Abstract
Accumulating in vitro evidence suggests that the p38 mitogen-activated protein kinase (MAPK) pathway is involved in endochondral ossification. To investigate the role of this pathway in endochondral ossification, we generated transgenic mice with expression in chondrocytes of a constitutively active mutant of MKK6, a MAPK kinase that specifically activates p38. These mice had a dwarf phenotype characterized by reduced chondrocyte proliferation, inhibition of hypertrophic chondrocyte differentiation, and a delay in the formation of primary and secondary ossification centers. Histological analysis with in situ hybridization showed reduced expression of Indian hedgehog, PTH/PTH-related peptide receptor (PTH, parathyroid hormone), cyclin D1, and increased expression of p21 in chondrocytes. In addition, both in vivo and in transfected cells, p38 signaling increased the transcriptional activity of Sox9, a transcription factor essential for chondrocyte differentiation. In agreement with this observation, transgenic mice that express a constitutively active mutant of MKK6 in chondrocytes showed phenotypes similar to those of mice that overexpress SOX9 in chondrocytes. These observations are consistent with the notion that increased activity of Sox9 accounts at least in part for the phenotype caused by constitutive activation of MKK6 in chondrocytes. Therefore, our study provides in vivo evidence for the role of p38 in endochondral ossification and suggests that Sox9 is a likely downstream target of the p38 MAPK pathway.
Keywords: ossification, p38, Sox9
Endochondral ossification, a process involving a cartilage intermediate, is responsible for the formation of most vertebrate skeletal elements. After the condensation of mesenchymal chondroprogenitor cells (1), cells differentiate into chondrocytes, which express cartilaginous matrix molecules and form cartilage that prefigures future skeletal elements. Endochondral bone growth takes place at the growth plate, where chondrocytes undergo unidirectional proliferation and then become hypertrophic chondrocytes. Hypertrophic chondrocytes eventually undergo apoptosis and are replaced by bone cells (2). This complex process of endochondral ossification is under the concerted regulation of various cytokines and growth factors, including fibroblast growth factors (FGFs), parathyroid hormone (PTH)-related peptide, Indian hedgehog (Ihh), and bone morphogenetic proteins (3–7).
Several transcription factors, including Sox9, Sox5, Sox6, Osterix, and Runx2, have critical roles in endochondral ossification (8–12). In particular, we previously showed that Sox9 has an essential role at sequential steps in the chondrocyte differentiation pathway (8, 9). Indeed, Sox9 is needed for the condensation of chondrogenic mesenchymal cells; it is also required for the overt differentiation of these cells into chondrocytes, in part because Sox9 is needed for the expression of Sox5 and Sox6, which are also needed at this step. A further role for Sox9 is inhibition of the proliferation of chondrocytes and of the transition of these cells into hypertrophic chondrocytes. Identification of the mechanisms that control expression and activity of Sox9 would provide important insights into the regulation of endochondral ossification.
We previously showed that the extracellular signal-regulated kinase (ERK)1/ERK2 mitogen-activated protein kinase (MAPK) pathway mediates the up-regulation of Sox9 expression by FGF in mouse primary chondrocytes (13). The MAPK pathways, which are activated by various stimuli in eukaryotic cells, transduce extracellular signals into cells, thereby coordinating appropriate responses. The MAPK pathways are generally organized into three kinase modules, i.e., MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). MAPKKK phosphorylates and activates MAPKK, which in turn phosphorylates and activates MAPK (14, 15). A number of reports have implicated the role of the MAPK pathways in chondrocyte differentiation. Indeed, we recently showed that constitutive activation of the MEK1–MAPK pathway in chondrocytes in transgenic mice inhibits hypertrophic chondrocyte differentiation and causes a dwarf phenotype without a decrease in cell proliferation (16). The p38 MAPK pathway, another subfamily of the MAPK pathways, has also been shown to affect chondrocyte differentiation in chondrocyte cultures (17–23). p38 MAPKs consist of four isoforms, all of which are activated by MKK6, an upstream MAPKK (24). A number of in vitro studies have suggested that p38 potentially regulates chondrocyte proliferation. However, both proliferative and antiproliferative effects have been reported based on the effects of p38 inhibitors in a number of culture systems (18, 21, 25, 26). Therefore, examining how increased or decreased p38 activity in chondrocytes affects chondrocyte differentiation, proliferation, and the entire process of endochondral ossification in intact animals is of considerable interest.
To study the role of p38 in chondrocyte differentiation in vivo,we generated transgenic mice that express in chondrocytes a constitutively active mutant of MKK6, a MAPKK that specifically activates p38. These mice had a dwarf phenotype characterized by reduced chondrocyte proliferation, inhibition of hypertrophic chondrocyte differentiation, and a delay in the formation of primary and secondary ossification centers. Signaling by p38 also increased the activity of a Sox9-dependent reporter. Therefore, our study provides in vivo evidence for the role of p38 in endochondral ossification and suggests that Sox9 is a likely downstream target of the p38 MAPK pathway.
Results
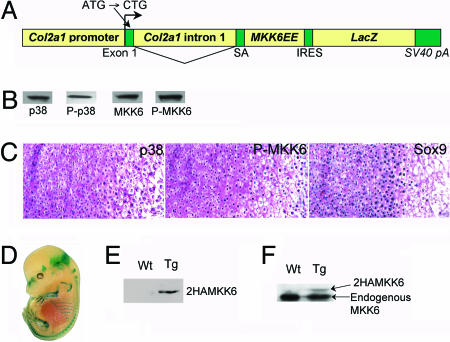
p38 and MKK6 Expression in Chondrocytes. Western blot analysis in primary chondrocytes showed that p38 and MKK6, as well as their phosphorylated forms, were present in primary chondrocytes (Fig. 1B). Immunohistochemistry further indicated that both p38 and phosphorylated MKK6 were present in chondrocytes throughout the growth plate, suggesting that the p38 MAPK pathway is active in these cells (Fig. 1C). As shown previously, Sox9 was expressed in chondrocytes throughout the growth plate, except hypertrophic chondrocytes (Fig. 1C); hence, MKK6, p38, and Sox9 are present in chondrocytes in the growth plate.
Fig. 1.
Generation of transgenic mice that express MKK6EE, a constitutive active MKK6, in chondrocytes. (A) Schematic representation of the construct driving expression of MKK6EE in chondrocytes. The original initiation codon of Col2a1 was mutated to CTG to facilitate translation from downstream cDNA. (B) Western blot analysis showed that p38, MKK6, and their phosphorylated forms were present in primary chondrocytes. The negative control using nonimmune IgG showed virtually no signal. (C) Immunohistochemistry showed that p38, phosphorylated MKK6, and Sox9 were present in growth-plate chondrocytes. (D) X-Gal staining of an embryo at E14.5 showed cartilage-specific expression of the transgene. (E) Western blot analysis using an anti-HA antibody showed that MKK6EE that is tagged with two copies of an HA epitope (2HAMKK6) is expressed in primary chondrocytes isolated from the ribs of transgenic mice at P1. (F) Western blot analysis using an anti-MKK6 antibody showed both endogenous MKK6 and transgene MKK6 (2HAMKK6) in primary chondrocytes from transgenic mice. Wt, wild type; Tg, transgenic.
Generation of Transgenic Mice. To examine the role of the p38 MAPK pathway in chondrocytes, we generated transgenic mice that express in chondrocytes a constitutively active MKK6 mutant (MKK6EE). The LacZ gene preceded by an internal ribosome entry site was placed downstream from the MKK6EE cDNA (Fig. 1A). Eight of 10 founder mice expressed the transgene based on X-Gal staining of tail cartilage. X-Gal staining of embryonic day 14.5 (E14.5) embryos further showed that the transgene was expressed in a cartilage-specific manner (Fig. 1D). Histological examination showed that the transgene was expressed in all chondrocytes with reduced staining in hypertrophic chondrocytes (data not shown). Western blot analysis of primary chondrocytes isolated from transgenic mice with an anti-hemagglutinin (HA) antibody showed that the MKK6EE that is tagged with two copies of the HA epitope (2HAMKK6) was expressed in chondrocytes (Fig. 1E), whereas Western blot analysis with an anti-MKK6 antibody showed that 2HAMKK6 migrated more slowly than the endogenous MKK6 (Fig. 1F). The expression level of the mutant MKK6 was 5–10% of that of the endogenous MKK6 in homozygous transgenic mice.
Expression of a Constitutively Active Mutant of MKK6 in Chondrocytes Delays Endochondral Bone Formation. Transgenic mouse lines were established from five founder mice that were selected based on the intensity of X-Gal staining of their tail cartilage. These heterozygous transgenic mice displayed normal growth; however, mice homozygous for the transgene from four of the five lines had a dwarf phenotype. One line with the most intense X-Gal staining and a marked dwarf phenotype was used for further analysis (Fig. 2A). Skeletal preparations showed that transgenic mice had a shortened axial and appendicular skeleton (Fig. 2B). At 3 weeks of age, the average length of long bones was decreased by ≈20% and 30% in male and female transgenic mice, respectively (Table 1, which is published as supporting information on the PNAS web site).
Fig. 2.
Transgenic mice that express a constitutively active MKK6 in chondrocytes showed a dwarf phenotype. (A) Transgenic (Tg) and wild-type (Wt) littermates at P21. (B) Skeletal preparation of the transgenic and wild type littermates at P21.
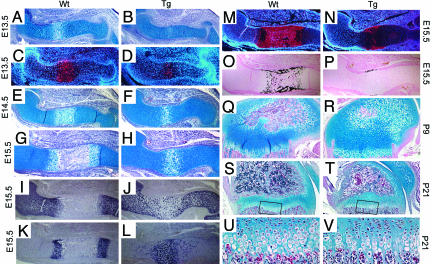
Epiphyseal growth plates of the long bones were examined histologically at various time points between E13.5 and postnatal day 21 (P21). At E13.5, hypertrophic chondrocytes were present in wild type but were absent in transgenic embryos (Fig. 3 A and B). At E14.5, hypertrophic chondrocytes were seen both in wild type and transgenic embryos at the center of long bones; however, the zone of hypertrophic chondrocytes was markedly reduced in the transgenic embryos (Fig. 3 E and F). These results strongly suggested a delay in formation of hypertrophic chondrocytes. This was further confirmed by in situ hybridization for Col10a1, a specific marker for hypertrophic chondrocytes (Fig. 3 C and D). We also examined expression of osteopontin, a marker for both terminally differentiated chondrocytes and osteoblasts, by in situ hybridization at E15.5. In wild-type embryos, osteopontin was expressed in terminally differentiated hypertrophic chondrocytes as well as in osteoblasts, whereas in transgenic embryos, much less osteopontin was expressed (Fig. 3 M and N), supporting the view that constitutive activation of MKK6 inhibits hypertrophic chondrocyte differentiation. At E15.5, the zone of hypertrophic chondrocytes was separated by the developing primary ossification center in wild-type embryos, whereas no bone tissues had replaced the hypertrophic chondrocytes in transgenic embryos, indicating a delay in the formation of the primary ossification center (Fig. 3 G and H). Consistent with the histological analysis, the expression domains of Col2a1 were separated by bone tissues at the center of long bones in wild-type embryos, whereas those in transgenic embryos were continuous with reduced expression at the center where hypertrophic chondrocytes were present (Fig. 3 I and J). Similarly, expression domains of Col10a1 were separated by bone tissues in the growth plates of wild-type embryos, whereas Col10a1 expression was observed at the center corresponding to the zone of hypertrophic chondrocytes in transgenic embryos (Fig. 3 K and L). von Kossa staining further demonstrated that cartilage mineralization and bone collar formation were delayed in transgenic embryos (Fig. 3 O and P). At P9, the secondary ossification center in the middle of the epiphysis was much smaller in transgenic than in wild-type mice, indicating a delay in the formation of the secondary ossification center (Fig. 3 Q and R). At P21, the overall size of the epiphysis and secondary ossification center was much reduced in transgenic mice (Fig. 3 S and T). In addition, the zone of hypertrophic chondrocytes in the growth plate was also reduced in transgenic mice (Fig. 3 U and V). These results indicated that constitutive activation of the p38 MAPK pathway in chondrocytes delays hypertrophic chondrocyte maturation as well as the formation of both primary and secondary ossification centers.
Fig. 3.
Delayed formation of both primary and secondary ossification centers in transgenic mice that express a constitutively active MKK6 in chondrocytes. Alcian blue staining of the humerus at E13.5 for Wt (A) and Tg (B); at E14.5 for Wt (E) and Tg (F); at E15.5 for Wt (G) and Tg (H). Alcian blue staining of femur at P9 for Wt (Q) and Tg (R), at P21 for Wt (S) and Tg (T). In situ hybridization at E13.5 for Col10a1 for Wt (C) and Tg (D), at E15.5 for Col2a1 for Wt (I) and Tg (J), Col10a1 for Wt (K) and Tg (L), and Osteopontin for Wt (M) and Tg (N). von Kossa staining of humerus at E15.5 for Wt (O) and Tg (P). Magnified view of the zone of hypertrophic chondrocytes at P21 for Wt (U) and Tg (V) corresponding to the boxed areas in S and T. For each time point, multiple endochondral bones were examined, and consistent results were obtained. Wt, wild type; Tg, transgenic.
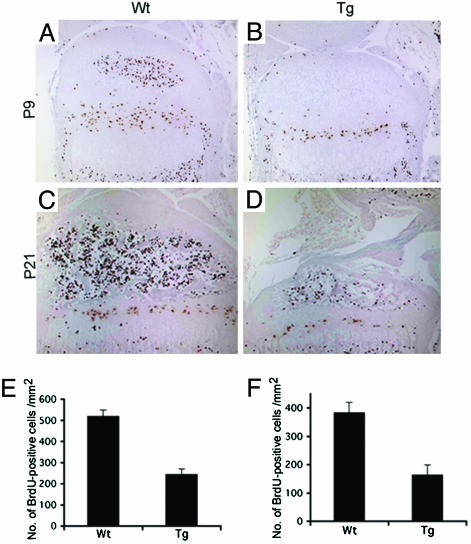
Chondrocyte Proliferation Is Reduced in Transgenic Mice. To examine the mechanisms that caused the dwarf phenotype, we examined chondrocyte proliferation and apoptosis. To examine chondrocyte proliferation, mice were killed 2.5 h after i.p. BrdUrd injection. Immunostaining was then performed to detect cells that incorporated BrdUrd in the proximal growth plates of the tibia at P9 and P21 (Fig. 4). At P9, the number of BrdUrd-incorporating cells in the proliferating zone was 520 ± 24 and 244 ± 20 cells/mm2 for wild-type and transgenic mice, respectively; the percentage of BrdUrd-incorporating cells was 18.6% and 10.6%, respectively, indicating statistically significant reduction in chondrocyte proliferation in transgenic mice (P < 0.01). At P21, chondrocyte proliferation was also reduced in transgenic mice (P < 0.01). At P21, the number of BrdUrd-incorporating cells in the proliferating zone was 385 ± 19 and 165 ± 31 cells/mm2 for wild-type and transgenic mice, respectively; the percentage of BrdUrd-incorporating cells was 12.0% and 5.1%, respectively. These observations suggested that reduced chondrocyte proliferation could account for the dwarf phenotype of the transgenic mice. To examine apoptosis, we performed TUNEL assay for mice at E14.5, E15.5, P9, and P21. No significant difference of apoptosis was detected between wild-type and transgenic mice (data not shown).
Fig. 4.
Reduced chondrocyte proliferation in transgenic mice that express a constitutively active MKK6 in chondrocytes. Immunostaining to detect cells that incorporated BrdUrd in the proximal tibial growth plates at P9 for Wt (A) and Tg (B) and at P21 for Wt (C) and Tg (D). Numbers of BrdUrd-incorporating cells in the proliferating zone were significantly reduced in transgenic mice (P < 0.01) at both P9 (E) and P21 (F). Wt, wild type; Tg, transgenic.
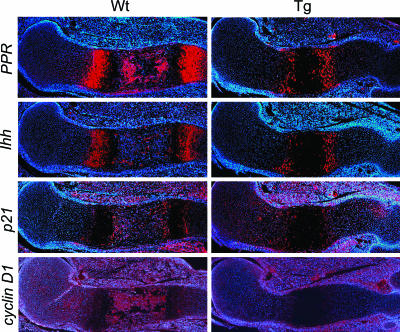
Expression of PTH/PTH-Related Peptide Receptor (PPR), Ihh, Cyclin D1, and p21. To further characterize the phenotype of transgenic mice, we examined the expression of PPR and Ihh, which have been shown to positively regulate chondrocyte proliferation (27, 28). We found that PPR and Ihh expression was reduced in transgenic embryos at E15.5 (Fig. 5), although it is possible that this reduction may be linked to the delay in hypertrophic chondrocyte differentiation. We further examined the cell cycle regulators cyclin D1 and p21. In wild-type embryos, cyclin D1 expression was detected in the resting and proliferating chondrocytes, with more intense signals in bone tissues. However, in transgenic embryos, cyclin D1 expression in chondrocytes was reduced (Fig. 5). This is consistent with previous reports of other cell types showing inhibitory effects of p38 on cyclin D1 expression (29, 30). In contrast, expression of p21 was increased in the zones of proliferating and resting chondrocytes in transgenic embryos (Fig. 5). The decreased expression of cyclin D1 and increased expression of p21 in transgenic embryos are consistent with the phenotype of reduced chondrocyte proliferation.
Fig. 5.
In situ hybridization at E15.5 for PPR, Ihh, p21, and cyclin D1. Expression of a constitutively active MKK6 down-regulated PPR, Ihh, cyclin D1, and up-regulated p21 in chondrocytes. Wt, wild type; Tg, transgenic.
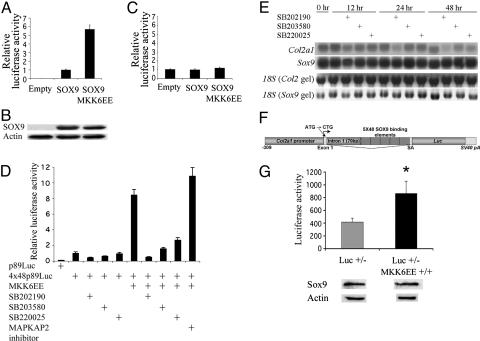
p38 MAPK Increases Sox9 Transcriptional Activity in Transient Transfection and in Transgenic Mice. Because Sox9 has an essential role in chondrocyte differentiation, we examined whether SOX9 activity is affected by the p38 MAPK pathway. COS7 cells, which do not express endogenous Sox9, were transfected with a SOX9 expression vector and a luciferase construct harboring four tandem copies of a SOX9-dependent 48-bp enhancer in Col2a1. Luciferase expression in COS7 cells transfected with this plasmid depends completely on SOX9. Coexpression of MKK6EE further increased the activity of the SOX9-dependent Col2a1 enhancer by ≈5-fold (Fig. 6A). Western blot analysis showed that SOX9 protein levels were not increased under these conditions (Fig. 6B). This increase in enhancer activity was not observed in a mutant enhancer construct harboring mutations that abolish SOX9 binding and virtually eliminate enhancer activity (Fig. 6C). These data strongly suggested that activation of the p38 pathway results in increased activity of SOX9. In addition, the increased activity of the SOX9-dependent Col2a1 enhancer by MKK6EE was also observed in primary chondrocytes; this increase was abolished by the p38-specific inhibitors, SB202190, SB203580, and SB220025 (Fig. 6D). Furthermore, a >5-fold increase in enhancer activity was also observed with the coexpression of a constitutively active mutant of MKK3, another MAPKK that specifically activates p38; this increase was abolished by the coexpression of the p38 phosphatase, MKP5 (data not shown). It is noteworthy that these p38-specific inhibitors decreased the activity of the SOX9-dependent Col2a1 enhancer in primary chondrocytes not transfected with MKK6EE (Fig. 6D), suggesting that the p38 pathway plays a role in maintaining Sox9 activity in these cells. Treatment of primary chondrocytes with p38 inhibitors decreased the expression of Col2a1, a downstream target of Sox9, without affecting Sox9 RNA levels (Fig. 6E). This observation is consistent with the hypothesis that p38 plays a role in regulating Sox9 activity in chondrocytes.
Fig. 6.
p38 MAPK increases Sox9 transcriptional activity in transient transfection and in transgenic mice. (A) In COS7 cells, coexpression of MKK6EE further increased the activity of the SOX9-dependent 48-bp enhancer by ≈5-fold. (B) Western blot analysis showed that SOX9 protein levels were not increased under these conditions. (C) This increase in enhancer activity was not observed in a mutant enhancer construct harboring mutations that abolish SOX9 binding. Note that in A (wild-type enhancer) and C (mutant enhancer), luciferase activity obtained in the presence of SOX9 was set to 1. The actual values of luciferase were >100 times higher with the wild-type enhancer than with the mutant enhancer in the presence of SOX9. (D) In primary chondrocytes, coexpression of MKK6EE further increased the activity of the SOX9-dependent 48-bp enhancer by >5-fold, and this increase was abolished by 10 μM of p38-specific inhibitors, SB202190, SB203580, and SB220025. In contrast, 10 μM of an inhibitor for MAPK-activated protein kinase 2 (MAPKAP2), a downstream kinase of p38, did not abolish the increased enhancer activity, suggesting that MAPKAP2 is not involved. (E) Northern blot analysis showed that treatment of primary chondrocytes with the p38 inhibitors decreased the expression of Col2a1, a downstream target of Sox9, without affecting Sox9 RNA levels, further supporting the hypothesis that p38 plays a role in regulating Sox9 activity in chondrocytes. (F) Schematic representation of the construct used to generate transgenic mice harboring a luciferase reporter under the control of a 309-bp Col2a1 promoter and five copies of the 48-bp Col2a1 enhancer. (G) In mice that were homozygous for the MKK6EE transgene (MKK6EE+/+) and also heterozygous for the luciferase transgene (Luc+/-), luciferase activity in cartilage extracts showed a 2-fold increase (P < 0.05) compared with cartilage extracts of newborn mice without the MKK6EE transgene, and levels of Sox9 were comparable. This strongly suggested that Sox9 activity was increased in transgenic mice that express MKK6EE in chondrocytes. Western blot analysis for Sox9 was performed for multiple litters, and consistent results were obtained. Luciferase activity was expressed as the value for 1 μg of total protein.
To examine whether Sox9 activity was increased in chondrocytes of transgenic mice that express MKK6EE in these cells, we generated mice harboring a luciferase reporter under the control of a 309-bp Col2a1 promoter and five copies of the 48-bp Col2a1 enhancer (Fig. 6F). The 309-bp Col2a1 promoter is not active by itself in chondrocytes of transgenic mice, and its activity is not affected by SOX9 in transient transfection experiments (31). In mice that were homozygous for the MKK6EE transgene and that also contained the luciferase transgene, luciferase activity in cartilage extracts showed a 2-fold increase compared with cartilage extracts from newborn mice without the MKK6EE transgene (P < 0.05), and the levels of Sox9 were comparable (Fig. 6G). These results are compatible with the notion that, as in DNA transfection experiments, Sox9 activity was increased in transgenic mice that express MKK6EE in chondrocytes.
To examine the mechanism of the increase in SOX9 activity, fusion polypeptides were generated that fused the SOX9 segment (residues 402–475) to the Gal4 DNA-binding domain (Fig. 7, which is published as supporting information on the PNAS web site). In cotransfection experiments in COS7 cells, the fusion protein still showed increased activity when coexpressed with MKK6EE (Fig. 7). This suggested that the segment of SOX9 from 402 to 475 was involved in the p38-mediated increased Sox9 activity. A mutant was generated in which serine-to-alanine mutations were introduced in all five of the potential MAPK phosphorylation sites within this segment. These five mutations in the mutant did not abolish the increased activity caused by MKK6EE (Fig. 7), suggesting that the increase in SOX9 activity was not due to direct phosphorylation of SOX9 by p38. These observations raised the possibility that the increase in SOX9 activity by p38 could be mediated through phosphorylation of a coactivator.
Discussion
The p38 MAPK pathway has recently emerged as another MAPK pathway that has a role in endochondral ossification based on evidence in tissue culture cells. To investigate the role of the p38 MAPK pathway in chondrocytes in vivo, we generated transgenic mice that express a constitutively active mutant of MKK6 in chondrocytes. Here we show that constitutive activation of MKK6 signaling in chondrocytes results in dwarfism and a delay in endochondral ossification, associated with reduced chondrocyte proliferation and inhibition of hypertrophic chondrocyte differentiation.
During chondrocyte differentiation, chondrocytes undergo unidirectional proliferation, resulting in parallel stacks of cells, and subsequently undergo hypertrophy. This process is largely responsible for the longitudinal growth of bones (32). Therefore, reduced chondrocyte proliferation could account, at least in part, for the dwarf phenotype in these mice. To gain an understanding of the mechanism of reduced chondrocyte proliferation, we examined expression of Ihh, PPR, and cell cycle regulators, cyclin D1 and p21, which have been demonstrated to have a role in chondrocyte proliferation (27, 28, 33–35). Signals for Ihh, PPR, and cyclin D1 RNAs were decreased, whereas p21 was up-regulated in the transgenic embryos that express MKK6EE in chondrocytes. Therefore, these results strongly suggest that the p38 MAPK pathway might coordinately regulate Ihh and PPR as well as the cell cycle regulators cyclin D1 and p21 to inhibit chondrocyte proliferation.
Because Sox9 has been demonstrated to play an essential role in chondrocyte differentiation and also functions to inhibit chondrocyte proliferation, we examined whether p38 signaling affects either Sox9 activity or expression. A role of p38 in regulating Sox9 activity has been suggested in limb mesenchymal cells in culture (23). In agreement with these observations, cotransfection experiments with SOX9 and MKK6EE, or a constitutively active mutant of MKK3 in COS7 cells and primary chondrocytes, suggested that SOX9 activity was increased in these cells. We also present evidence consistent with an increase in Sox9 activity in chondrocytes of transgenic mice that express MKK6EE in these cells. Interestingly, the phenotypes of these transgenic mice were similar to those of mice with increased expression of SOX9 in chondrocytes (36). Mice from both lines showed similar dwarfism characterized by a delay in hypertrophic chondrocyte differentiation and endochondral ossification, decreased chondrocyte proliferation, and reduced cyclin D1 expression. Therefore, the similarity in phenotypes between the two mouse lines supports the notion that increased Sox9 activity has a role in the decreased chondrocyte proliferation in transgenic mice that express MKK6EE. Our previous work indicated that the transcriptional activity of SOX9 was increased by PTH-related peptide (PTHrP) (46), supporting the notion that Sox9 might mediate, at least in part, the effect of PTHrP in inhibiting hypertrophic chondrocyte differentiation. Transgenic mice that express MKK6EE showed a delay in hypertrophic chondrocyte differentiation, despite a reduced expression of PPR. We hypothesize that the increase in the transcriptional activity of Sox9 by MKK6 will, in large part, cause the inhibition of proliferation and the delay in hypertrophy, despite the apparent decrease in the expression of PPR.
Previous in vitro studies have implicated both the p38 and ERK1/ERK2 MAPK pathways in chondrocyte differentiation. Recently, we generated transgenic mice that express a constitutively active mutant of MEK1 in chondrocytes (16). These mice showed a dwarf phenotype characterized by inhibition of hypertrophic chondrocyte differentiation and a delay in endochondral ossification. Interestingly, these mice did not show reduced chondrocyte proliferation, which is in remarkable contrast with the phenotype of transgenic mice that express a constitutively active mutant of MKK6. These observations strongly suggest that the ERK1/ERK2 and p38 MAPK pathways play distinct roles in endochondral ossification.
Various cytokines and growth factors have been shown to activate the p38 MAPK pathway in chondrocytes. These include FGFs, bone morphogenetic proteins, TGF-β, and IL-1 (18, 37–39). Among these, FGFs have been shown to negatively regulate chondrocyte proliferation and bone growth. Activating mutations of FGF receptor 3 (FGFR3) cause the most common forms of human dwarfism: achondroplasia, hypochondroplasia, and thanatophoric dysplasia (40–42). Our previous study of transgenic mice that express a constitutively active mutant of MEK1 in chondrocytes strongly suggested that the ERK1/ERK2 MAPK pathway plays an important role in FGFR3 signaling (16). However, despite the presence of an achondroplasia-like dwarfism characterized by inhibition of hypertrophic chondrocyte differentiation and a delay in endochondral ossification, these mice did not show reduced chondrocyte proliferation, indicating the presence of additional pathways that act downstream from FGFR3 to inhibit chondrocyte proliferation. Consistent with these observations, the reduced chondrocyte proliferation in mice that express a human achondroplasia mutant of FGFR3 was at least partially rescued by loss of Stat1, another signaling molecule implicated in FGFR3 signaling. Because FGF also activates the p38 MAPK pathway in chondrocytes (ref. 18; data not shown), it is possible that antiproliferative effects of FGFR3 are mediated by both p38 and Stat1. In addition, p38 may also have a role in the delayed endochondral ossification caused by increased FGFR3 signaling.
Taken together, our results provide in vivo evidence that the p38 MAPK pathway delays endochondral ossification and reduces chondrocyte proliferation. Histological analysis using in situ hybridization showed that p38 signaling in chondrocytes down-regulates Ihh, PPR, and cyclin D1 and up-regulates p21. In addition, p38 signaling increases the activity of Sox9, a master transcription factor in chondrocyte differentiation. Further biochemistry and genetic experiments to analyze the role of the p38 MAPK pathway may provide important insights into the mechanisms of endochondral ossification and human skeletal disorders.
Materials and Methods
Mice. To express the transgene in chondrocytes, the cDNA encoding an HA-tagged constitutively active mutant of MKK6 (S207E/T211E) was cloned into a vector containing 3 kb of the Col2a1 promoter and 3.02 kb of intron 1 sequences (31). The LacZ gene preceded by an internal ribosome entry site was placed downstream from the MKK6 mutant cDNA (Fig. 1A). The construct was microinjected into the pronuclei of fertilized B6D2F1 hybrid eggs to generate transgenic mice. X-Gal staining of tail cartilage was used to identify transgene-expressing founder mice. Copy numbers of the LacZ gene were estimated by using real-time PCR and used to identify transgenic mice that were homozygous for the transgene.
Transgenic mice containing a Sox9-dependent luciferase reporter gene were generated to examine the activity of endogenous Sox9 in chondrocytes. This transgene contains a 309-bp Col2a1 promoter, followed by Col2a1 exon 1, part of intron 1, in which five copies of a 48-bp chondrocyte enhancer were inserted before a splice acceptor and the luciferase reporter. Homogenized cartilage obtained from newborn mice was used to perform the luciferase assays.
Histological Analysis. Tissues were fixed in 10% formalin and embedded in paraffin. Seven-micrometer-thick sections were stained with hematoxylin/eosin and alcian blue. Immunohistochemical staining was performed by using Peroxidase-conjugated polymer (Zymed) and TrueBlue substrate (KPL). The following antibodies were used: Sox9 (43), p38, and phospho-MKK3/MKK6 (Cell Signaling Technology). RNA in situ hybridization was performed by using digoxigenin- or 35S-UTP labeled riboprobes. To examine cell proliferation, BrdUrd staining kit (Zymed) was used to stain BrdUrd-incorporated cells for the mice killed 2.5 h after i.p. BrdUrd injection.
Plasmids, Cell Culture, and Transient Transfections. The cDNA encoding human SOX9 was cloned into the pGL3-control plasmid (Promega) to replace the luciferase gene. The 4 × 48-p89 luciferase construct and its MA6 mutant were described previously (44, 45). For some experiments, cells were incubated with 10 μM of one of the p38 inhibitors, SB202190, SB203580, and SB220025 or the inhibitor of MAPK-activated protein kinase 2 (Calbiochem) for the last 12 h. Cell culture and transient transfections were described previously (13).
Supplementary Material
Acknowledgments
We thank Eisuke Nishida (Kyoto University, Kyoto) for MKK6EE and MKP5 cDNA and Haruhiko Akiyama (Kyoto University), Andrew McMahon (Harvard University, Boston), Henry Kronenberg (Massachusetts General Hospital, Boston), Tamayuki Shinomura (Tokyo Medical and Dental University, Tokyo), and Stephen Elledge (Baylor College of Medicine, Waco, TX) for probes of cyclin D1, Ihh, PPR, Col10a1, and p21, respectively. We also thank Chad Smith and Zhaoping Zhang for pronuclear injections. This work was funded by National Institutes of Health Grant P01 AR42919 (to B.d.C.) and an Arthritis Investigator Award from the Arthritis Foundation (to S.M.). We also acknowledge National Institutes of Health Grant CA16672 for DNA sequence analysis.
Author contributions: R.Z., S.M., and B.d.C. designed research; R.Z. and Y.W. performed research; F.C. contributed new reagents/analytic tools; R.Z., S.M., and B.d.C. analyzed data; and R.Z., S.M., and B.d.C. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FGF, fibroblast growth factor; PTH, parathyroid hormone; Ihh, Indian hedgehog; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; ERK, extracellular signal-regulated kinase; En, embryonic day n; Pn, postnatal day n; PPR, PTH/PTH-related peptide receptor; FGFR3, FGF receptor 3; HA, hemagglutinin.
References
- 1.Tacchetti, C., Tavella, S., Dozin, B., Quarto, R., Robino, G. & Cancedda, R. (1992) Exp. Cell Res. 200, 26-33. [DOI] [PubMed] [Google Scholar]
- 2.Cancedda, R., Descalzi Cancedda, F. & Castagnola, P. (1995) Int. Rev. Cytol. 159, 265-358. [DOI] [PubMed] [Google Scholar]
- 3.de Crombrugghe, B., Lefebvre, V. & Nakashima, K. (2001) Curr. Opin. Cell Biol. 13, 721-727. [DOI] [PubMed] [Google Scholar]
- 4.Olsen, B. R., Reginato, A. M. & Wang, W. (2000) Annu. Rev. Cell Dev. Biol. 16, 191-220. [DOI] [PubMed] [Google Scholar]
- 5.Cancedda, R., Castagnola, P., Cancedda, F. D., Dozin, B. & Quarto, R. (2000) Int. J. Dev. Biol. 44, 707-714. [PubMed] [Google Scholar]
- 6.DeLise, A. M., Fischer, L. & Tuan, R. S. (2000) Osteoarthritis Cartilage 8, 309-334. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima, K. & de Crombrugghe, B. (2003) Trends Genet. 19, 458-466. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama, H., Chaboissier, M. C., Martin, J. F., Schedl, A. & de Crombrugghe, B. (2002) Genes Dev. 16, 2813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. (1999) Nat. Genet. 22, 85-89. [DOI] [PubMed] [Google Scholar]
- 10.Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L. & Karsenty, G. (1997) Cell 89, 747-754. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R. & de Crombrugghe, B. (2002) Cell 108, 17-29. [DOI] [PubMed] [Google Scholar]
- 12.Smits, P., Li, P., Mandel, J., Zhang, Z., Deng, J. M., Behringer, R. R., de Crombrugghe, B. & Lefebvre, V. (2001) Dev. Cell 1, 277-290. [DOI] [PubMed] [Google Scholar]
- 13.Murakami, S., Kan, M., McKeehan, W. L. & de Crombrugghe, B. (2000) Proc. Natl. Acad. Sci. USA 97, 1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobb, M. H. & Goldsmith, E. J. (1995) J. Biol. Chem. 270, 14843-14846. [DOI] [PubMed] [Google Scholar]
- 15.Robinson, M. J. & Cobb, M. H. (1997) Curr. Opin. Cell Biol. 9, 180-186. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, S., Balmes, G., McKinney, S., Zhang, Z., Givol, D. & de Crombrugghe, B. (2004) Genes Dev. 18, 290-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura, K., Shirai, T., Morishita, S., Uchida, S., Saeki-Miura, K. & Makishima, F. (1999) Exp. Cell Res. 250, 351-363. [DOI] [PubMed] [Google Scholar]
- 18.Shimoaka, T., Ogasawara, T., Yonamine, A., Chikazu, D., Kawano, H., Nakamura, K., Itoh, N. & Kawaguchi, H. (2002) J. Biol. Chem. 277, 7493-7500. [DOI] [PubMed] [Google Scholar]
- 19.Stanton, L. A., Sabari, S., Sampaio, A. V., Underhill, T. M. & Beier, F. (2004) Biochem. J. 378, 53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanton, L. A., Underhill, T. M. & Beier, F. (2003) Dev. Biol. 263, 165-175. [DOI] [PubMed] [Google Scholar]
- 21.Halawani, D., Mondeh, R., Stanton, L. A. & Beier, F. (2004) Oncogene 23, 3726-3731. [DOI] [PubMed] [Google Scholar]
- 22.Zhen, X., Wei, L., Wu, Q., Zhang, Y. & Chen, Q. (2001) J. Biol. Chem. 276, 4879-4885. [DOI] [PubMed] [Google Scholar]
- 23.Weston, A. D., Chandraratna, R. A., Torchia, J. & Underhill, T. M. (2002) J. Cell Biol. 158, 39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nebreda, A. R. & Porras, A. (2000) Trends Biochem. Sci. 25, 257-260. [DOI] [PubMed] [Google Scholar]
- 25.Raucci, A., Laplantine, E., Mansukhani, A. & Basilico, C. (2004) J. Biol. Chem. 279, 1747-1756. [DOI] [PubMed] [Google Scholar]
- 26.Krejci, P., Bryja, V., Pachernik, J., Hampl, A., Pogue, R., Mekikian, P. & Wilcox, W. R. (2004) Exp. Cell Res. 297, 152-164. [DOI] [PubMed] [Google Scholar]
- 27.Amizuka, N., Warshawsky, H., Henderson, J. E., Goltzman, D. & Karaplis, A. C. (1994) J. Cell Biol. 126, 1611-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St-Jacques, B., Hammerschmidt, M. & McMahon, A. P. (1999) Genes Dev. 13, 2072-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awad, M. M., Enslen, H., Boylan, J. M., Davis, R. J. & Gruppuso, P. A. (2000) J. Biol. Chem. 275, 38716-38721. [DOI] [PubMed] [Google Scholar]
- 30.Page, K., Li, J. & Hershenson, M. B. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 280, L955-64. [DOI] [PubMed] [Google Scholar]
- 31.Zhou, G., Garofalo, S., Mukhopadhyay, K., Lefebvre, V., Smith, C. N., Eberspaecher, H. & de Crombrugghe, B. (1995) J. Cell Sci. 108, 3677-3684. [DOI] [PubMed] [Google Scholar]
- 32.Wilsman, N. J., Farnum, C. E., Leiferman, E. M., Fry, M. & Barreto, C. (1996) J. Orthop. Res. 14, 927-936. [DOI] [PubMed] [Google Scholar]
- 33.Lanske, B., Karaplis, A. C., Lee, K., Luz, A., Vortkamp, A., Pirro, A., Karperien, M., Defize, L. H., Ho, C., Mulligan, R. C., et al. (1996) Science 273, 663-666. [DOI] [PubMed] [Google Scholar]
- 34.Beier, F., Ali, Z., Mok, D., Taylor, A. C., Leask, T., Albanese, C., Pestell, R. G. & LuValle, P. (2001) Mol. Biol. Cell. 12, 3852-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su, W. C., Kitagawa, M., Xue, N., Xie, B., Garofalo, S., Cho, J., Deng, C., Horton, W. A. & Fu, X. Y. (1997) Nature 386, 288-292. [DOI] [PubMed] [Google Scholar]
- 36.Akiyama, H., Lyons, J. P., Mori-Akiyama, Y., Yang, X., Zhang, R., Zhang, Z., Deng, J. M., Taketo, M. M., Nakamura, T., Behringer, R. R., et al. (2004) Genes Dev. 18, 1072-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ionescu, A. M., Schwarz, E. M., Zuscik, M. J., Drissi, H., Puzas, J. E., Rosier, R. N. & O'Keefe, R. J. (2003) Exp. Cell Res. 288, 198-207. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, S., Votta, B. J., Rieman, D. J., Badger, A. M., Gowen, M. & Lee, J. C. (2001) J. Cell Physiol. 187, 294-303. [DOI] [PubMed] [Google Scholar]
- 39.Nishihara, A., Fujii, M., Sampath, T. K., Miyazono, K. & Reddi, A. H. (2003) Biochem. Biophys. Res. Commun. 301, 617-622. [DOI] [PubMed] [Google Scholar]
- 40.Rousseau, F., Bonaventure, J., Legeai-Mallet, L., Pelet, A., Rozet, J. M., Maroteaux, P., Le Merrer, M. & Munnich, A. (1994) Nature 371, 252-254. [DOI] [PubMed] [Google Scholar]
- 41.Rousseau, F., Saugier, P., Le Merrer, M., Munnich, A., Delezoide, A. L., Maroteaux, P., Bonaventure, J., Narcy, F. & Sanak, M. (1995) Nat. Genet. 10, 11-12. [DOI] [PubMed] [Google Scholar]
- 42.Bellus, G. A., McIntosh, I., Smith, E. A., Aylsworth, A. S., Kaitila, I., Horton, W. A., Greenhaw, G. A., Hecht, J. T. & Francomano, C. A. (1995) Nat. Genet. 10, 357-359. [DOI] [PubMed] [Google Scholar]
- 43.Lefebvre, V., Huang, W., Harley, V. R., Goodfellow, P. N. & de Crombrugghe, B. (1997) Mol. Cell. Biol. 17, 2336-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, G., Lefebvre, V., Zhang, Z., Eberspaecher, H. & de Crombrugghe, B. (1998) J. Biol. Chem. 273, 14989-14997. [DOI] [PubMed] [Google Scholar]
- 45.Lefebvre, V., Zhou, G., Mukhopadhyay, K., Smith, C. N., Zhang, Z., Eberspaecher, H., Zhou, X., Sinha, S., Maity, S. N. & de Crombrugghe, B. (1996) Mol. Cell. Biol. 16, 4512-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang, W., Chung, U. I., Kronenberg, H. M. & de Crombrugghe, B. (2001) Proc. Natl. Acad. Sci. USA 98, 160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.