Abstract
Nuclear-cytoplasmic partitioning and traffic between cytoplasmic and nuclear compartments are fundamental processes in eukaryotic cells. Nuclear pore complexes mediate transport of proteins, RNAs and ribonucleoprotein particles in and out of the nucleus. Here we present positional cloning of a plant nucleoporin gene, Nup133, essential for a symbiotic signal transduction pathway shared by Rhizobium bacteria and mycorrhizal fungi. Mutation of Nup133 results in a temperature sensitive nodulation deficient phenotype and absence of mycorrhizal colonization. Root nodules developing with reduced frequency at permissive temperatures are ineffective and electron microscopy show that Rhizobium bacteria are not released from infection threads. Measurement of ion fluxes using a calcium-sensitive dye show that Nup133 is required for the Ca2+ spiking normally detectable within minutes after application of purified rhizobial Nod-factor signal molecules to root hairs. Localization of NUP133 in the nuclear envelope of root cells and root hair cells shown with enhanced yellow fluorescent protein fusion proteins suggests a novel role for NUP133 nucleoporins in a rapid nuclear–cytoplasmic communication after host–plant recognition of symbiotic microbes. Our results identify a component of an intriguing signal process requiring interaction at the cell plasma membrane and at intracellular nuclear and plastid organelle-membranes to induce a second messenger.
Keywords: legume symbiosis, nucleoporin, nuclear pore, plant–microbe interaction
Two of the nutrients plants require in high amounts for optimal growth are nitrogen and phosphate. Plant roots mine the soil for nutrients, and 80–90% of all land plants have symbiotic interaction with arbuscular mycorrhizal fungi that increase the root surface and supply plants with phosphate (1, 2). Extending the repertoire for nutrient acquisition, legumes also enter into symbiosis with nitrogen fixing bacteria, collectively called rhizobia (3). Nitrogen fixed in the root nodule organs that house the nitrogen-fixing bacteria is estimated to be equal to the amount used around the world in chemical fertilizers. Genetic studies in the model legume Lotus japonicus have, in addition to Nup133, defined six loci (SymRK, Castor, Pollux, Sym15, Sym24, and Sym30) required for both mycorrhizal colonization and rhizobial induced root nodule development (4, 5). The fungal symbiotic signal molecule triggering this signal transduction pathway is unknown. In contrast, both the rhizobial Nod-factors and corresponding plant receptor kinases required for perception have been identified. In Lotus, NFR1 and NFR5 are predicted receptor kinases that operate upstream of the common pathway. Presumably they funnel Nod-factor signaling into the signal tranduction pathway shared with mycorrhizal fungi (6, 7). Nod-factor perception is a key feature in rhizobial symbiosis determining host–bacteria recognition, initiation of infection thread formation, and root nodule inception. Purified Nod-factors trigger a variety of responses in roots, such as cell divisions forming nodule primordia, activation of gene expression, root hair deformation, and calcium oscillations in root hair cells (8, 9). Functional NFR1/NFR5 receptor kinases are required for these responses, whereas common pathway mutants usually display an attenuated response to Nod-factor. Several lines of evidence suggested that calcium acts as secondary messenger, and the analysis of mutants of Lotus and/or Medicago indicates that calcium spiking is a component of the common pathway. Analysis of mutants has shown that a LRR protein kinase (SYMRK/NORK) and a cation channel(s) (CASTOR/POLLUX/DMI1) are required for the induction of calcium spiking. A predicted calcium calmodulin protein kinase (DMI3) probably integrates the calcium fluctuations to activate induction of downstream genes (10–15). Interestingly, CASTOR and POLLUX proteins were localized to root cell plastids, implicating ion signaling through plastids in the early signal transduction after signal perception (14). Characterization of nodulation deficient nup133 mutants reported here add a unique component to this pathway and focus attention on the role of nuclear associated calcium spiking in Nod-factor mediated signaling.
Materials and Methods
Plant Material. Isolation of nup133-1, nup133-2 mutants previously called sym3-1 and sym3-2 were described in ref. 16, and nup133-3 and nup133-4 (previously called sym3-3 and sym45) were also isolated in the ecotype Gifu B-129 background. Plants were grown with or without M. loti strain NZP2235, TONO, and R7A. The root hair curling procedure has been described (6).
Electrophysiology. Seedlings of L. japonicus were germinated, mounted, and microinjected with Oregon Green-488 BAPTA-1 (Molecular Probes) essentially as described (9). Fluorescence was imaged by using a Nikon TE2000 inverted microscope coupled to a Hamamatsu Photonics digital charge-coupled device (CCD) camera. The excitation wavelength of 488 nm with an 11-nm bandpass was selected by using an Optoscan Monochromator (Cairn, Faversham, Kent, U.K.), and an emission filter of 545 (±15) nm was used. Images covering the protruding part of the root hair, including the entire nuclear region, were collected every 5 s with a 200-ms exposure using metafluor software, and derivative traces were generated by using Microsoft excel. The data presented were transformed to first derivative traces (in arbitrary units) as described by Wais et al. (17) using the formula Y = X(n+1) - Xn, where Y is the change in fluorescence and X(n+1) and Xn are the fluorescence intensity measurements at time points n and n + 1 separated by 5-s intervals. After microinjection, root hairs were left at least 20 min before Nod-factor addition, and only cells showing active cytoplasmic streaming were used for analysis; at least 95% of such cells in wild-type plants induce calcium spiking in response to Nod-factor. Nod factors, isolated from a reverse phase C18 column, were added directly to the incubation chamber to give an estimated final concentration of 10-8 M. In the experiments done with the mutants, seven, four, six, and three cells were analyzed all with separate seedlings of the nup133-1, nup133-2, nup133-3, and nup133-4 mutants, respectively. An additional 10 cells were assayed by using three seedlings of the nup133-3 mutant grown at 10°C and assayed at 15°C.
Map-Based Cloning and cDNA Isolation. An F2 mapping population was established by crossing a nup133-1 mutant and a wild-type L. japonicus ecotype “MG-20.” F2 plants homozygous for the nup133-1 mutant allele were identified after screening for the nonnodulation mutant phenotype. In total, 822 homozygous F2 mutant plants were analyzed. Microsatellite markers and single nucleotide polymorphism developed from BAC and TAC clones anchored to the general genetic map of the region were used for fine mapping and for building the physical TAC/BAC contig. Five TAC/BAC clones from MG-20 and two BAC clones from Gifu were assembled to cover the Nup133 region between the two flanking markers 1F24R and T47f06. On the basis of sequence differences between the two parents, additional PCR markers were developed inside this contig, and the region was narrowed down to 22 kb between markers 1F24-7 and T45d07-3. Finally, clone LjT44M23 from MG-20 containing the entire Nup133 region was sequenced as a part of the Lotus genome sequence program as was Nup133 from Gifu. A full-length cDNA clone of Nup133 with two in frame stop codons 5′to an ORF was isolated by screening a λ ZAPII cDNA library prepared from mRNA extracted from leaves (Stratagene).
Complementation Experiment and LjNUP133 Localization in Yeast. LjNup133 cDNA was PCR amplified with the following primers: 5′-GGGGGGGTCGACCTATTCCATGGGAGAAGGCC-3′ and 5′-GGGGGGGTCGACATGTTTTCGTGTGGAACGAAGAA-3′. The fragment was cloned into the SalI site of the pPS808 plasmid (2μ) in frame with the GFP gene, under the control of a galactose-inducible promoter. A WT and a ΔNUP133 yeast strain (18) were transformed with the plasmid expressing the fusion protein and the empty plasmid. Cells were grown on plates with galactose at different temperatures. For localization, the ΔNUP133 yeast strain was transformed with either a plasmid expressing a GFP–LjNup133 fusion protein or an empty plasmid expressing GFP. Transformed strains were grown in selective media without glucose containing 2% lactate/2% glycerol. Expression of GFP proteins was induced by incubation with 2% galactose for 4 h, and cells were subsequently analyzed by fluorescent microscopy.
In Planta Complementation. In planta complementation of all four mutant alleles was accomplished by using the Agrobacterium rhizogenes transformed “hairy roots” methodology (19). A 12-kb genomic region containing the Nup133 gene from BAC clone 10M23 was inserted into pIV10 plasmid and conjugated to A. rhizogenes AR12. Transgenic hairy roots and nodulation tests were done as described (6). Nodulated nup133-1 plants were transferred to new pots to cocultivate with Allium schoenoprasum for mycorrhization tests and incubated at 24°C for 5 weeks (5).
Expression Analysis. Total RNA was isolated by using a CsCl cushion and mRNA was isolated from root, leaf, flower, pod, and nodule tissue using DynaBeads. Twenty micrograms total RNA was used for Northern analysis with a Nup133 specific probe. The probe was produced by PCR with primers 5′-CTTCTGGCATTCATCGAACAG-3′ (forward) and 5′-CAATGATTGATTTGCTCATCCTC-3′ (reverse) covering part of exon 3 (not NUP-motif). Quantitative real-time PCR was done on three independent RNA preparations as described (6). Primers used for LjNup133 transcript amplification were: 5′-TCCCTGGACTCCTCGCTTATCTGT-3′ (forward) and 5′-ACGCGCAGCCTTCAACGGG-3′ (reverse). The identity of the product was confirmed by sequencing.
Localization of NUP133 Protein. For localization of NUP133 protein, a full length Nup133 cDNA was fused to enhanced yellow fluorescent protein (eYFP) coding regionn to create an N-terminal in frame fusion. The recombinant gene was inserted between the 35S promoter and pANOS 3′ terminal, and cloned into the pIV10 plasmid. Transgenic hairy roots were observed by using confocal laser microscopy (LSM510 META, Zeiss).
Computer Analysis. blastp and psi-blast (www.ncbi.nlm.nih.gov/BLAST) were used to identify homologues of NUP133. Sequences were aligned in clustalw (www.ebi.ac.uk/clustalw) and the alignment was edited in jalview 2005. Fold recognition was done with phyre (www.sbg.bio.ic.ac.uk/~\phyre). The secondary structure prediction used is a consensus prediction based on the results of psipred, jpred, profsec, sspro, and target99, all of which can be reached through the PredictProtein server (www.embl-heidelberg.de/predictprotein/predictprotein.html). For the prediction of protein localization sites in cells, psort (http://psort.nibb.ac.jp) was carried out. Multiple alignment of the Lotus NUP133 toward yeast and human Nup133 sequences was done by using clustalx (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html). Nup133 gene sequence is GenBank accession no. AJ890252, and Nup133 mRNA is GenBank accession no. AJ890251.
Results
nup133 Mutants Are Nodulation Deficient. We have phenotypically characterized Lotus nup133 mutants and cloned the Nup133 gene belonging to the common symbiotic pathway. Four monogenic recessive mutant alleles, nup133-1, nup133-2, nup133-3, and nup133-4 (Table 2, which is published as supporting information on the PNAS web site), were originally found in a screen for symbiotic mutants and assigned to the same locus by diallelic crosses. The nup133-1 allele has the strongest effect on nodulation, and nup133-1 mutants do not nodulate when inoculated at 22°C with the Lotus microsymbiont, Mesorhizobium loti strain NZP2235 (Table 1). Two other M. loti strains, TONO and R7A, trigger formation of a few small ineffective nodules (Fig. 1 A and B). On average nup133-1 mutants developed one nodule on four of 25 plants and six of 28 plants inoculated with TONO and R7A, respectively. Compared to wild-type plants, this a >30-fold reduction in nodulation frequency (Table 1). The nup133-2, nup133-3, and nup133-4 mutants are nodulation deficient and developed small ineffective nodules with a frequency reduced from 5- to 30-fold after inoculation with one of the three M. loti strains (Table 1). When inoculated at 26°C, nup133-2, nup133-3, and nup133-4 were essentially nonnodulating, revealing a temperature-sensitive nodulation deficiency (Table 1).
Table 1. Nodulation phenotype of nup133 alleles inoculated with three different M. loti strains at two temperatures.
| 22°C
|
26°C
|
||||
|---|---|---|---|---|---|
| Plant | Rhizobium strain | Nodulation ratio | No. of nodules per plant | Nodulation ratio | No. of nodules per plant |
| Wild type | NZP2235 | 24/24 | 6.4 | 41/41 | 6.1 |
| TONO | 36/36 | 4.88 | 22/24 | 5.67 | |
| R7A | 33/33 | 4.48 | 26/26 | 5.0 | |
| nup133-1 | NZP2235 | 0/29 | 0 | 0/41 | 0 |
| TONO | 4/25 | 0.16 | 1/36 | 0.03 | |
| R7A | 6/28 | 0.36 | 0/34 | 0 | |
| nup133-2 | NZP2235 | 6/32 | 0.23 | 0/27 | 0 |
| TONO | 11/34 | 0.68 | 1/36 | 0.03 | |
| R7A | 13/37 | 0.49 | 1/29 | 0.03 | |
| nup133-3 | NZP2235 | 6/31 | 0.23 | 0/28 | 0 |
| TONO | 11/23 | 0.61 | 0/20 | 0 | |
| R7A | 14/31 | 0.77 | 0/26 | 0 | |
| nup133-4 | NZP2235 | 5/29 | 0.24 | 0/11 | 0 |
| TONO | 7/16 | 0.50 | 0/20 | 0 | |
| R7A | 7/27 | 0.26 | 0/20 | 0 | |
Fig. 1.
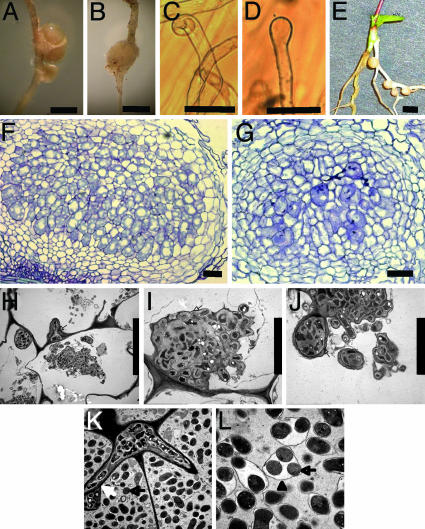
Phenotype of nup133 mutants and genetic complementation. (A) Wild-type root nodules. (B) Ineffective nodule on nup133-1 inoculated with the M. loti TONO strain. (C) Root hair curling on a wild-type seedling inoculated with M. loti. (D) Root hair swelling on nup133-1 inoculated with M. loti. (E) Nodules on transgenic root showing complementation of nup133-2. (F) Section of mature wild-type root nodule. (G) Section of nup133-2 nodule. (H–J) Electron micrographs of an ineffective nodule showing infection threads (arrow) and M. loti bacteria within enlarged infection droplet structures (arrow). (K and L) Electron micrographs of a wild-type nodule showing a branched infection thread releasing an M. loti containing infection droplet (white arrow) and bacteroids (black arrow) surrounded by peribacteroid membrane (arrowhead). The symbiosome consists of bacteroids enclosed within a peribacteroid membrane. (Scale bars: 1 mm in A, B, and G and 50 μm in C–F and H.)
Nod-Factor Perception and Calcium Oscillations. The earliest visible cellular change in wild-type plants after inoculation is root hair deformation and root hair curling (Fig. 1C). In the nup133-1 mutants inoculated with NZP2235, root hair swelling and in nup133-4 also root hair branching were observed (Fig. 1D), whereas root hair curling was not observed. These observations indicate that Nod-factor perception did occur in the mutants. This conclusion was supported by microelectrode measurements of Nod-factor induced alkalinization in the root hair space of nup133-1, nup133-2 and nup133-3 mutants (data not shown). In wild type, this response is associated with rapid calcium influx (6, 15). Nodules that developed on the nup133-1 mutants with TONO were further characterized by microscopy. Sections of mutant nodules show that they are developmentally arrested, either empty nodules or nodules with only few cells containing bacteria (Fig. 1 F and G). Electron microscopy of mutant nodule sections shows infection threads and bacteria inside plant cells contained in structures resembling enlarged infection droplets (Fig. 1 H–J). Symbiosomes containing endocytosed bacteria were not observed in these sections. To investigate whether infection of these ineffective nodules occurred through root hairs, we examined nup133 mutant root hairs in the infection zone for presence of infection threads. After inoculation with strain NZP2235 or TONO constitutively expressing LacZ, root hair infection threads of nup133-1, nup133-2, nup133-3, and nup133-4 mutants were visualized by X-Gal staining. With this technique, up to 20 infection threads are visible in the infection zone of wild-type plants. At the permissive temperature, one infection thread was found on 14 nup133-1 mutant plants; in nup133-2, no infection threads were found in 14 plants; in nup133-3, no infection threads were found in 12 plants; in nup133-4, one infection thread was found in four plants. These rare infection threads are most likely associated with the occasional formation of ineffective root nodules observed on nup133 mutants.
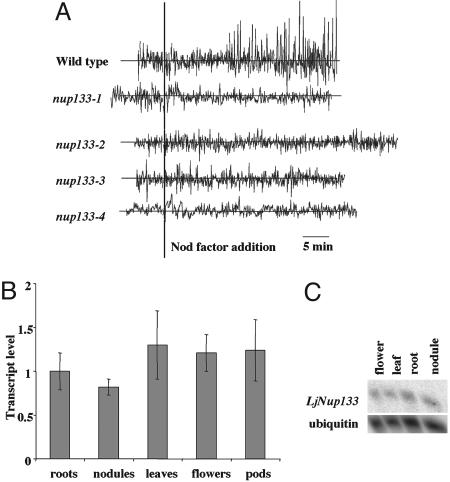
Measurements of ion fluxes in wild type root hairs using the calcium-sensitive dye Oregon Green-488 BAPTA-1 show Ca2+ spiking ≈10 min after the addition of Nod-factor (Fig. 2A). In contrast, calcium spiking was not detected in the four allelic nup133 mutants (Fig. 2A). Sixteen individual root hairs from nine seedlings of the nup133-3 mutant were tested and no Ca2+ spiking was detected, even though 10 of these cells were from plants grown and assayed at the lower temperature that permits low level nodulation. An additional 14 root hairs (each from a separate seedling) from the nup133-1, nup133-2, and nup133-4 mutants were also tested, and none induced Ca2+ spiking after Nod factor addition. Because our success rate for observing Ca2+ spiking in wild type is 94% of those root hairs chosen for assay after microinjection, it is absolutely clear that mutations in the nup133 gene block Nod-factor induced Ca2+ spiking in young root hairs, because 30 individual root hairs from four independent mutants all lacked this response. Microinjection of L. japonicus root hairs is laborious and technically demanding, so this technique does not lend itself to identification of rare cells that might induce Ca2+ spiking. Furthermore, we are unable to microinject older root hairs. Therefore, the occasional infection and nodulation events observed with these mutants could be due to either a lack of requirement for Ca2+ spiking or that Ca2+ spiking is induced occasionally in some cells, possibly even among cell types where root-hair calcium measurements are not possible using the microinjection approach. Nevertheless, it is very clear that nup133 mutations can block Ca2+ spiking in those young root-hair cells normally assayed by us and others (8, 9, 15, 17) for this phenotype.
Fig. 2.
Calcium oscillations in root hairs in response to purified Nod-factor. (A) Each trace is from a single root hair using seedlings of wild-type L. japonicus or mutants carrying the nup133-1, nup133-2, nup133-3 or nup133-4 alleles. Seven, four, six, and three root hairs were analyzed for each allele, respectively. An additional 10 analyses were done with nup133-3 grown at 10°C and assayed at 15°C, but the traces were similar to that shown. Root hairs were injected with the calcium-sensitive dye Oregon green-488 BAPTA-1 and, after ≈20 min, Nod factor was added at 10-8 M. The data are graphed showing typical traces of the first derivatives of fluorescence intensity (in arbitrary units) between 5 s sequential time points. There was no observed calcium spiking in any of the mutants under the conditions tested. (B and C) Transcript level of Nup133 in wild-type plants. (B) Quantitative RT-PCR results are shown as fold increase compared to roots. Error bars represent a 95% confidence interval. (C) Northern hybridization showing similar expression levels of Nup133 in different Lotus tissues
Taken together, the phenotypic characterization of a temperature-dependent nonnodulation phenotype, the impaired mycorrhizal colonization (4, 5), the lack of normal Nod-factor induced alkalinization in the root hair space, and the lack of Nod-factor induced calcium spiking in developing root hairs, lead us to conclude that NUP133 is part of the common pathway required to mount a response to signals from rhizobial and mycorrhizal symbionts.
Positional Cloning of Nup133. To characterize the gene molecularly, Nup133 was isolated by using a positional cloning approach. On the genetic map of Lotus,the Nup133 locus is located on the short arm of chromosome I (20). Subsequent fine mapping in an F2 population and genotyping of 822 mutant plants identified markers delimiting Nup133 to a 0.06 cM region of 22 kb (Fig. 5, which is published as supporting information on the PNAS web site). Located within this region are four genes predicted to encode a pentatricopeptide repeat-containing protein, a chlorophyll a/b binding protein, a Ca2+ transporting ATPase, and a homologue of an Arabidopsis expressed protein, respectively (Fig. 5). Considering the suggested role for calcium as second messenger, the search for mutant alleles was initially focused on sequencing the Ca2+ transporting ATPase gene. Contrary to our expectations, this gene was not mutated. Subsequent sequencing of gene regions corresponding to the homologue of the Arabidopsis expressed protein in nup133-1, nup133-2, nup133-3, and nup133-4 identified the Nup133 gene. Short deletions shifting the reading frame, leading to premature stop codons, were found in nup133-1, nup133-2, and nup133-4 (Table 2). A retrotransposon is inserted in nup133-3. All four mutant alleles were complemented by the cloned Nup133 using Agrobacterium rhizogenes to generate transgenic roots (19). The wild-type gene, including a 1.8-kb promoter region and a 3.6-kb 3′ region was introduced into nup133-1, nup133-2, nup133-3, and nup133-4 mutant plants via A. rhizogenes and the nodulation phenotype scored after inoculation with strain NZP2235 of M. loti. All mutants were complemented with high efficiency for nodulation (Fig. 1E and Table 3, which is published as supporting information on the PNAS web site). Complementation for mycorrhizal colonization was tested and obtained in the nup133-1 mutant. Complementation was scored as normal colonization and abundant arbuscule formation in transgenic roots.
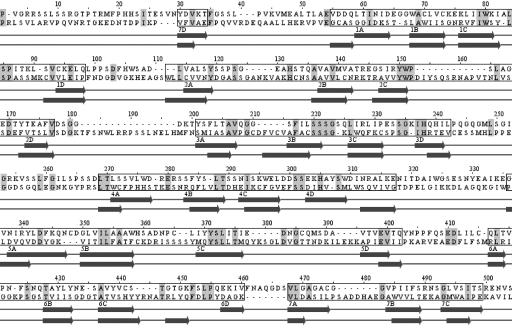
Sequencing of full-length cDNAs isolated from a Lotus leaf library determined the transcription start site at least 62 bp upstream of the start codon and a 3′ untranslated region of 210 nucleotides (Fig. 5). Alignment of genomic and cDNA sequences defined eight exons in Nup133 (Fig. 5). The Lotus Nup133 cDNA encodes a conceptual protein of 1,309 aa, corresponding to 146.5 kDa. Southern hybridization indicates that NUP133 is encoded by a single gene in both the small genome of Lotus japonicus and the large genome of pea (data not shown). In the fully sequenced Arabidopsis and rice genomes, Nup133 homologs are single functionally uncharacterized genes in each species. The Arabidopsis predicted protein At2g05120 is 54% identical, and the predicted rice protein (AAN52748) is 47% identical to NUP133. Less similar, with 20% global identity, are yeast and human Nup133 nucleoporins (Fig. 6, which is published as supporting information on the PNAS web site). Three-way alignment of human, yeast, and Lotus proteins identifies a set of 59 conserved amino acids positioned along the length of the proteins suggesting common origin of corresponding genes. This finding is supported by conservation of the two first intron positions among 25 introns of the human gene and eight introns of plant Nup133 genes. The Lotus NUP133 protein was identified as a Nup133 nucleoporin by psi-blast. After two iterations, the Lotus NUP133 amino acid sequence aligned to the entire length of Nup133 proteins from mouse and human with E values of 0 and overall identities of ≈20%. The N-terminal domain of human Nup133 consists of an α/β domain with a seven bladed β-propeller fold, and the C-terminal domain was predicted to be all α-helical (21). Secondary structure prediction and fold recognition on Lotus NUP133 suggest a similar overall structure and recognize the β-propeller structure of human Nup133 within the first 600 N-terminal residues of Lotus NUP133. From secondary structure predictions and an alignment with human Nup133, the N-terminal α/β domain and C-terminal α helical domain of Lotus NUP133 were estimated to localize between residues 50–535 and 555-1309, respectively. The positions of β-propeller blades suggested by secondary structure prediction are shown on the alignment of N-terminal domains of Lotus NUP133 and human Nup133 proteins in Fig. 3.
Fig. 3.
Alignment of the N-terminal 50–535 residues of Lotus NUP133 to human Nup133 (top sequence). Amino acid residues marked in gray are identical. Propeller blades are boxed and β strands named from 1 (A–D) to 7 (A–D) after position in the human Nup133 propeller are shown below. Human Nup133 strand annotations are depicted above the Lotus NUP133 strand predictions. Although Lotus NUP133 has quite a few insertions compared to human Nup133, the N-terminal domain of Lotus NUP133 is predicted to adopt a seven-bladed β-propeller fold, similar to the human Nup133 propeller.
Expression of Nup133 in Different Plant Organs. The symbiotic mutant phenotype suggests a function for Nup133 in root tissues. To test this prediction, expression of Nup133 in different plant organs was determined by Northern and quantitative RT-PCR analysis. Results of both analyses demonstrate that Nup133 transcripts are present in all organs tested (Fig. 2 B and C). No significant induction was detected in roots several days after inoculation with M. loti (data not shown). The observed expression in all organs tested and the similarity to a nucleoporin would predict a general function for Nup133 and a mutant phenotype affecting overall plant development. For example, mutation of the PAUSED gene (encoding an homolog of nuclear export receptor for tRNA) in Arabidopsis affected shoot and apical meristem growth, leaf development, and lateral root formation (22). No such general effect was observed with the nup133 mutants, but generally the four mutant lines have a lower number of seeds in mature pods than wild type.
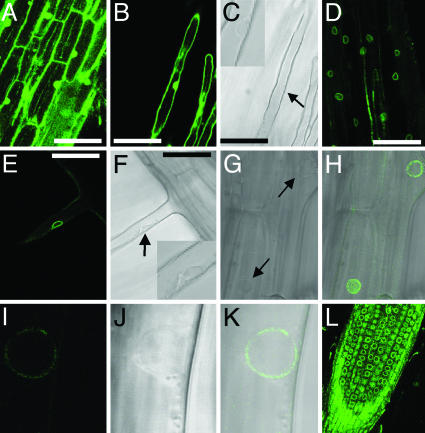
Localization of NUP133 Protein in the Nuclear Rim. Yeast and mammalian nuclear pore complexes contain ≈30 proteins identified mainly by proteomics. Although nuclear pores serve the same purpose in nuclear–cytoplasmic trafficking, global conservation of nucleoporin sequences is within a range of 10–20% identities, which is surprisingly low. Corroborating this observation, a search of the Arabidopsis genome using human nucleoporins and nuclear pore associated protein sequences, has so far identified only four Arabidopsis encoded proteins with ≈20% identities and 30–40% similarity to nucleoporins (23). Neglecting this lack of conservation between organisms, we attempted functional characterization of NUP133 protein in yeast. A GFP–NUP133 N-terminal fusion expressed from a galactose-inducible promoter was transformed into a temperature-sensitive yeast strain deleted for the NUP133 gene. Complementation was not achieved, and the GFP fusion protein was not localized to the nuclear rim as expected for a nucleoporin (Fig. 7, which is published as supporting information on the PNAS web site). These results suggest that Lotus NUP133 and yeast Nup133p are too diverse for functional complementation. In contrast, an eYFP–NUP133 N-terminal fusion protein expressed in transgenic Lotus roots complemented nup133-1 mutants, and the fusion protein was clearly localized to the nuclear rim in root cells (Fig. 4 D and G–L) as well as nuclear rim in root hair cells (Fig. 4 E and F). In Fig. 4 E–K, the nuclear rim localization is illustrated by corresponding sets of light microscopy and confocal images. Analysis of confocal stack images of consecutive sections of nuclei shows nuclear rim localization of the fusion protein in the complete nuclear sphere. In accordance with the NUP133 similarity to nucleoporins, the punctuate appearance of eYFP–NUP133 fluorescence (Fig. 4 H, I, and K) is comparable to the punctuate localization characteristic for proteins residing in nuclear pore complexes of yeast and mammalian cells. In control experiments, expression of eYFP alone resulted in an even distribution of fluorescent protein within nuclei (Fig. 4 A–C).
Fig. 4.
Subcellular localization of eYFP–NUP133 fusion protein in the nuclear rim. (A and B) Fluorescent image of a control eYFP localization in nup133-1.(C) Light microscopy of B with nucleus marked by arrow and highlighted in insert. (D, E, H, I, K, and L) Fluorescent images of eYFP–NUP133 fusion protein showing nuclear rim localization in nup133-1 mutant background. (E–K) Single cell localisation of eYFP–NUP133 in the nuclear rim. (E) Root hair cell showing nuclear rim localization of eYFP–NUP133. (F) Corresponding light microscopy with nucleus marked by arrow and highlighted in Inset. (H) Fluorescent overlay of G light microscopy with nuclei marked by arrows. Note the surface view of one nucleus showing punctate localization of eYFP–NUP133 fusion protein. (K) Overlay of corresponding fluorescent image of eYFP–NUP133 (I) and light microscopy image (J). Note the punctate eYFP–NUP133 localization. (L) Localization of eYFP–NUP133 in nuclear rims of cells in a transgenic root tip; note the cell files. (A and D) Epidermal cells. (B, C, E, and F) Root hair cells. (Scale bar: 50 μm.)
Discussion
The collective evidence obtained by genetic and physical mapping, sequencing of mutant alleles, and successful complementation unequivocally identified the nucleoporin Nup133 gene. The presence of only one gene copy in Lotus, pea, and the fully sequenced Arabidopsis and rice genomes, together with conservation of two intron positions, suggest that plant Nup133 genes are closest homologs of yeast and mammalian Nup133 genes. Further evidence from protein sequence conservation in Lotus NUP133, comparable to the general level of conservation in the NUP133 protein family and the overall α/β–α domain structure together with the predicted N-terminal seven-bladed β-propeller domain, support the identification of Lotus NUP133 as a member of the Nup133 nucleoporin family. Localization of eYFP fusion proteins at the nuclear rim makes it most likely that NUP133 and its homologs are components of the nuclear pore complex in plants. The punctuate localization of eYFP–NUP133 fluorescence, characteristic for nuclear pore proteins, supports this interpretation (Fig. 4 H, I, and K).
In yeast, Nup133p is a constituent of the Nup84p complex and in mammals the corresponding protein is a component of the Nup107–160 complex residing in the nuclear basket structure of the nuclear pore (24–27). Studies in yeast and HeLa cells suggest a function in export of mRNA, nuclear pore assembly and distribution (18, 28). Such a general function for the Lotus NUP133 nucleoporin is difficult to reconcile with the predominantly symbiotic phenotype observed. Interestingly, Drosophila has one example of cell-specific developmental effects in a nucleoporin mutant that is also affected in the response to fungal and bacterial pathogens. Mutation of members only, the gene encoding a nucleoporin homologous to mammalian Nup88, leaves Dorsal and Dif transcription factors in the cytoplasm upon pathogen infection, and innate immune response is compromised in Drosophila larvae (29). In Arabidopsis, recently published results show that a putative nucleoporin 96 protein is required for both basal resistance against bacterial pathogens and for R-gene-mediated pathogen resistance (30). These parallels are tantalizing and might have implications for the emerging comparative studies of IL-1/Toll receptor-mediated innate immune response in mammals/flies, inducible disease resistance in plants, and symbiotic interactions (31). A general role in plant–microbe interaction may also explain the absence of widespread developmental changes in Lotus nup133 mutants. In yeast, Nup133p is involved in controlling the RanGTPase gradient across the nuclear envelope that, in turn, regulates import processes and nuclear pore assembly (32). Deletion of NUP133 leads to reduced binding of the RanGTPase nuclear carrier Ntf2p protein to nuclear pore complexes. In light of these results, RanGTPase interaction could be the key to understanding NUP133 function during symbiosis and a target to take into consideration in pharmacological studies using agonist and antagonists to study early signal transduction downstream of Nod-factor perception (33). An alternative, more speculative, possibility is a NUP133 involvement in nuclear calcium oscillations (34). Atomic force microscopy of Xenopus nuclear pores showed that the distal ring of the nuclear basket opens and closes in a calcium-dependent fashion and was suggested to serve as a calcium-sensitive iris-like diaphragm (35). Cryoelectronic tomography studies of Xenopus nuclear pore architecture also revealed an interesting temperature-dependent correlation between plugging of nuclear pores and nucleocytoplasmic transport (36).
Functional analysis of nuclear pores, nucleoporins, and nuclear trafficking in plants is in its infancy and we expect Lotus nup133 mutants to provide inroads to these studies. Further impact is expected from characterization of the first Arabidopsis nucleoporin (Nup96) mutant isolated in a parallel effort (30). We find it very interesting that both the Lotus and the Arabidopsis nucleoporin genes identified are involved in plant–microbe interaction. This finding indicates an essential function for nuclear–cytoplasmic trafficking in plant responses to microbes and directs attention toward a role for a plant complex corresponding to the Nup107–160 subcomplex that, in mammalian nuclear pores, contains both Nup133 and Nup96 proteins. In symbiotic studies, one of the future challenges is to explain the NUP133-dependent calcium spiking mounted by wild-type plants 10 min after Nod-factor application and the intriguing paucity of Ca2+ spiking in nup133 mutants carrying weaker alleles that unexpectedly initiate nodule development albeit at reduced frequency. Signaling involving both plastids through CASTOR/POLLUX and the nucleus through NUP133 makes the early symbiotic signal transduction pathway unusual. To our knowledge, a developmental signaling process dependent on participation of these two subcellular compartments in the first stages of signal amplification has not previously been found in plants. We expect that further studies of this pathway will provide insights into eukaryotic processes coordinating nuclear and organellar activities as well as a better understanding of plant–microbe interactions.
Supplementary Material
Acknowledgments
We thank Giles Oldroyd and Jongho Sun for help with establishing and using microinjection and epifluorescence microscopy. The work in the laboratory of J.A.D. was supported by the Biotechnology and Biological Sciences Research Council; H.M. was supported by a John Innes Foundation studentship and an award from Universities UK, Overseas Research Student's Awards Scheme. N.K. was supported by The Scandinavia–Nippon Sasakawa Foundation, The Sumitomo Foundation, and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Author contributions: T.H.J., S.T., and J.S. designed research; N.K., L.H.M., S.R., M.F., H.M., E.K.J., H.H.F., L.L.H., S.S., Y.N., and N.S. performed research; N.K., L.H.M., S.R., M.F., E.M.H.Q., H.M., J.A.D., E.K.J., H.H.F., L.L.H., S.S., N.S., and J.S. analyzed data; and J.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: eYFP, enhanced yellow fluorescent protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ890251, AJ890252, and AP008949–AP008952).
References
- 1.Harrison, M. J. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 361-389. [DOI] [PubMed] [Google Scholar]
- 2.Parniske, M. (2000) Current Opin. Plant Biol. 3, 320-328. [DOI] [PubMed] [Google Scholar]
- 3.Stougaard, J. (2000) Plant Physiol. 124, 531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stougaard, J. (2001) Curr. Opin. Plant Biol. 4, 328-335. [DOI] [PubMed] [Google Scholar]
- 5.Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Webb, J. K., et al. (2005) Plant Cell 17, 2217-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., et al. (2003) Nature 425, 585-592. [DOI] [PubMed] [Google Scholar]
- 7.Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., et al. (2003) Nature 425, 637-640. [DOI] [PubMed] [Google Scholar]
- 8.Ehrhardt, D. W., Wais, R. & Long, S. R. (1996) Cell 85, 673-681. [DOI] [PubMed] [Google Scholar]
- 9.Harris, J. M., Wais, R. & Long, S. R. (2003) Mol. Plant–Microbe Interact. 16, 335-341. [DOI] [PubMed] [Google Scholar]
- 10.Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., et al. (2002) Nature 417, 959-962. [DOI] [PubMed] [Google Scholar]
- 11.Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P. & Kiss, G. B. (2002) Nature 417, 962-966. [DOI] [PubMed] [Google Scholar]
- 12.Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E., Ayax, C., Lévy, J., Debelle, F., Baek, J. M., Kalo, P., et al. (2004) Science 303, 1364-1367. [DOI] [PubMed] [Google Scholar]
- 13.Lévy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., Journet, E. P., Ané, J. M., Lauber, E., Bisseling, T., et al. (2004) Science 303, 1361-1364. [DOI] [PubMed] [Google Scholar]
- 14.Imaizumi-Anraku, H. Takeda, N., Charpentier, M., Perry, J., Miwa, H., Umehara, Y., Kouchi, H., Murakami, Y., Mulder, L., Vickers, K., et al. (2004) Nature 433, 527-531. [DOI] [PubMed] [Google Scholar]
- 15.Oldroyd, G. E. & Downie, J. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 566-576. [DOI] [PubMed] [Google Scholar]
- 16.Schauser, L., Handberg, K., Sandal, N., Stiller, J., Thykjær, T., Pajuelo, E., Nielsen, A. & Stougaard, J. (1998) Mol. Gen. Genet. 259, 414-423. [DOI] [PubMed] [Google Scholar]
- 17.Wais, R. J., Keating, D. H. & Long, S.R. (2002) Plant Physiol. 129, 211-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doye, V., Wepf, R. & Hurt, E. C. (1994) EMBO J. 13, 6062-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stougaard, J. (1995) Methods Mol. Biol. 49, 49-61. [DOI] [PubMed] [Google Scholar]
- 20.Sandal, N., Krusell, L., Radutoiu, S., Olbryt, M., Pedrosa, A., Stracke, S., Sato, S., Kato, T., Tabata, S., Parniske, M., et al. (2002) Genetics 161, 1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berke, I. C., Boehmer, T., Blobel, G. & Schwartz, T. U. (2004) J. Cell Biol. 167, 591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter, C. A., Aukerman, M. J., Sun, H., Fokina, M. & Poethig, R. S. (2003) Plant Physiol. 132, 2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose, A., Patel, S. & Meier, I. (2004) Planta 218, 327-336. [DOI] [PubMed] [Google Scholar]
- 24.Lutzmann, M., Kunze, R., Buerer, A., Aebi, U. & Hurt, E. (2002) EMBO J. 21, 387-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen, N. P., Patel, S. S., Huang, L., Chalkley, R. J., Burlingame, A., Lutzmann, M., Hurt, E. C. & Rexach, M. (2002) Mol. Cell Proteomics 1, 930-946. [DOI] [PubMed] [Google Scholar]
- 26.Walther, T. C., Alves, A., Pickersgill, H., Loiodice, I., Hetzer, M., Galy, V., Hulsmann, B. B., Kocher, T., Wilm, M., Allen, T., et al. (2003) Cell 113, 195-206. [DOI] [PubMed] [Google Scholar]
- 27.Vasu, S. K. & Forbes, D. J. (2001) Curr. Opin. Cell Biol. 13, 363-375. [DOI] [PubMed] [Google Scholar]
- 28.Li, O., Heath, C. V., Amberg, D. C., Dockendorff, T. C., Copeland, C. S., Snyder, M. & Cole, C. N. (1995) Mol. Biol. Cell 6, 401-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uv, A. E., Roth, P., Xylourgidis, N., Wickberg, A., Cantera, R. & Samakovlis, C. (2000) Genes Dev. 14, 1945-1957. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y. & Li, X. (2005) Plant Cell 17, 1306-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nürnberger, T., Brunner, F., Kemmerling, B. & Piater, L. (2004) Immunol. Rev. 198, 249-266. [DOI] [PubMed] [Google Scholar]
- 32.Gao, H., Sumanaweera, N., Bailer, S. M. & Stochaj, U. (2003) J. Biol. Chem. 278, 25331-25340. [DOI] [PubMed] [Google Scholar]
- 33.Charron, D., Pingret, J. L., Chabaud, M., Journet, E. P. & Barker, D. G. (2004) Plant Physiol. 136, 3582-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Echevarria, W., Leite, M. F., Guerra, M. T., Zipfel, W. R. & Nathanson, M. H. (2003) Nat. Cell Biol. 5, 440-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoffler, D., Goldie, K. N., Feja, B. & Aebi, U. (1999) J. Mol. Biol. 287, 741-752. [DOI] [PubMed] [Google Scholar]
- 36.Stoffler, D., Feja, B., Fahrenkrog, B., Walz, J., Typke, D. & Aebi, U. (2003) J. Mol. Biol. 328, 119-130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.