Abstract
PA-824 is a promising new compound for the treatment of tuberculosis that is currently undergoing human trials. Like its progenitors metronidazole and CGI-17341, PA-824 is a prodrug of the nitroimidazole class, requiring bioreductive activation of an aromatic nitro group to exert an antitubercular effect. We have confirmed that resistance to PA-824 (a nitroimidazo-oxazine) and CGI-17341 (a nitroimidazo-oxazole) is most commonly mediated by loss of a specific glucose-6-phosphate dehydrogenase (FGD1) or its deazaflavin cofactor F420, which together provide electrons for the reductive activation of this class of molecules. Although FGD1 and F420 are necessary for sensitivity to these compounds, they are not sufficient and require additional accessory proteins that directly interact with the nitroimidazole. To understand more proximal events in the reductive activation of PA-824, we examined mutants that were wild-type for both FGD1 and F420 and found that, although these mutants had acquired high-level resistance to PA-824 (and another nitroimidazo-oxazine), they retained sensitivity to CGI-17341 (and a related nitroimidazo-oxazole). Microarray-based comparative genome sequencing of these mutants identified lesions in Rv3547, a conserved hypothetical protein with no known function. Complementation with intact Rv3547 fully restored sensitivity to nitroimidazo-oxazines and restored the ability of Mtb to metabolize PA-824. These results suggest that the sensitivity of Mtb to PA-824 and related compounds is mediated by a protein that is highly specific for subtle structural variations in these bicyclic nitroimidazoles.
Keywords: comparative genome sequencing, F420, nitroimidazole
Despite the availability of curative chemotherapy and a widely used vaccine, the World Health Organization estimates that two billion people are infected with Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), and that two million people die of the disease each year (www.who.int/gtb). Curative TB chemotherapy consists of a 6-month-long regimen with at least four drugs. No new drug has been introduced into this regimen since the mid-1970s, when rifampicin was first used (1).
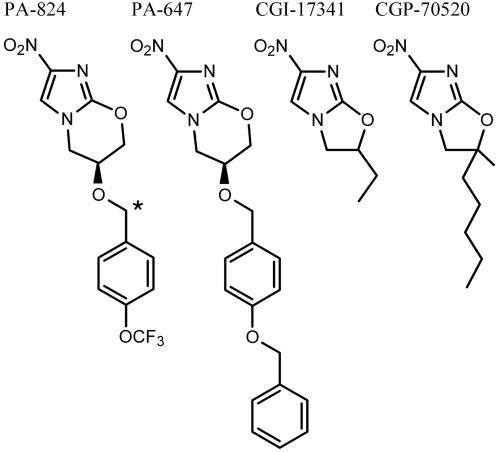
Nitroimidazoles are widely used drugs in humans for a variety of primarily anaerobic microbial infections. Metronidazole, a 5-nitroimidazole, is an important bactericidal agent for the treatment of anaerobic infections (2) and shows excellent selective toxicity toward anaerobic bacterial and protozoal pathogens (3). This class of compounds has only recently begun to be explored for Mtb, because only anaerobic activity of metronidazole against Mtb has been reported (4, 5). Bicyclic 4-nitroimidazoles such as PA-824 (a nitroimidazo-oxazine) and CGI-17341 (a nitroimidazo-oxazole) (Fig. 1) have inhibitory activity against aerobically growing and nonreplicating anaerobic Mtb (6). Although anaerobic conditions have not been demonstrated during TB disease in humans, various authors have suggested that an anaerobic microenvironment may contribute to a nonreplicating state that may be linked with latent disease in humans (7). Thus, PA-824 has been developed, in part, because it may be a promising lead for therapy against latent disease that may be linked to anaerobically persisting bacilli. The Global Alliance for TB Drug Development has recently initiated phase-I clinical trials with PA-824 (www.tballiance.org).
Fig. 1.
Chemical structures of the 4-nitroimidazoles used in this study. Asterisk denotes the position of 14C in radiolabeled PA-824.
In all cases where mechanistic information is available, the reductive activation of an aromatic nitro group is a prerequisite step for biological function in this class of molecules. Even though the biological target(s) of 5-nitroimidazoles is not entirely clear, it is known that the nitro group is reduced within the cell, forming a reactive intermediate that can damage DNA and other cellular components (3). Bioreductive activation of PA-824 has been shown to depend on Rv0407, which encodes an F420-dependent, glucose-6-phosphate dehydrogenase (FGD1) (6). Cofactor F420 has 7,8-didemethyl-8-hydroxy-5-deazariboflavin, a phospholactyl moiety, and a variable number (two to six) of glutamate residues (8). F420 has a limited distribution among the archaea and GC-rich Gram-positive bacteria, shows a low redox potential (E′=-350 mV), and functions as a two-electron redox-carrier coenzyme (9).
Results
Generation and Characterization of PA-824-Resistant Mutants. The in vitro frequency of PA-824-resistant mutation is ≈6.5 × 10-7, similar to isoniazid, and does not vary significantly with growth phase of the organism (data not shown). To determine the in vivo frequency of emergence of resistance, 46 mice with established Mtb infections were treated with PA-824 monotherapy for 2 weeks at 25 and 50 mg/kg. Organ homogenates from 9 of these animals yielded PA-824-resistant mutants. These infected animals had total colony-forming-unit burdens of ≈6.5 × 106 bacilli. The minimal inhibitory concentration for all characterized mutants was at least 100 times higher than that of wild-type H37Rv.
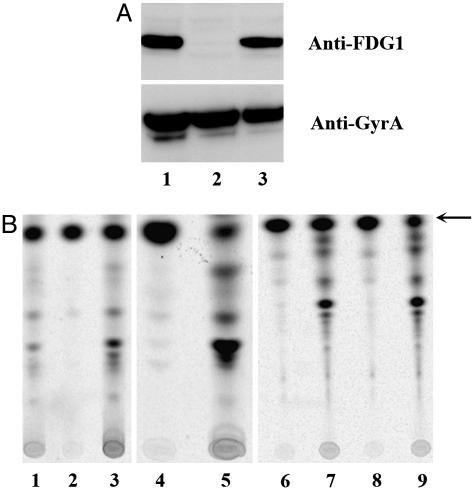
Ten in vitro and 10 in vivo mutants of Mtb strain H37Rv were further characterized for the presence of FGD1 protein by Western blotting and the presence of cofactor F420 or its biosynthetic precursor FO by HPLC. FO is a biosynthetic intermediate containing a deazaflavin ring and ribityl sugar but lacking both the phospholactyl group and glutamate moieties. Twenty percent of the in vivo PA-824-resistant mutants failed to produce FGD1 protein (Fig. 2A and Table 1), and 60% lacked cofactor F420 (Fig. 3 B and C). Ten percent of these mutants had an accumulation of FO (Fig. 3C). The remaining 20% of the mutants were FGD1+ and F420+. Mutants of all three phenotypes (FGD1-, F420-, and FGD1+/F420+) were found to be unable to chemically convert 14C-labeled PA-824 into more polar metabolites (Fig. 2B).
Fig. 2.
FGD and PA-824 activation assays of PA-824-resistant mutants. (A) Western blot analysis of cell lysates from FGD1- mutant with polyclonal anti-FGD1 antibody (Upper) and polyclonal antibody to GyrA (Lower). (B) TLC analysis of 14C-labeled PA-824 metabolism. Whole bacterial cells were exposed to 1.8 μCi [14C]-PA-824 for 3 h and then lysed mechanically. Lysates were analyzed by silica gel TLC. Unconverted [14C]-PA-824 is denoted by the arrow. Strains are detailed in Table 1: Lanes 1-9 are H37Rv, H37Rv-T3, H37Rv-T3-fgd1, H37Rv-5A1, H37Rv-5A1-fbiC, H37Rv-T2, H37Rv-T2-Rv3547, H37Rv-14A1, and H37Rv-14A1-Rv3547, respectively.
Table 1. Characterization of a few representative Mtb H37Rv PA-824-resistant mutants.
| Minimal inhibitory concentration, μM
|
|||||||
|---|---|---|---|---|---|---|---|
| No. | Name | Phenotype | Genotype* | PA-824 | PA-647 | CGI-17341 | CGP-70520 |
| 1 | H37Rv | FGD1+; F420+; 824+ | – | 0.4–0.8 | 0.1–0.2 | 0.4 | 0.1 |
| 2 | H37Rv-T3 | FGD1-; F420+; 824- | na | >100 | >100 | >100 | >100 |
| 2a | H37Rv-T3-fgd1 | FGD1+; F420+; 824+ | fgd1::attP | 0.8 | 0.2 | 0.8 | 0.2 |
| 3 | H37Rv-5A1 | FGD1+; F420-; 824- | Δ1fbiC; Δ2PDIM | >100 | >100 | >100 | >100 |
| 3a | H37Rv-5A1-fbiC | FGD1+; F420+; 824+ | Δ1fbiC; Δ2PDIM; fbiC::attP | 0.4 | 0.2 | 0.8 | 0.2 |
| 4 | H37Rv-5A2 | FGD1+; F420- | fbiCV630E; fadD26D76A | >100 | >100 | >100 | >100 |
| 5 | H37Rv-7A2 | FGD1+; F420-; FO++; 824- | Unknown | >100 | >100 | >100 | >100 |
| 5a | H37Rv-7A2-fbiAB | FGD1+; F420+; 824+ | fbiAB::attP | 0.8 | 0.2 | 0.8 | 0.2 |
| 6 | H37Rv-T2 | FGD1+; F420+; 824- | Rv3547A76E | >100 | >100 | 0.8 | 3.2 |
| 6a | H37Rv-T2–3547 | FGD1+; F420+; 824+ | Rv3547A76E; Rv3547::attP | 1.6 | 0.2–0.4 | 0.4 | na |
| 7 | H37Rv-14A1 | FGD1+; F420+; 824- | Δ3Rv3547; Δ4ppsB | >100 | >100 | 0.8 | 3.2 |
| 7a | H37Rv-14A1–3547 | FGD1+; F420+; 824+ | Δ3Rv3547; Δ4ppsB; Rv3547::attP | 0.8–1.6 | 0.2 | 0.4 | na |
| 8 | H37Rv-Tn-824R#1 | FGD1+, F420+; 824- | Rv3547Y89ochre; 2258008::Tn | >100 | >100 | na | na |
| 9 | H37Rv-Tn-824R#4 | FGD1+, F420+; 824- | Rv3547::Tn | >100 | >100 | 0.8 | na |
| 9a | H37Rv-Tn-824R#4–3547 | FGD1+, F420+ | Rv3547::Tn; Rv3547::attP | 0.8 | 0.8 | 0.8 | na |
na, Not analyzed; 824+/- refers to PA-824 metabolism; Δ1, 400-bp deletion; Δ2, 66-kbp deletion; Δ3, a single bp deletion; Δ4, ≈1-kbp deletion.
Characterized by whole-genome CGS analysis, Tn-mutagenesis approach or complementation experiments
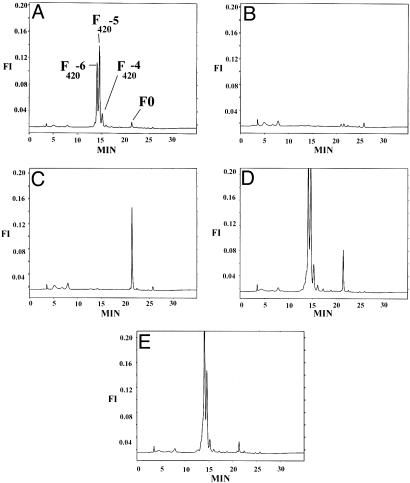
Fig. 3.
F420 and FO analysis. HPLC elution profiles of extracts from H37Rv (A), H37Rv-5A1 (B), H37Rv-7A2 (C), H37Rv-5A1-fbiC (D), and H37Rv-7A2-fbiAB (E). Strains are detailed in Table 1. The x axis is elution time in minutes, and the y axis is fluorescence intensity (FI).
Complementation of FGD1- and F420- Mutants Restores PA-824 Sensitivity. Complementation of wild-type FGD1 on an integrative plasmid vector in FGD1- mutant T3 fully restored FGD1 protein levels (Fig. 2A), PA-824 metabolism (Fig. 2B, lanes 2 and 3), and PA-824 sensitivity (Table 1). It has been reported that insertional inactivation of fbiC (Rv1173) caused an F420- phenotype in Mycobacterium bovis (10). Complementation of mutant 5A1, which entirely lacks F420, with wild type fbiC on an integrative plasmid vector, restored F420 biosynthesis (Fig. 3 B and D), PA-824 metabolism (Fig. 2B, lanes 4 and 5) and PA-824 sensitivity (Table 1). It has also been reported that inactivation of the fbiA (Rv3261) or fbiB (Rv3262) genes in M. bovis lead to an accumulation of FO with concomitant loss of F420 (11). Therefore, mutant 7A2 was complemented with the fbiAB operon, restoring normal levels of F420 biosynthesis (Fig. 3 C and E) and PA-824 sensitivity (Table 1).
Reduced F420 Is Necessary but Not Sufficient for PA-824 Activation. Studies had established that loss of FGD1 or the loss of biosynthetic potential for F420 individually resulted in resistance to PA-824 (6, 10, 11). To establish whether FGD1 or reduced F420 interacted with PA-824 directly, we partially purified recombinant Mtb-FGD1 from Escherichia coli and incubated this with oxidized F420 purified from Mycobacterium smegmatis (see Supporting Text, which is published as supporting information on the PNAS web site). Mtb-FGD1 was fully competent at reducing F420 in a glucose-6-phosphate-dependent fashion; however, addition of PA-824 to this reaction at a range of concentrations showed neither metabolism of PA-824 nor inhibition of F420-reducing activity by FGD1 (see Fig. 7, which is published as supporting information on the PNAS web site). Likewise, overexpression of Mtb-FGD1 in M. smegmatis, which naturally produces cofactor F420, did not result in sensitivity to PA-824 (data not shown).
Mutants with Wild-Type Levels of FGD1 and F420 Retain Sensitivity to Nitroimidazo-Oxazoles. To attempt to understand the molecular basis for PA-824-resistance in the FGD1+/F420+ mutants, we analyzed the cross-resistance of these mutants with CGI-17341, a structurally related oxazole (Fig. 1). Surprisingly, all of the FGD1+ and F420+ mutants examined remained fully sensitive to CGI-17341 (Table 1). To see whether this resistance was specific for this bicyclic nitroimidazole structure, we synthesized a different oxazine (PA-647, Fig. 1) that had been reported to be the most active molecule of this series in vitro (but not in vivo) (12) and a different oxazole, CGP-70520, that had a longer hydrophobic substituent on the oxazole ring (Fig. 1) (5). All FGD1- and F420- mutants were completely cross-resistant to all four compounds, whereas FGD1+ and F420+ PA-824-resistant mutants were resistant to PA-647 but fully sensitive to CGI-17341 and showed only slight resistance to CGP-70520 (Table 1). This oxazine-specific resistance pattern suggests the involvement of discrete F420-dependent nitro-reductases for activation of both oxazines and oxazoles.
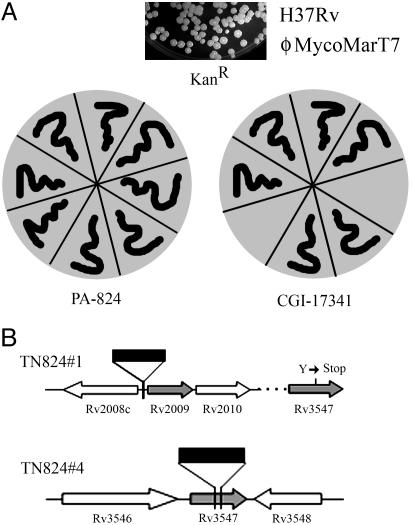
Transposon-Mediated Screening for a Nitroimidazo-Oxazine-Specific Activator. Because the nature of the genomic lesion in the PA-824R FGD1+/F420+ mutants remained elusive, we adopted a phenotype-based screening method to select transposon mutants that acquired PA-824 resistance while retaining sensitivity to CGI-17341 (Fig. 4). A library of transposon mutants was selected for PA-824 resistance and subsequently screened for CGI-17341 sensitivity (Fig. 4A). Ten percent to 15% of PA-824-resistant mutants were sensitive to CGI-17341, similar to the observed in vitro and in vivo frequency of spontaneous oxazine-specific-resistant mutants. Two independent PA-824R/CGI-17341S mutants (Tn824R#1 and Tn824R#4) were further characterized and shown to be FGD1+ and F420+ (Table 1). Sequencing of genomic DNA by using transposon-specific primers allowed the identification of the insertion sites. In Tn824R#4, the transposon insertion was localized in the C terminus of conserved hypothetical protein Rv3547, whereas, in Tn824R#1, the transposon was inserted in an intergenic region near Rv2009 (Fig. 4B). Because the spontaneous mutation frequency to PA-824R approached the frequency of transposon-mediated PA-824R and because the transposon in the intergenic region was difficult to interpret, neither of these mutants was pursued in more depth.
Fig. 4.
Phenotype-based screening method for isolating oxazine-specific mutants. (A) Schematic representation of the protocol for identifying oxazine-specific resistant mutants. A pool of ΦMycoMarT7-infected H37Rv was selected for PA-824R (2 μg/ml). Isolated PA-824R mutants were counterscreened for CGI-17341S sensitivity. (B) Identification of transposon-insertion sites in two PA-824R/CGI-17341S mutants. The point mutation subsequently discovered in Rv3547 in Tn824R#1 is also indicated.
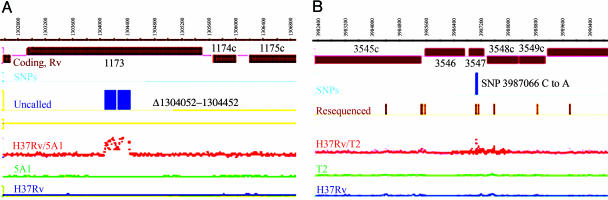
Comparative Genome Sequencing of PA-824R Mutants. Because of the apparent failure of conventional genetic strategies to identify the oxazine-specific activator, a comparative genome sequencing (CGS) technique developed by NimbleGen Systems was applied. CGS employs DNA microarray-based DNA sequencing to identify and characterize SNPs and insertion-deletion sites in the genome (13). The first (mapping) phase of CGS is a genome-wide mutational analysis by hybridization of a mutant and reference-strain genomic DNA to an array tiling the entire H37Rv reference genome at 7- to 8-bp probe spacing. In the second phase of CGS, customized resequencing arrays are used to confirm and identify putative SNPs. These arrays contain oligonucleotides representing all four potential sequences for each putative SNP position on both DNA strands. The mapping-phase hybridization-intensity ratio of genomic DNA from mutants T2 and 5A1 to genomic DNA from H37Rv are plotted as the red traces on Fig. 5A and B. Large deletions are apparent in mutant 5A1, whereas mutant T2 shows potential, and then confirmed, SNPs.
Fig. 5.
SignalMap (NimbleGen) representation of CGS analysis of H37Rv-5A1 (A) and H37Rv-T2 (B) mutants. The lower two traces are the signal intensity for the wild type and the mutant hybridizations, and third from the bottom is the ratio of these. The fourth trace shows genomic regions that were subjected to resequencing by using custom arrays, and, in B, the fifth line from the bottom illustrates SNPs confirmed by resequencing. In A, the fifth line shows the genomic deletion boundaries suggested by CGS in Rv1173 (fbiC). Only portions of the full genome hybridizations and resequenced regions are shown.
To validate this method, we first performed CGS on two known F420- mutants. Based on the initial mapping array results, both strains displayed potential genomic anomalies. An additional 97.2 kb and 48.6 kb of genomic DNA was resequenced by customized-array synthesis for mutants 5A1 and 5A2, respectively. This analysis revealed a 400-bp genomic deletion in the fbiC gene in mutant 5A1 (Fig. 5A) and a point mutation in a highly conserved region of FbiC in mutant 5A2 (see Fig. 8, which is published as supporting information on the PNAS web site). CGS also revealed unpredicted differences in the phthiocerol dimycocerosate (PDIM) locus in both mutants (a point mutation in mutant 5A2 and a genomic deletion mutant 5A1). The PDIM mutations were confirmed biochemically but were not associated with PA-824 resistance, because complementation of 5A1 with a wild-type fbiC allele restored sensitivity. Mutant H37Rv-7A2 (F420- and FO++) was also analyzed by CGS, but no unique mutations were detected.
Analysis of two independent PA-824R FGD1+/F420+ resistant mutants (T2 and 14A1) was then performed. Based on initial mapping-array results, 97.2- and 48.6-kb genomic regions were resequenced by microarray in T2 and 14A1, respectively. These data confirmed 21 and 9 SNPs in the T2 and 14A1 mutants, respectively. The only gene altered in both mutants was Rv3547, encoding a conserved hypothetical protein. In mutant T2, there was a transversion mutation in Rv3547, resulting in an Ala76-Glu substitution (Fig. 5B), and, in mutant 14A1, a frame-shift induced by a single base deletion in the 13th codon of the Rv3547 gene was discovered (Table 1). Conventional sequencing of Rv3547 PCR products from both strains confirmed the genome-wide mutation-mapping results. Because one of the two transposon mutants (Tn824R#4) also contained an insertion in Rv3547, we sequenced the Rv3547 gene from Tn824R#1 with an insertion in the intergenic region near Rv2009. This strain also showed a lesion in Rv3547 (Table 1); thus, in all four mutants, this protein was altered. Native Rv3547 fully restored PA-824 and PA-647 sensitivity (Table 1) as well as metabolism of [benzyl-14C] PA-824 (Fig. 2B, lanes 6-9) in T2, 14A1, and Tn824R#4 mutants. However, complementation with the mutated Rv3547-A76E from the T2 mutant did not restore PA-824 sensitivity as expected (data not shown).
Discussion
The 5-nitroimidazoles, such as metronidazole, are activated only by anaerobic enzyme systems that generate very low redox potentials (-480 mV), such as pyruvate-ferrodoxin oxidoreductases and hydrogenases (14). However, neither redox potential nor hydrophobicity correlates with the activity of various nitroimidazoles against Trichomonas vaginalis, suggesting a specific protein-ligand interaction (15). The crystal structure of the T. vaginalis ferrodoxin allowed modeling of the fit of various nitroimidazoles to the active site (2Fe-2S) cluster and supports the argument that the molecular shape of the drug appeared to play an important role in determining sensitivity to metronidazole (16). More recently, stopped-flow kinetic analysis of the reduction of a small group of nitroimidazoles also reported a correlation between activity and the size of the substituent on the N-1 position (17). These results suggest that the molecular fit of the nitroimidazole ligand at the ferrodoxin active site was the factor driving the structure-activity relationship.
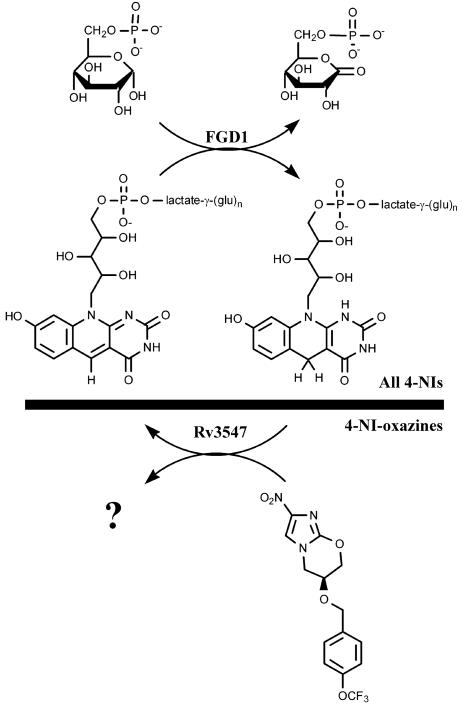
PA-824 is a 4-nitroimidazole that is reductively activated in Mtb in a manner that depends on the low-redox-potential (-350 mV) hydride-transfer coenzyme F420 (6). Perhaps because this cofactor does not appear to be essential in vitro, and the biosynthetic coding capacity required for its production is substantial, mutation of Mtb to PA-824R occurs with a high frequency, approaching that of INH, another prodrug that requires an apparently nonessential activating system (18). Although this phenomenon allowed the rapid implication of coenzyme F420 in the mechanism of activation of PA-824, it has not been previously possible to identify the specific protein that mediates electron transfer to the nitroimidazole moiety of this prodrug. In this article, we have fully characterized an extensive panel of PA-824R mutants and discovered a small class of mutants with lesions in Rv3547, a previously undescribed protein that is likely to interact directly with the prodrug (Fig. 6).
Fig. 6.
Summary of the role of FGD1, cofactor F420, and Rv3547 in the bioreductive activation of nitroimidazo-oxazines, including PA-824. Mtb acquires resistance to all bicyclic 4-nitroimidazoles (4-NIs) upon loss of the ability to produce, or reduce, the deazaflavin cofactor F420. Loss or mutation of Rv3547, in contrast, results in resistance only to 4-nitroimidazo-oxazines while retaining sensitivity to even closely related 4-nitroimidazo-oxazoles. This suggests that Rv3547, either alone or as part of a complex, is an F420-dependent nitro-reductase with a high degree of ligand selectivity.
Consistent with previous studies, loss of F420 biosynthesis or the loss of the ability to reduce this cofactor by FGD1 were causal for PA-824R. However, reduced F420 is not sufficient either in vivo or in vitro to reduce PA-824; instead, reduced F420 appears to simply provide electrons to a protein that directly interacts with PA-824. Identifying this protein was facilitated by the recognition of an oxazine-specificity of the drug resistance. However, because the frequency of spontaneous mutation approached the efficiency of transposition events, a straightforward counterscreen for transposon mutants that retained oxazole susceptibility while becoming oxazine resistant did not, at first, appear to have been successful. We therefore resorted to a whole-genome microarray-based sequencing technology that had not been applied to high-GC-content organisms. This technique resulted in the rapid identification of multiple SNPs and deletions in various mutants. In the two oxazine-specific mutants that were analyzed, T2 and 14A1, 21 and 9 SNPs, respectively, were confirmed by microarray-based sequencing. Sequence analysis revealed that 3 SNPs were identical and likely represent real differences between the sequenced H37Rv strain (19) and the H37Rv strain used in our laboratory. Of the remaining SNPs, only Rv3547 was mutated in common between the two isolates. Because we did not examine non-drug-treated organisms, it is not possible to define a basal mutation frequency or to say conclusively whether this frequency was increased in the nitroimidazole-selected mutants. Transcriptional profiling of short-term treatment of Mtb with PA-824 does not suggest that error-prone DNA replication or repair systems are induced (data not shown); however, other nitroimidazoles have been shown to have mutagenic potential in bacteria, and DNA damage has been proposed to be one toxic effect of prodrug activation (4).
Three of the five mutants analyzed by whole-genome CGS analysis also showed mutations not associated with PA-824R in the PDIM locus. We have observed spontaneous PDIM mutants frequently in other experiments as well, perhaps because of the large coding capacity required to produce this unique mycobacterial wax. These results suggest that it would be prudent to examine all mutants selected in Mtb for the presence of PDIM, because loss of this molecule has been associated with attenuation (20).
Rv3547 is a conserved hypothetical protein of 151 amino acid residues with no detectable sequence homology with any other proteins of known function. An equivalent gene is present in M. bovis, and homologues are also present in Mycobacterium avium (86% homology) and M. smegmatis (72% homology) but not in Mycobacterium leprae. Mtb has three homologues of Rv3547 showing 55-56% sequence similarity, namely Rv1261c, Rv1558, and Rv3178 (see Fig. 9, which is published as supporting information on the PNAS web site). Consistent with saturating-transposon-essentiality analysis, three of the four mutants in Rv3547 obtained in this study were found to be unable to produce Rv3547, confirming this protein to be nonessential (21). The fourth mutant (T2) showed a SNP resulting in an Ala76-Glu substitution in a highly conserved hydrophobic region adjacent to invariant Ser and Gly residues (Fig. 9). The fold-recognition program phyre predicted that Rv3547 is structurally related to Rv1155 (PDB 1XXO and 1W9A) with 90% precision and to Rv2991 (PDB 1RFE) with 75% precision. Based on the three-dimensional structure, Rv1155 is structurally related to the superfamily of flavin mononucleotide-binding proteins, and Rv3547 shows a predicted flavin-nucleotide-binding site (22). Rv2991 is homologous to several Nim-proteins that confer resistance to 5-nitroimidazole antibiotics in Bacteroides. Nim-proteins encode reductases that reductively convert the nitro group on metronidazole to a nontoxic amine in Bacteroides fragilis (23). Thus, we propose that Rv3547 is, or constitutes an essential part of, an F420H2-dependent nitro-reductase that donates electrons specifically to oxazines (Fig. 6). It is possible that other Rv3547 homologues may play a role in activation of oxazole analogues. Rv3547 and its homologues may, therefore, form a previously uncharacterized class of F420H2-dependent nitro-reductases in mycobacteria.
Complementation of the T3 mutant with fgd1, the 5A1 mutant with fbiC, and the T2 and 14A1 mutants with Rv3547, completely restored PA-824 activation and sensitivity, suggesting that the resistance was mediated in all cases by mutations causing lack of PA-824 activation. Because all of the mutants characterized so far are defective in activation, these results do not identify the proximal target responsible for cell death. As with other prodrugs that are activated to highly reactive species, it is possible that there is no single target. However, because Rv3547 appears to be capable of significant structure-based specificity, understanding the details of the protein-ligand interaction would be highly informative for ongoing attempts to optimize these molecules as backup clinical candidates for PA-824. Finally, the CGS technique used in this study has tremendous potential for streamlining the target-identification process for any antitubercular with whole-cell activity and is an enabling technology for a chemical genetics approach to target identification.
Materials and Methods
Bacteria, Culture Conditions, Plasmids, and Primers. The bacteria, plasmids, and primers used in this study are described in Table 2, which is published as supporting information on the PNAS web site. Mtb culture conditions and minimal inhibitory concentration determination have been described in ref. 24. Mycobacterial genomic DNA was isolated and transformation conducted as described in refs. 25 and 26.
Isolation of PA-824-Resistant Mutants. Mtb was cultured in 7H9 broth to midlog phase, and aliquots containing 107 to 108 colony-forming units (cfu) were plated on 7H11 agar plates containing 2 μg/ml PA-824 and incubated at 37°C for 3 wk. Spontaneous PA-824-resistant colonies were observed at a frequency of 6.5 × 10-7, and independent colonies were picked for further characterization. For in vivo PA-824-resistant mutants, 6- to 8-week-old female C57BL/6 mice (Taconic Farms) were infected with 50-100 cfu of Mtb H37Rv by aerosol. Nine weeks after infection, two groups of 23 mice were treated by gavage for 2 wk (100 μl/day per animal) with 25 mg/kg and 50 mg/kg PA-824 in CM-2 formulation (6). Before and after drug treatment, organs of 4 mice were plated to monitor efficiency of bacterial clearance. From the remaining 15 animals, whole lung and spleen homogenates were plated on PA-824 (2 μg/ml) to monitor emergence of drug resistance. Mutants were propagated in PA-824-free medium.
Analogues and Analysis of PA-824 Metabolism. PA-824 and PA-647 were prepared as described in U.S. patent 5,668,127 (12). CGI-17341 and CGP-70520 were prepared by using the method described in ref. 27. [benzyl-14C] PA-824 (Fig. 1) was synthesized by condensation of [benzyl-14C]4-(trifluoromethoxy)benzyl bromide (American Radiolabeled Chemicals, specific activity 55 mCi/mmol) (1 Ci = 37 GBq) with (6S)-2-nitro-6,7-dihydro-5H-imidazo[2-1b][1,3]oxazin-6-ol. Early log-phase (A650 nm 0.2) cultures (5 ml) were harvested by centrifugation and then resuspended in 200 μl of fresh medium with 5 μg of 1.8 μCi [benzyl-14C] PA-824 for 3 h. The cells were lysed by bead-beating and centrifuged at 10,000 × g for 10 min. Ten microliters was applied to a normal-phase silica gel TLC plate that was developed in 1-butanol/H2O/acetic acid (4:1:1). Radioactivity was recorded by PhosphorImaging.
FO/F420 Analysis and Western Blotting. For FO/F420 analysis, cell lysate was prepared by boiling cells for 20 min before analyzing on HPLC with fluorescence detection (11). For Western blotting, cell lysates were prepared as described in ref. 28. Polyclonal antibodies to purified Mtb-FGD1-His were raised in rabbits (Biosource, Worcester, MA) and serum used at 1:20,000 dilution.
Screening of ΦMycoMar Insertion Library and Mapping Transposon Insertion. A transposon-insertion library was made as described in ref. 21, and resistant clones were selected on 7H11 agar supplemented with 2 μg/ml PA-824. To screen for oxazine-specific mutants, clones were replica-plated on 7H11 agar supplemented with 2 μg/ml CGI-17341. For localization of the transposon-insertion site, chromosomal DNA from transposition mutants was digested with BamHI, purified, self-ligated, and transformed into DH5α pir116 strain (29). A kanamycin-resistant plasmid was isolated, and the chromosomal junction sequence was determined by using transposon-specific primers (21).
Comparative Genome Sequencing. Five micrograms of genomic DNA was digested with 0.005 units of DNase I (Amersham Pharmacia), and digested fragments were labeled with Biotin-N6-ddATP by using 25 units of terminal transferase. Mutation-mapping microarrays were designed to include a 29-mer oligonucleotide every seven bases for both strands of the complete H37Rv genome (GenBank accession no. AL123456), with each probe overlapping 22 bases of each adjacent probe. The arrays were synthesized as described in ref. 30 by using a random probe layout. Separate arrays were hybridized with labeled genomic DNA from reference and test strains in 1× hybridization buffer (NimbleGen) for 16 h at 45°C. These arrays were washed with nonstringent wash buffer [6× standard saline phosphate/EDTA/0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA (SSPE) and 0.01% (vol/vol) Tween-20], followed by two 5-min washes in stringent wash buffer [100 mM Mes/0.1 M NaCl/0.01% (vol/vol) Tween-20] at 47.5°C. The arrays were stained with Cy3-streptavidin conjugate (Amersham Pharmacia), and Cy3 signal was amplified by secondary labeling of the DNA with biotinylated goat anti-streptavidin (Vector Laboratories) and restained with the Cy3-streptavidin solution. The arrays were washed in nonstringent wash buffer, followed by two 30-s washes in 0.5× SSC and a 15-s wash in 70% ethanol. Microarrays were scanned at 5-μm resolution by using the Genepix 4000b scanner, and pixel intensities were extracted by using the program nimblescan. Probes that spanned potential mutations from both the strands were identified by using custom software. An array design was generated to sequence these positions by using a strategy similar to that described in ref. 13, with the exception of an algorithm to optimize the length, melting temperature, and mismatch position of each probe. These microarray-based sequencing arrays were synthesized and hybridized with genomic DNA from each isolate and scanned as above. Sequence base assignments were made by using a machine learning algorithm (31).
Supplementary Material
Acknowledgments
We acknowledge Thomas Keller (Novartis Institute for Tropical Diseases, Singapore) for valuable discussions. We thank Eric Rubin (Harvard University, Cambridge, MA) for mycomar mariner transposon phage; Gregory Philips (Iowa State University, Ames, IA) for providing DH5α pir plus strain; V. Nagaraja (IISc, Bangalore, India) for GyrA antibodies; and Taegwon Oh, Jacqueline Gonzales, and Michael Goodwin [National Institute of Allergy and Infectious Disease (NIAID)] for discussions and technical assistance. This work was supported by the Intramural Research Program of the National Institues of Health, NIAID.
Author contributions: U.H.M., T.J.A., and C.E.B. designed research; U.H.M., J.E.N., and L.D. performed research; U.H.M., C.S.D., L.Z., T.J.A., J.E.N., L.D., T.D., and S.S.P. contributed new reagents/analytic tools; U.H.M., H.B., T.D., S.S.P., and C.E.B. analyzed data; and U.H.M. and C.E.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CGS, comparative genome sequencing; FGD1, F420 dependent Glc-6-P dehydrogenase; Mtb, Mycobacterium tuberculosis, PDIM, phthiocerol dimycocerosate; TB, tuberculosis.
References
- 1.Duncan, K. & Barry, C. E., III (2004) Curr. Opin. Microbiol. 7, 460-465. [DOI] [PubMed] [Google Scholar]
- 2.Edwards, D. I. (1993) J. Antimicrob. Chemother. 31, 9-20. [DOI] [PubMed] [Google Scholar]
- 3.Samuelson, J. (1999) Antimicrob. Agents Chemother. 43, 1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wayne, L. G. & Sramek, H. A. (1994) Antimicrob. Agents Chemother. 38, 2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barry, C. E., III, Boshoff, H. I. & Dowd, C. S. (2004) Curr. Pharm. Des. 10, 3239-3262. [DOI] [PubMed] [Google Scholar]
- 6.Stover, C. K., Warrener, P., VanDevanter, D. R., Sherman, D. R., Arain, T. M., Langhorne, M. H., Anderson, S. W., Towell, J. A., Yuan, Y., McMurray, D. N., et al. (2000) Nature 405, 962-966. [DOI] [PubMed] [Google Scholar]
- 7.Wayne, L. G. (1994) Eur. J. Clin. Microbiol. Infect. Dis. 13, 908-914. [DOI] [PubMed] [Google Scholar]
- 8.Bair, T. B., Isabelle, D. W. & Daniels, L. (2001) Arch. Microbiol. 176, 37-43. [DOI] [PubMed] [Google Scholar]
- 9.Purwantini, E., Gillis, T. P. & Daniels, L. (1997) FEMS Microbiol. Lett. 146, 129-134. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. P., Kendrick, N. & Daniels, L. (2002) J. Bacteriol. 184, 2420-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, K. P., Bair, T. B., Bae, Y. M. & Daniels, L. (2001) J. Bacteriol. 183, 7058-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker, W. R. Shaopei, C. & Keeler, E. L. (1997) U.S. Patent 5,668,127.
- 13.Wong, C. W., Albert, T. J., Vega, V. B., Norton, J. E., Cutler, D. J., Richmond, T. A., Stanton, L. W., Liu, E. T. & Miller, L. D. (2004) Genome Res. 14, 398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sisson, G., Goodwin, A., Raudonikiene, A., Hughes, N. J., Mukhopadhyay, A. K., Berg, D. E. & Hoffman, P. S. (2002) Antimicrob. Agents Chemother. 46, 2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarlett, N., Gorrell, T. E., Marczak, R. & Muller, M. (1985) Mol. Biochem. Parasitol. 14, 29-40. [DOI] [PubMed] [Google Scholar]
- 16.Crossnoe, C. R., Germanas, J. P., LeMagueres, P., Mustata, G. & Krause, K. L. (2002) J. Mol. Biol. 318, 503-518. [DOI] [PubMed] [Google Scholar]
- 17.Vidakovic, M., Crossnoe, C. R., Neidre, C., Kim, K., Krause, K. L. & Germanas, J. P. (2003) Antimicrob. Agents Chemother. 47, 302-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slayden, R. A. & Barry, C. E., III (2000) Microbes Infect. 2, 659-669. [DOI] [PubMed] [Google Scholar]
- 19.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S. & Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 20.Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. (1999) Mol. Microbiol. 34, 257-267. [DOI] [PubMed] [Google Scholar]
- 21.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2001) Proc. Natl. Acad. Sci. USA 98, 12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canaan, S., Sulzenbacher, G., Roig-Zamboni, V., Scappuccini-Calvo, L., Frassinetti, F., Maurin, D., Cambillau, C. & Bourne, Y. (2005) FEBS Lett. 579, 215-221. [DOI] [PubMed] [Google Scholar]
- 23.Carlier, J. P., Sellier, N., Rager, M. N. & Reysset, G. (1997) Antimicrob. Agents Chemother. 41, 1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domenech, P., Reed, M. B. & Barry, C. E., III (2005) Infect. Immun. 73, 3492-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelicic, V., Jackson, M., Reyrat, J. M., Jacobs, W. R., Jr., Gicquel, B. & Guilhot, C. (1997) Proc. Natl. Acad. Sci. USA 94, 10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T. & Jacobs, W. R., Jr. (1990) Mol. Microbiol. 4, 1911-1919. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajan, K., Shankar, R. G., Rajappa, S., Shenoy, S. J. & Costa-Pereira, R. (1989) Eur. J. Med. Chem. 24, 631-633. [Google Scholar]
- 28.Manjunatha, U. H., Mahadevan, S., Visweswariah, S. S. & Nagaraja, V. (2001) Eur. J. Biochem. 268, 2038-2046. [DOI] [PubMed] [Google Scholar]
- 29.Platt, R., Drescher, C., Park, S. K. & Phillips, G. J. (2000) Plasmid 43, 12-23. [DOI] [PubMed] [Google Scholar]
- 30.Albert, T. J., Norton, J., Ott, M., Richmond, T., Nuwaysir, K., Nuwaysir, E. F., Stengele, K. P. & Green, R. D. (2003) Nucleic Acids Res. 31, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molla, M., Shavlik, J., Albert, T., Richmond, T. & Smith, S. (2004) The Proceedings of the IEEE Conference on Computer Systems Bioinformatics, Stanford, CA. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.