Ka = 1.44 × 1010 M-1) complex between type II Doc, together with its neighboring X module from the cellulosome scaffold of Clostridium thermocellum, and a type II Coh module associated with the bacterial cell surface. Identification of X module-Doc and X module-Coh contacts reveal roles for the X module in Doc stability and enhanced Coh recognition. This extremely tight interaction involves one face of the Coh and both helices of the Doc and comprises significant hydrophobic character and a complementary extensive hydrogen-bond network. This structure represents a unique mechanism for cell-surface attachment in anaerobic bacteria and provides a rationale for discriminating between type I and type II Coh modules.
Keywords: cellulose degradation, cellulosome, protein-protein interaction, calcium
Anaerobic bacteria rely on secreted hydrolytic enzymes for the breakdown of extracellular polysaccharides into carbon sources, which are readily taken up by the microbe and metabolized. Attachment of these enzymes to the bacterial cell surface, either as independent entities or components of multienzyme complexes, is a key mechanism for the optimal uptake of byproducts and, thus the viability of the microbe, through maintaining its close proximity to extracellular substrates and byproducts. One such complex is the cellulosome, which is responsible for the degradation of crystalline cellulose and associated plant cell-wall polysaccharides (1-6). The cellulosome from Clostridium thermocellum has been the most extensively studied of those identified to date and comprises various cellulases and hemicellulases anchored to a large, multimodular, noncatalytic scaffoldin subunit (CipA). An enzyme-associated calcium (Ca2+)-binding module termed type I Doc mediates enzyme attachment to the scaffoldin subunit through high affinity noncovalent interactions with its nine highly conserved type I cohesin (Coh) modules yet displays very little preference for particular CipA Coh modules (7, 8). The balance of the scaffoldin modular architecture includes a cellulose-binding domain, an X module of unknown function, and a C-terminal type II dockerin (Doc) module. Type II Doc tethers the cellulosome to the proteoglycan layer of the bacterial cell surface through high-affinity interactions with type II Coh modules of the surface-layer homology-containing cell-surface proteins SdbA, OlpB, and Orf2p (9-11). The specificity displayed between the type I and type II Coh-Doc interactions is thus critical to cellulosome assembly and attachment.
Identification of the molecular determinants dictating Coh binding and specificity for the Coh-Doc interaction has been aided by extensive structural and mutagenesis studies primarily focused on the type I interaction. The crystal structures of type I Coh modules from C. thermocellum (12, 13) and Clostridium cellulolyticum (14) display elongated β-barrel jellyroll topologies, whereas the NMR solution structure of type I Doc revealed a unique fold comprising two EF-hand-like Ca2+-binding motifs (15). Both Ca2+-binding segments of type I Doc are required for Coh recognition. Mutation of the conserved hydroxyl-containing residues at positions 10 and 11 in both Ca2+-binding loops resulted in a significant decrease in affinity (16-20). The type I complex structure showed that the hydroxyl groups of the serine and threonine residues in the second Ca2+-binding loop are part of an extensive hydrogen-bond network with a face of the Coh module comprising β-strands 8, 3, 6, and 5 (21). A structural rationale for mutations within the Coh module that affected Doc binding (22, 23) was also provided by the type I complex structure.
Biophysical and structural studies have begun to provide insight into the type II Coh-Doc interaction. Binding and Doc-mutagenesis studies have revealed an association constant (Ka) of 5.6 × 108 M-1 and the importance of positions 10 and 11 in the second Ca2+-binding loop for recognition of type II Coh (20, 24). Type II Doc undergoes a Ca2+-induced exposure of a hydrophobic surface and homodimerization of the X module-type II Doc modular pair (XDoc) (25). Addition of the type II Coh causes dissociation of the XDoc homodimer and formation of a stable 1:1 heterodimeric type II Coh-XDoc complex. Crystal structures of type II Coh modules from C. thermocellum (26), Acetivibrio cellulolyticus (27), and Bacteroides cellulosolvens (28) exhibit the β-jellyroll topology common to the type I Coh modules. A unique crowning α-helix, first identified by solution NMR studies (29), and two regions disrupting strands 4 and 8, termed”β-flaps,” were also observed and have been proposed to play roles in the type II interaction and specificity (26, 28). However, to unambiguously identify the underlying structural elements responsible for the type II interaction and type I-type II specificity, a type II Coh-Doc complex is needed.
Here, we describe the 2.1-Å resolution crystal structure of the heterodimeric SdbA type II Coh-XDoc complex from the cellulosome of C. thermocellum. This structure of an ultra-high-affinity type II Coh-Doc complex reveals intimate hydrophobic interface between type II Doc and the Ig-like X-module fold, giving the C-terminal region of the CipA scaffoldin subunit a rigid, elongated conformation. The complex interface has significant hydrophobic character and an extensive hydrogen-bonding network that involves residues from the X module, both Doc helices, and the 8-3-6-5 face of the Coh module. A structural rationale for type I-type II specificity is revealed by observed differences in Doc orientation, the physiochemical surface properties of the type I and II interfaces, and the role of the X module in the type II interaction. The type II Coh-XDoc structure also serves as a structural model for a unique mechanism of bacterial cell-surface attachment.
Results and Discussion
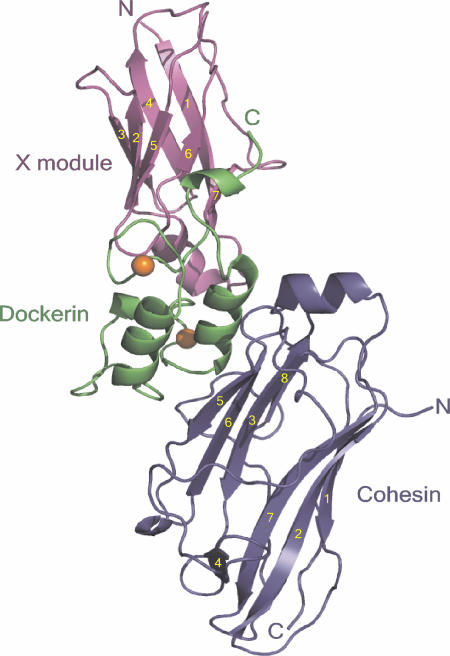
The crystal structure of the 1:1 type II Coh-XDoc complex was solved by using single-wavelength anomalous diffraction (SAD) data from a single selenomethionyl crystal (30). The atomic model refined at 2.1-Å resolution and the final statistics are summarized in Table 1, which is published as supporting information on the PNAS web site. The complex structure has overall dimensions of 88 Å × 45 Å × 31 Å and includes residues 17-182 from SdbA type II Coh (residues 28-193 from SWISSPROT accession P71143), the 163 C-terminal residues of the C. thermocellum CipA scaffoldin subunit (XDoc modular pair; residues 1697-1852 from SWISSPROT accession Q06851), 309 water molecules, and two Ca2+ ions (Fig. 1).
Fig. 1.
Structure of the type II Coh-XDoc complex. Ribbon representation of the complex with the type II Coh module in blue, type II Doc in green, and X module in magenta. The β-strands of the X module and type II Coh are numbered in yellow. The N and C termini are labeled accordingly, and the Ca2+ ions are depicted as orange spheres.
Type II Coh Structure in the Complex. The type II Coh module in the complex forms an elongated nine-stranded β-sandwich in a classical jellyroll topology with an extensive hydrophobic core (Fig. 1, blue). The two faces of the β-sandwich comprise strands 8, 3, 6, and 5 (8-3-6-5 face) and 9, 1, 2, 7, and 4 (9-1-2-7-4 face), respectively, all of which are aligned in an antiparallel arrangement, with the exception of strands 1 and 9, which are parallel to one another. The unique crowning α-helix between strands 6 and 7 and two β-flaps, regions that disrupt the normal course of strands 4 and 8 recently identified in the isolated type II Coh modules from A. cellulolyticus (27), B. cellulosolvens (27, 28) and C. thermocellum (26), are maintained in the complex. Comparison of the SdbA type II Coh structure in the absence and presence of the XDoc modular pair indicates that this module undergoes very little conformational change upon binding to the Doc module (backbone root mean square deviation (rmsd) of 0.99 Å), an observation that can be extended to all type II Coh modules, given the structural similarities of SdbA type II Coh from the complex and the type II Coh modules from A. cellulolyticus (backbone rmsd of 0.91 Å) and B. cellulosolvens (backbone rmsd of 0.96 Å).
Type II XDoc Modular Pair Structure in the Complex. The C-terminal construct from the scaffoldin subunit of the C. thermocellum cellulosome comprises an X module from the X60 family with unknown function and a type II Doc module (XDoc). The X module (residues 1-98) contains seven β-strands arranged in two sheets and a short α-helical region connecting strands 1 and 2. Strands 1, 4, and 7 and 2, 3, 5, and 6 form the two sheets of the β-sandwich, which is stabilized by a hydrophobic core and is reminiscent of an Ig-like fold (Fig. 1, magenta). The β-strand topology and overall fold of the X module is most similar to the Ig-like module of avian carboxypeptidase D domain II (31), displaying a backbone rmsd value of 1.64 Å to this module. The structure of the type II Doc module (residues 99-163) constitutes two loop-helix motifs, termed F-hand motifs (16), separated by a 14-residue linker region (Fig. 1, green). The 12-residue Ca2+-binding loop of each motif coordinates one Ca2+ ion in the typical pentagonal bipyramid configuration of EF-hand Ca2+-binding proteins (32, 33). Positions 1 (Asp-102; Asp-135), 3 (Asn-137), 5 (Asp-106; Asn-139), and 12 (Asp-113; Asp-146) provide side-chain carboxylate oxygen ligands to the Ca2+ ion. The backbone carbonyl oxygen of Val-104 at position 3 of the first loop directly coordinates the Ca2+, thereby compensating for the lack of the traditional Asn/Asp side-chain Ca2+-coordinating group at this position. The backbone carbonyl oxygen from residues at position 7 (Ser-108; Ala-141) and a bridged water molecule at position 9 (Asn-110; Asn-143) provide the final two coordinating ligands in both loops.
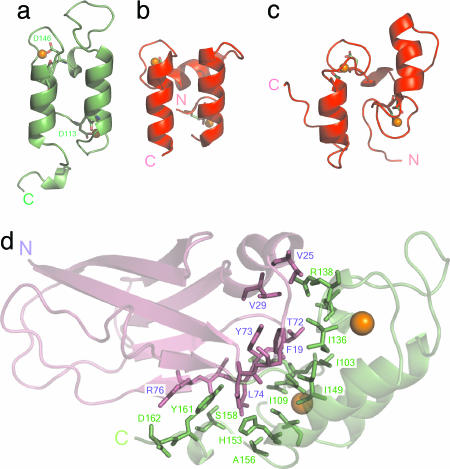
The two helices of the Doc module (I, Leu-111-Cys-120; II, Met-144-His-153) are arranged in an antiparallel orientation and form a large planar surface on one face of the XDoc structure. Because of this antiparallel organization, the two Ca2+-binding sites are located at opposite ends of the type II Doc module, similar to that observed for the type I Doc module (Fig. 2 a-c) (15, 21) yet dramatically different from typical EF-hand proteins, such as calmodulin, troponin C, and the S100 family (32, 34). In these latter cases, the Ca2+-binding loops of sequentially adjacent EF-hands form intimate contacts with one another, including a short antiparallel β-sheet. Short, extended interacting regions do exist between each Ca2+-binding loop and the exiting loop regions of the adjacent helices in type II Doc. These interactions were not observed in the type I-Doc structure and lead to notably different loop conformations at both ends of the helices (Fig. 2 a-c). The linker region is less structured than that of the Doc in the type I complex structure (21), comprising only a single helical turn (Glu-132-Ile-136), as opposed to the three turns of helix in the type I Doc, whereas the C-terminal region (Phe-154-Ala-163) is well ordered and folds back to form contacts with the X module.
Fig. 2.
Doc conformations and XDoc interface. The structures of type II Doc from the type II Coh-XDoc complex (a); type I Doc from the type I Coh-Doc complex (21) (b); and isolated type I Doc (15) (c) illustrate the differences in loop conformations. Coordinating residues at position 1 and 12 of each Ca2+-binding loop as stick models, with the Asp at position 12 in each loop of the type II-Doc structure labeled with one-letter code and the corresponding residue number. (d) The XDoc illustration shows the module-module contacts. Interface residues from the X module and type II Doc are depicted as magenta and green stick models, respectively, on the backbone ribbon representation of the XDoc structure.
The structure of the XDoc modular pair within the complex has an elongated conformation, consistent with previous sedimentation velocity studies of isolated Ca2+-XDoc (25) and due to an intimate interface between the X module and type II Doc. The interface is primarily hydrophobic in nature and involves Phe-19, Val-25, Val-29, Thr-72, Tyr-73, and Leu-74 from the X module and Gly-101, Ile-103, Ile-109, Ile-136, Arg-138, Ile-149, Ala-156, and Tyr-161 from type II Doc (Fig. 2d). Additional ionic- and hydrogen-bonding contacts stabilize the interface and include residues Asp-18, Asp-75, and Arg-76 and Asp-102, Lys-105, Glu-133, His-153, Ser-158, and Asp-162 from the X module and type II Doc, respectively.
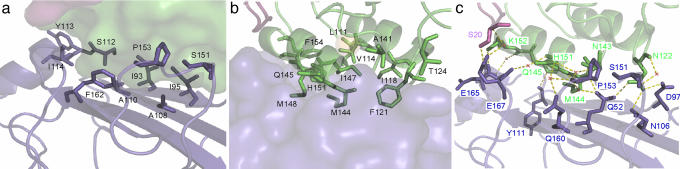
The Complex Interface. The type II Coh-Doc interface in the complex structure is located on the 8-3-6-5 face and the loop region leading into the helix between β-strands 6 and 7 of type II Coh and the planar surface formed by both loop-helix motifs of type II Doc. Residues Ile-93, Ile-95, Ala-108, Ala-110, Ser-112, Tyr-113, Ile-114, Pro-153, and Phe-162 from the type II Coh (Fig. 3a) and Leu-111, Val-114, Ile-118, Phe-121, Thr-124, Ala-141, Met-144, Gln-145, Ile-147, Met-148, and Phe-154 of the type II Doc (Fig. 3b) contribute to the pronounced hydrophobic character of the type II interaction. A significant hydrogen-bonding network also exists at the interface and includes both direct and water-mediated contacts between charged and polar, uncharged residues from the cognate modules (Fig. 3c; and see Table 2, which is published as supporting information on the PNAS web site). Interestingly, the X module also directly contributes to the complex interaction, where two hydrogen bonds were identified between Ser-20 of the X module and Glu-167 of type II Coh (Fig. 3c).
Fig. 3.
Type II Coh-XDoc complex interface contacts. (a) Ribbon representation of type II Coh, displaying hydrophobic interface residues as stick models on the molecular-surface representation of XDoc. (b) Ribbon representation of XDoc, displaying hydrophobic interface residues as stick models on the molecular-surface representation of type II Coh. (c) Interface hydrogen-bond network, with water molecules shown as red X and hydrogen-bond contacts as yellow dashed lines. Type II Coh, Doc, and X module are colored blue, green, and magenta, respectively. Residues depicted as stick models are labeled accordingly.
Comparison of Doc sequences has suggested that residues at positions 10 and 11 of the two Ca2+-binding loops play a role in type I-type II specificity. The side chain of Leu-111 at position 10 of the first Ca2+-binding loop in the type II Doc contributes to the hydrophobic content of the interaction through nonpolar van der Waals contacts with Ile-93 of type II Coh. Met-144 at position 10 of the second Ca2+-binding loop is hydrogen-bonded to Gln-52 (Oε2) and makes nonpolar contacts with the side chains of Ala-108, Ala-110, and Phe-162, whereas Gln-145 at position 11 forms hydrogen bonds with Gln-52 (Oε2) and Pro-153 (backbone O) and nonpolar contacts with Phe-162. These observations provide an explanation for the results of a recent site-directed mutagenesis study, which observed that mutating residues at both positions 10 and 11 (Met → Ser and Gln → Ser) in the second Ca2+-binding loop in type II Doc abrogates Coh recognition (20). Mutating the Met and Gln residues at sites 10 and 11 in the second loop would have a dramatic effect, because these residues are both located directly within the interface and form numerous nonpolar and hydrogen-bonding contacts with type II Coh.
Affinity of the Type II Coh-XDoc Interaction. Isothermal titration calorimetry and differential scanning calorimetry (DSC) were used to assess the binding affinity of the type II Coh-XDoc interaction in solution (see Supporting Methods, which is published as supporting information on the PNAS web site). Titration of XDoc into type II Coh at both 30°C and 55°C (see Fig. 5a, which is published as supporting information on the PNAS web site) showed that these proteins bind with a 1:1 stiochiometry and ΔH values of -21.7 and -25.5 kcal·mol-1, respectively. Given Wiseman's rule (35), the detection limits of this technique, and the very high affinity of the interaction, an accurate Ka could not be determined, and only a qualitative value of ≥109 M-1 could be assigned at both temperatures. DSC thermograms of the isolated type II Coh and XDoc constructs and the Coh-XDoc complex showed single transitions with melting temperatures of 80.37°C, 80.26°C, and 94.77°C, respectively (see Fig. 5b). By using these melting temperatures, the derived ΔH and ΔCp thermodynamic parameters for the individual type II Coh (54.1 kJ·mol-1; 2.1 kJ·K-1·mol-1) and XDoc (83.2 kJ·mol-1; 5.9 kJ·K-1·mol-1) constructs and an equation for a high-affinity protein-protein interaction that takes into account the temperature shift of both transitions (36), a Ka of 1.44 × 1010·M-1 for the type II Coh-XDoc interaction was determined. The affinity of this interaction is markedly higher than that for type II Doc lacking the X module, which was reported to have a Ka of 5.6 × 108·M-1 (24).
A Role for the X Module in Doc Stability and Cell-Surface Attachment. The elongated conformation of the C-terminal region of the C. thermocellum scaffoldin subunit, comprising the X module and type II Doc, would allow the cellulosome to extend away from the bacterial envelope when in contact with type II Coh. Roles in structural stability and enhanced solubility of cellulosomal components have been proposed for X modules (25, 37-39). Indeed, the interface between the X module and type II Doc provides a structural rationale for both of these functions in the CipA scaffoldin structure. Sequence analysis has also identified X modules directly adjacent to type II Doc modules in the A. cellulolyticus ScaA scaffoldin subunit and a putative secreted gene product from C. thermocellum (see Fig. 6, which is published as supporting information on the PNAS web site), which suggests that the intimate X module-Doc pairing is not reserved to the C. thermocellum CipA scaffoldin subunit but, rather, is a more common modular association in anaerobic bacteria.
The reported difference in affinity of type II Coh for the XDoc modular pair versus that for the isolated type II Doc also suggests a role for the X module in enhanced Coh recognition. Based on structural data presented here and conformational differences observed for type I Doc in the absence (15) and presence (21) of type I Coh, we propose that the increased affinity of the type II interaction is due to the X-module-mediated stabilization of the type II Doc structure in solution combined with the hydrogen-bond contacts that exist directly between the X module and type II Coh. A more rigid type II Doc structure in solution would lessen the entropic cost that would arise from a tightening of the isolated type II Doc structure upon type II Coh binding, as seen for the type I Coh-Doc interaction (15, 21). Furthermore, the increased hydrophobic character of the type II interaction would also suggest that desolvation of the binding surfaces upon complex formation will lead to a larger gain in entropy when compared with the type I interaction. The functional enhancement of adjacent modules also appears to be a more general role of the Ig-like X modules, because these modules conferred increased catalytic activity on endoglucanase CelD and cellobiohydrolase CbhA from C. thermocellum (38-40).
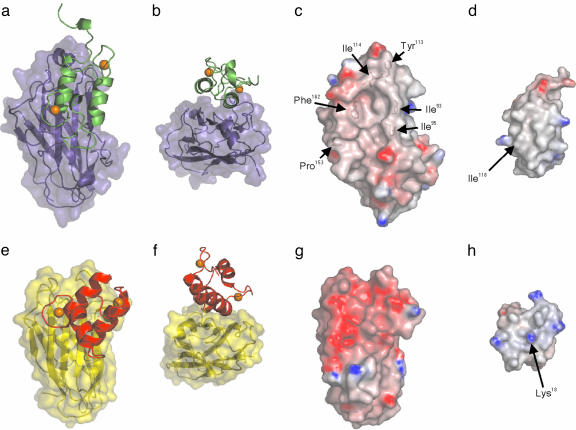
Structural Rationale for Specificity Among Coh-Doc Pairs. The specificity of the type I and type II Coh-Doc interactions is critical to the precise assembly of a functional cellulosome and its attachment to the bacterial surface. The type II Coh-XDoc complex structure allows for identification of structural elements dictating specificity through direct comparison with the type I complex. The Doc modules on the surfaces of type I and type II Coh have markedly distinct orientations. Type II Doc is in a parallel/antiparallel arrangement with the Coh module, whereas the orientation of the type I Doc is rotated clockwise ≈20° on the surface of type I Coh (Fig. 4 a and e). Furthermore, in the type II complex, both Doc helices contact the Coh surface over their entire length, whereas the Coh in the type I complex comes into contact with the entire length of helix II but only the C terminus of helix I from the type I Doc (Fig. 4 b and f). The N terminus of helix I from the type I Doc is diverted away from the Coh surface such that the analogous region of the type II Doc is 4 Å closer to the Coh surface. These orientational differences increase the occluded surface area of the interface from 1,612 Å2 in the type I complex to 1,696 Å2 in the type II complex. In their analysis of the type I Coh-Doc complex structure, Carvalho et al. (21) suggested the possibility of two type I Coh modules binding simultaneously to type I Doc, given the orientation of the Doc on the Coh surface and the two-fold symmetry that exists within the type I Doc structure. A similar mechanism for the type II Coh-Doc interaction can be discounted based on the multiple contacts made with the Coh module by both helices of the type II Doc and the lack of symmetry of type II Doc interface residues.
Fig. 4.
Interaction surfaces of type I- and type II Coh-Doc complexes. Ribbon representations of type II Doc (green) (a) on the molecular surface of the type II Coh (blue) and type I Doc (red) (e) on the molecular surface of type I Coh (yellow) (21). Representations in b and f have been rotated clockwise 90° around the x axis, followed by a 180° clockwise rotation around the z axis. Electrostatic surface potential representations of C. thermocellum type II Coh (c), C. thermocellum type II Doc (d), C. thermocellum type I Coh (21) (g), and C. thermocellum type I Doc (21) (h). Positive regions are shown in blue and negative regions in red. Residues contributing to the hydrophobic surface character of C. thermocellum type II Coh are labeled accordingly. The location of Ile-118 on the surface of the type II Doc and the analogous residue in the type I Doc (Lys-18) are identified. The electrostatic surface potentials were calculated in grasp (47) and are contoured from -14 (red) to +14 (blue). Ca2+ ions are shown as orange spheres.
Comparison of the electrostatic surface potentials of the recognition surfaces also reveals conspicuous differences between the type I and II interactions. The interacting surfaces in the type II complex are less charged than the analogous surfaces from the type I complex. Type II Coh has more bulky aliphatic side-chain groups, such as branched Ile, a cyclic Pro ring, and Phe and Tyr phenyl rings exposed at the Doc-recognition site, whereas the Doc-binding site on type I Coh is primarily negatively charged (Fig. 4 c and g). Several contacts with type II Doc occur in the C-terminal region of β-strand 8 (Gln-160; Phe-162; Asp-165, Gly-166, Glu-167) and the preceding type II-specific β-flap (Ser-151; Pro-153), whereas only a single contact is made to the C terminus of the Coh module (Glu-131) in the type I complex (21). The Coh-recognition site on type II Doc displays pronounced hydrophobic character, whereas the analogous surface on the type I Doc is more positively charged, made necessary by the negatively charged type I Coh (Fig. 4 d and h). This observation is prompted by the larger number of aliphatic side chains from both helices of type II Doc making direct contacts with type II Coh. The prominent positively charged residue Lys-18, which is located at the center of the Coh-recognition site of the type I Doc and makes a water-mediated hydrogen bond with Thr-66 from type I Coh (21), is highly conserved throughout the documented type I Doc sequences (12). In contrast, an Ile occupies the analogous position in the CipA type II Doc, leading to a contiguous aliphatic Coh-recognition surface and suggests that this residue has a role in type I-type II specificity.
Given the involvement of both Doc helices at the interface with type II Coh and the extent of the hydrophobic and hydrogen bond contacts identified in this study, we propose that, unlike the type I interaction, mutation of a small number of residues will not have as dramatic an effect on the affinity of the type II interaction. Size-exclusion chromatography and isothermal titration calorimetry (ITC) studies of the type II Coh-XDoc interaction, where mutations at positions 10 and 11 in either Ca2+-binding loop of XDoc (LL → ST and MQ → SS) or in both, have indicated that these mutants did not have an observable effect on the affinity of the interaction (data not shown). However, we cannot rule out that these mutations do, in fact, significantly decrease the affinity of Coh-XDoc interaction, although not to an extent that would be accurately detected by ITC or size-exclusion chromatography. These results are also in contrast to those of Schaeffer et al., where nondenaturing gel electrophoresis showed that the MQ → SS mutations in the second Ca2+-binding site abrogated the type II recognition (20). This discrepancy can be accounted for by the presence of the X module in the current study, which we have suggested imparts structural stability on type II Doc and enhances binding to type II Coh.
Carvalho et al. recently used amino acid sequence and structural comparisons of the Coh module from the type I Coh-Doc complex with type II Coh modules from C. thermocellum, A. cellulolyticus, and B. cellulosolvens to predict residues that would play a role in type II Doc recognition (26). The structure of the type II complex presented here reveals that, whereas several of the Coh-interface residues were correctly predicted, others were not (Asn-54, Ile-104, and Thr-149). These later residues are located at the C terminus of strand 3 (Asn-54), at the N terminus of strand 6 (Ile-104), and at the beginning of the β-flap preceding strand 8 (Thr-149) and form a contiguous surface adjacent to the Doc-binding site on the 8-3-6-5 face. Only a conformational reorientation of the β-flap or a complete shift of the type II Doc would put these residues in a position to take part in intermolecular contacts. Furthermore, these structural comparisons were not able to predict the increased number of contacts at the type II Coh-XDoc interface, or the unique orientation of the type II Doc on the Coh surface. Given these discrepancies, sequence- and structural-based models of Coh-Doc complexes are, at best, limited, and only x-ray or NMR structures of such complexes from various species will provide unambiguous structural insight into species-specificity.
The crystal structure of the type II Coh-XDoc complex has provided insight into the role of the X module in the interaction and identified the structural and physiochemical elements dictating this high-affinity interaction and their implications for type I-II specificity. The high affinity and specificity of the Coh-Doc interaction also suggests that this mechanism for cell-surface attachment might be conserved within anaerobic bacterial species. The identification of these modules in recently completed bacterial genomes, such as the pathogenic anaerobe Clostridium perfringens (41), indicate that this is, in fact, the case and that the type II Coh-XDoc structure serves as a general structural model for bacterial cell-surface attachment.
Methods
Crystallization and Structure Determination. Cloning, protein expression, and crystallization of the SdbA type II Coh-type II-XDoc module modular-pair complex and the collection, indexing, and scaling of diffraction data sets were performed as described in ref. 42. The positions of six selenium sites of a possible eight sites were identified and refined by the program solve (43). Phase calculation and density modification were carried out by using the program resolve (31). The resulting map was readily interpretable, with a single molecule of type II Coh-XDoc complex in the asymmetric unit. The model was built by using the program xfit (44) and refined by using the program cns (45) (see Table 1). The N-terminal His tag, the first three residues (Arg-14, Ala-15, and Asp-16), and the five C-terminal residues (Gly-183, Asp-184, Glu-185, Pro-186, and Phe-187) of the Coh module are disordered and, hence, not observed. Disordered residues in the XDoc structure include the first seven residues (Met-1, Asn-2, Lys-3, Pro-4, Val-5, Ile-6, and Glu-7), the final residue (Gln-164), and the C-terminal His tag. Structural figures were prepared by using the programs xfit (44), pymol, (46) and grasp (47). Sequence alignments were created by using the program clustalx (48). Rmsd of the various structures were determined by using pymol (46). Molecular surface areas were calculated by using areaimol from the Collaborative Computing Project (CCP)4 crystallography suite (49).
Supplementary Material
Acknowledgments
We thank Kim Munro from the Protein Function Discovery Facility at Queen's and Melanie Adams, Rob Campbell, and Olivier Julien fortechnical assistance. We acknowledge X6A and X8C beam lines at Brookhaven National Laboratory (Upton, NY) and F2 station at Cornell High Energy Synchrotron Source (Ithaca, NY) for synchrotron data collection. This work was supported by Canadian Institutes of Health Research Operating Grants MOP-7776 (to S.P.S.) and MOP-77810 (to Z.J.). Infrastructure used in these studies was funded through the Canadian Foundation for Innovation and Ontario Innovation Trust. J.J.A. was supported by a NSERC PGSD scholarship. Z.J. is the recipient of the Natural Sciences and Engineering Research Council (Canada) Steacie Fellowship and is a Canadian Research Chair in Structural Biology.
Author contributions: S.P.S. designed research; J.J.A. performed research; G.P.P. and Z.J. contributed new reagents/analytic tools; J.J.A., G.P.P., Z.J., and S.P.S. analyzed data; and J.J.A. and S.P.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Coh, cohesin; Doc, dockerin; Ka, association constant; rmsd, root mean square deviation; XDoc, X module-type II Doc modular pair.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2B59).
References
- 1.Bayer, E. A., Belaich, J. P., Shoham, Y. & Lamed, R. (2004) Annu. Rev. Microbiol. 58, 521-554. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., Chanzy, H., Lamed, R. & Shoham, Y. (1998) Curr. Opin. Struct. Biol. 8, 548-557. [DOI] [PubMed] [Google Scholar]
- 3.Beguin, P. & Lemaire, M. (1996) Crit. Rev. Biochem. Mol. Biol. 31, 201-236. [DOI] [PubMed] [Google Scholar]
- 4.Demain, A. L., Newcomb, M. & Wu, J. H. (2005) Microbiol. Mol. Biol. Rev. 69, 124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, R. H. & Kosugi, A. (2004) Nat. Rev. Microbiol. 2, 541-551. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz, W. H. (2001) Appl. Microbiol. Biotechnol. 56, 634-649. [DOI] [PubMed] [Google Scholar]
- 7.Fierobe, H. P., Mechaly, A., Tardif, C., Belaich, A., Lamed, R., Shoham, Y., Belaich, J. P. & Bayer, E. A. (2001) J. Biol. Chem. 276, 21257-21261. [DOI] [PubMed] [Google Scholar]
- 8.Fierobe, H. P., Mingardon, F., Mechaly, A., Belaich, A., Rincon, M. T., Pages, S., Lamed, R., Tardif, C., Belaich, J. P. & Bayer, E. A. (2005) J. Biol. Chem. 280, 16325-16334. [DOI] [PubMed] [Google Scholar]
- 9.Lemaire, M., Ohayon, H., Gounon, P., Fujino, T. & Beguin, P. (1995) J. Bacteriol. 177, 2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leibovitz, E. & Beguin, P. (1996) J. Bacteriol. 178, 3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leibovitz, E., Ohayon, H., Gounon, P. & Beguin, P. (1997) J. Bacteriol. 179, 2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavares, G. A., Beguin, P. & Alzari, P. M. (1997) J. Mol. Biol. 273, 701-713. [DOI] [PubMed] [Google Scholar]
- 13.Shimon, L. J., Bayer, E. A., Morag, E., Lamed, R., Yaron, S., Shoham, Y. & Frolow, F. (1997) Structure 5, 381-390. [DOI] [PubMed] [Google Scholar]
- 14.Spinelli, S., Fierobe, H. P., Belaich, A., Belaich, J. P., Henrissat, B. & Cambillau, C. (2000) J. Mol. Biol. 304, 189-200. [DOI] [PubMed] [Google Scholar]
- 15.Lytle, B. L., Volkman, B. F., Westler, W. M., Heckman, M. P. & Wu, J. H. (2001) J. Mol. Biol. 307, 745-753. [DOI] [PubMed] [Google Scholar]
- 16.Pages, S., Belaich, A., Belaich, J. P., Morag, E., Lamed, R., Shoham, Y. & Bayer, E. A. (1997) Proteins 29, 517-527. [PubMed] [Google Scholar]
- 17.Lytle, B. & Wu, J. H. (1998) J. Bacteriol. 180, 6581-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mechaly, A., Yaron, S., Lamed, R., Fierobe, H. P., Belaich, A., Belaich, J. P., Shoham, Y. & Bayer, E. A. (2000) Proteins 39, 170-177. [DOI] [PubMed] [Google Scholar]
- 19.Mechaly, A., Fierobe, H. P., Belaich, A., Belaich, J. P., Lamed, R., Shoham, Y. & Bayer, E. A. (2001) J. Biol. Chem. 276, 9883-9888. [DOI] [PubMed] [Google Scholar]
- 20.Schaeffer, F., Matuschek, M., Guglielmi, G., Miras, I., Alzari, P. M. & Beguin, P. (2002) Biochemistry 41, 2106-2114. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho, A. L., Dias, F. M., Prates, J. A., Nagy, T., Gilbert, H. J., Davies, G. J., Ferreira, L. M., Romao, M. J. & Fontes, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 13809-13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miras, I., Schaeffer, F., Beguin, P. & Alzari, P. M. (2002) Biochemistry 41, 2115-2119. [DOI] [PubMed] [Google Scholar]
- 23.Nakar, D., Handelsman, T., Shoham, Y., Fierobe, H. P., Belaich, J. P., Morag, E., Lamed, R. & Bayer, E. A. (2004) J. Biol. Chem. 279, 42881-42888. [DOI] [PubMed] [Google Scholar]
- 24.Jindou, S., Kajino, T., Inagaki, M., Karita, S., Beguin, P., Kimura, T., Sakka, K. & Ohmiya, K. (2004) Biosci. Biotechnol. Biochem. 68, 924-926. [DOI] [PubMed] [Google Scholar]
- 25.Adams, J. J., Webb, B. A., Spencer, H. L. & Smith, S. P. (2005) Biochemistry 44, 2173-2182. [DOI] [PubMed] [Google Scholar]
- 26.Carvalho, A. L., Pires, V. M., Gloster, T. M., Turkenburg, J. P., Prates, J. A., Ferreira, L. M., Romao, M. J., Davies, G. J., Fontes, C. M. & Gilbert, H. J. (2005) J. Mol. Biol. 349, 909-915. [DOI] [PubMed] [Google Scholar]
- 27.Noach, I., Lamed, R., Xu, Q., Rosenheck, S., Shimon, L. J., Bayer, E. A. & Frolow, F. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1670-1673. [DOI] [PubMed] [Google Scholar]
- 28.Noach, I., Frolow, F., Jakoby, H., Rosenheck, S., Shimon, L. W., Lamed, R. & Bayer, E. A. (2005) J. Mol. Biol. 348, 1-12. [DOI] [PubMed] [Google Scholar]
- 29.Smith, S. P., Beguin, P., Alzari, P. M. & Gehring, K. (2002) J. Biomol. NMR 23, 73-74. [DOI] [PubMed] [Google Scholar]
- 30.Adams, J. J., Pal, G., Yam, K., Jia, Z. & Smith, S. P. (2005) Acta Crystallogr. F 61, 46-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomis-Ruth, F. X., Companys, V., Qian, Y., Fricker, L. D., Vendrell, J., Aviles, F. X. & Coll, M. (1999) EMBO J. 18, 5817-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikura, M. (1996) Trends Biochem. Sci. 21, 14-17. [PubMed] [Google Scholar]
- 33.Strynadka, N. C. & James, M. N. (1989) Annu. Rev. Biochem. 58, 951-998. [DOI] [PubMed] [Google Scholar]
- 34.Lewit-Bentley, A. & Rety, S. (2000) Curr. Opin. Struct. Biol. 10, 637-643. [DOI] [PubMed] [Google Scholar]
- 35.Wiseman, T., Williston, S., Brandts, J. F. & Lin, L. N. (1989) Anal. Biochem. 179, 131-137. [DOI] [PubMed] [Google Scholar]
- 36.Brandts, J. F. & Lin, L. N. (1990) Biochemistry 29, 6927-6940. [DOI] [PubMed] [Google Scholar]
- 37.Mosbah, A., Belaich, A., Bornet, O., Belaich, J. P., Henrissat, B. & Darbon, H. (2000) J. Mol. Biol. 304, 201-217. [DOI] [PubMed] [Google Scholar]
- 38.Kataeva, I. A., Uversky, V. N., Brewer, J. M., Schubot, F., Rose, J. P., Wang, B. C. & Ljungdahl, L. G. (2004) Protein Eng. Des. Sel. 17, 759-769. [DOI] [PubMed] [Google Scholar]
- 39.Schubot, F. D., Kataeva, I. A., Chang, J., Shah, A. K., Ljungdahl, L. G., Rose, J. P. & Wang, B. C. (2004) Biochemistry 43, 1163-1170. [DOI] [PubMed] [Google Scholar]
- 40.Juy, M., Amit, A. G., Alzari, P. M., Poljak, R. J., Claeyssens, M., Beguin, P. & Aubert, J. P. (1992) Nature 357, 89-91. [Google Scholar]
- 41.Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., Ogasawara, N., Hattori, M., Kuhara, S. & Hayashi, H. (2002) Proc. Natl. Acad. Sci. USA 99, 996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams, J. J., Jang, C. J., Spencer, H. L., Elliott, M. & Smith, S. P. (2004) Protein Expr. Purif. 38, 258-263. [DOI] [PubMed] [Google Scholar]
- 43.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D Biol Crystallogr. 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McRee, D. E. (1999) J. Struct. Biol. 125, 156-165. [DOI] [PubMed] [Google Scholar]
- 45.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 46.DeLano, W. L. (2002) The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 47.Petrey, D. & Honig, B. (2003) Methods Enzymol. 374, 492-509. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D 53, 240-255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.