Abstract
During humoral immune responses, two distinct genetic modification events diversify the Ig genes in germinal center (GC) B cells: somatic hypermutation and class switch recombination (CSR). Both processes require the activity of activation-induced cytidine deaminase (AID), an enzyme expressed specifically in GC B cells. However, the mechanisms that regulate AID activity are largely unknown. Here we report that protein kinase A (PKA) phosphorylates AID and regulates its activity in GC B cells. AID physically interacts with the PKA holoenzyme in the cytoplasm and is phosphorylated by the PKA catalytic subunit at specific residues. AID phosphorylation is required for CSR, because substitution of the two phosphorylation targets impairs its ability to rescue CSR in AID-deficient B cells. Pharmacologic inhibition of PKA prevents isotype class switching in a murine B-cell lymphoma cell line; conversely, B cells from mice where PKA activity is made constitutive by conditional deletion of the PKA regulatory subunit gene display enhanced CSR. These findings implicate PKA in the regulation of AID function and suggest that the control of T cell-dependent immune responses may be modulated, via AID, by signals that activate PKA.
Keywords: class switch recombination, phosphorylation
During T cell-dependent immune responses, the antibody genes are diversified in germinal center B cells by two orderly regulated DNA modification mechanisms. Somatic hypermutation (SHM) introduces single-nucleotide substitutions into the variable region of the Ig genes, allowing the selection of B cells that produce high-affinity antibodies (1, 2). Class switch recombination (CSR) is an intrachromosomal deletion/recombination event that replaces the heavy-chain constant region (CH) from Cμ to downstream CH segments, thereby conferring distinct effector functions to the antibodies without affecting their specificity for the antigen (3, 4).
Both SHM and CSR depend on activation-induced cytidine deaminase (AID), a B cell-specific deaminase whose expression is restricted to germinal center B cells and activated B cells (5-8). However, the mechanism by which AID initiates these DNA modification events remains a matter of debate. Although initial studies suggested that AID functions as an RNA-editing enzyme (8-10), substantial evidence indicates that AID can exert cytidine deaminase activity directly on DNA (11-16). Analysis of loss-of-function AID mutants suggests that interaction with specific co-factors is required to differentially regulate its activity on such independent processes: SHM and CSR (17, 18). Posttranslational modifications of AID have also been evoked as an essential requirement for its deaminase activity. Indeed, it was recently shown that AID interacts specifically with replication protein A in a manner that is phosphorylation-dependent when targeting double-stranded DNA on in vitro-transcribed SHM substrates (19). The identification of signals regulating AID is critical to understand how SHM and CSR are controlled and will also help in elucidating the mechanism by which these two functions operate aberrantly in B cell neoplasms displaying CSR-mediated chromosomal translocations or aberrant SHM (20, 21).
To identify AID-interacting proteins that may regulate its function, we have attempted the isolation of AID-containing complexes from B cells. Using this approach, we identified the type I regulatory subunit of PKA as one of the AID binding partners and provide evidence that PKA is the kinase responsible for AID phosphorylation in B cells. These data define an important physiologic role for PKA in CSR and possibly SHM, with relevant implications for the understanding of the mechanisms regulating immune responses.
Results
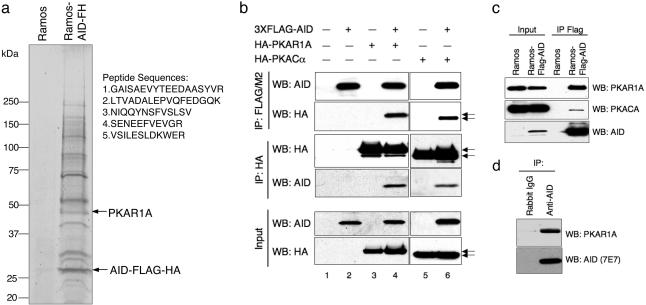
AID Interacts with PKA in the Cytoplasm of B Cells. To identify proteins associated with AID in B cells, we used a biochemical purification approach based on epitope tagging and affinity chromatography. We first engineered the Burkitt lymphoma line Ramos to stably express a C-terminal double-tagged AID-FLAG-HA (AID-FH) protein (HA, hemagglutinin). As previously described for endogenous AID, ≈90% of exogenous AID was found in the cytoplasm in steady-state conditions, whereas only trace amounts could be detected in the nuclear fraction (data not shown; ref. 6). We therefore subjected cytoplasmic extracts from Ramos-AID-FH and control cells transduced with empty vectors to sequential immunoaffinity purification with anti-FLAG antibodies, followed by anti-HA antibodies, and recovered the bound complex by elution with HA peptide. The double-step affinity purification method has the advantage of removing possible contaminants due to nonspecific binding of cellular proteins to the FLAG/M2 beads, because they will not bind to anti-HA antibodies. Thus, complexes can be isolated in virtually pure form (22). Colloidal Coomassie blue staining of purified complexes after SDS/PAGE analysis showed that a number of polypeptides were reproducibly seen in Ramos-AID-FH, but not in Ramos cells transduced with an empty vector (Fig. 1a), indicating that they represent proteins specifically interacting with AID.
Fig. 1.
Identification of PKAR1A as a cytoplasmic AID-interacting protein in vivo. (a) Colloidal blue staining of affinity-purified AID-FH complexes from cytoplasmic extracts of Ramos-AID-FH and control Ramos cells transduced with pBABEpuro retroviral vectors. Specific AID-associated bands were sequenced by mass spectrometry, and the PKAR1A peptide sequences are indicated. (b) Immunoblot analysis of AID and HA expression in FLAG/M2 (Top) and HA (Center) precipitates from 293T cells untransfected (lane 1) or transfected with vectors expressing 3XFLAG-AID, HA-PKAR1A, and HA-PKACA. In the experiments from lanes 2, 3, and 5, control empty vectors containing the 3XFLAG or HA tags only were present to monitor for nonspecific binding between the two tags. Total inputs before immunoprecipitation (30 μg) are shown at Bottom. Each set of arrows point to the PKAR1A (upper) and PKACA (lower) polypeptides. (c) In vivo interaction between endogenous PKA and AID in B cells. Cytoplasmic extracts from stable Ramos-FLAG-AID cells were immunoprecipitated by using FLAG/M2 antibodies, followed by peptide elution, and analyzed with PKAR1A, PKACA, and AID antibodies. Input corresponds to 2% of the lysate used for immunoprecipitation. (d) Physical interaction between AID and PKAR1A in the B cell line VAL. Cytoplasmic extracts were immunoprecipitated by using rabbit polyclonal anti-AID antibodies or control rabbit IgG and analyzed by Western blot with anti-AID (7E7) and anti-PKAR1α mouse monoclonal antibodies.
Peptide sequencing by mass spectrometry identified the c-AMP-dependent protein kinase A regulatory subunit Iα (PKAR1A) as a component of the AID complex (Fig. 1a). PKA is a serine/threonine protein kinase that controls a variety of key cellular processes (23-25). In unstimulated conditions, PKA exists predominantly in the cytoplasm as an inactive tetrameric holoenzyme, consisting of two regulatory subunits that serve as a receptor for cAMP, and two catalytic subunits that dissociate upon binding of cAMP, leading to enzyme activation (23). The identification of this kinase in the AID-containing complex was of particular interest because it has been recently shown that AID must be phosphorylated to interact with replication protein A and exert its activity on double-stranded DNA substrates (19). Thus, we first tested whether AID physically interacts with the PKA holoenzyme by cotransfecting 293T cells with vectors expressing 3XFLAG-AID and either PKA catalytic subunit α (PKACA) or PKAR1A, tagged with HA. Immunoblot analysis of total extracts after FLAG/M2 immunoprecipitation showed that both subunits interact with AID in cotransfected cells (Fig. 1b Top Lower, lanes 4 and 6), but not in cells transfected with 3XFLAG-AID alone (lane 2) or the HA-PKA subunits only (lanes 3 and 5). Reciprocally, AID was immunoprecipitated by HA antibodies in cells cotransfected with HA-PKACA or HA-PKAR1A (Fig. 1b Center Lower, lanes 4 and 6) but not in cells transfected with AID and a vector containing the HA tag (Fig. 1b Center Lower, lane 2). To confirm that this complex is formed in B cells, coimmunoprecipitation experiments were performed from cytoplasmic extracts of Ramos cells stably expressing a FLAG-AID protein. As shown in Fig. 1c, anti-PKAR1A antibodies after FLAG/M2 precipitation detected a specific band in Ramos-FLAG-AID, but not in control Ramos cells (Top), documenting that the endogenous PKAR1A subunit interacts with AID in B cells in vivo. The PKACA subunit was also detected in the immunoprecipitate, but at significantly lower levels as compared to the total input (Middle) (see Discussion). Finally, physical interaction between AID and PKAR1α was observed in the cytoplasm of native B cells by using anti-AID antibodies and the B cell line VAL, which expresses high levels of endogenous AID (Fig. 1d). Thus, AID is present in a complex containing the regulatory subunit Iα and, possibly, the catalytic subunit of PKA in the cytoplasm of B cells, suggesting that PKA itself may contribute in part to the localization of AID in this compartment.
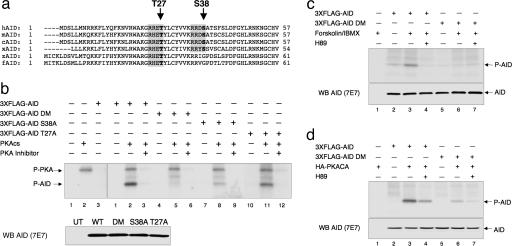
PKA Phosphorylates AID in Vitro and in Vivo. A search for potential PKA phosphorylation sites in the human AID protein revealed the presence of two putative target residues corresponding to threonine 27 (T27) and serine 38 (S38). In humans and chickens, both sites are embedded within PKA canonical consensus motifs (RRXS/T), whereas an amino acid substitution is present at position -2 in the T27 consensus from other species, and the S38 residue is not conserved in AID orthologs from zebra fish and Japanese puffer fish (Fig. 2a). To assess whether AID can serve as a target for PKA-mediated phosphorylation, we first performed in vitro phosphorylation assays by using purified PKACA and partially purified 3XFLAG-AID proteins. Immunoblotting with anti-AID antibodies controlled that comparable amounts of protein were used in the assay (Fig. 2b Lower). The results showed that AID is phosphorylated by PKACA (Fig. 2b Right, lane 2) and this modification is specific, because it can be blocked by addition of a PKA inhibitor peptide (lane 3), but not by protein kinase C and calcium/calmodulin kinase inhibitors that were used throughout the assay. In contrast, combined substitution of T27 and S38 to alanine completely abrogated phosphorylation (Fig. 2b Right, lane 5), demonstrating that they are the critical residues. A significant decrease in the ability to be phosphorylated was also observed in the AID-S38A single mutant and, to a lesser extent, in AID-T27A (Fig. 2b Right, lanes 8 and 11; see also Fig. 6, which is published as supporting information on the PNAS web site), suggesting an additive effect between these two sites, with S38 representing the major target (see Discussion). Overall, these findings indicate that AID is a substrate for PKA-mediated phosphorylation in vitro and that phosphorylation targets specifically T27 and S38.
Fig. 2.
AID is phosphorylated by PKA in vitro and in vivo at S38 and T27. (a) Conservation of AID peptide sequences (AA 1-57) from human (h), mouse (m), chicken (c), Xenopus (x), zebrafish (z), and Japanese puffer fish (f). Shaded amino acids correspond to the consensus PKA phosphorylation motifs; the T27 and S38 residues are in bold. (b Upper Right) Semipurified 3XFLAG-AID or its derivatives were incubated with recombinant PKA catalytic subunit (cs) and [γ-32P]ATP. Eluates from untransfected 293T cells precipitated with FLAG antibodies were used as control (Left, lane 1). The reactions were loaded on SDS/PAGE and analyzed by autoradiography. Data shown are representative of three independent experiments. P-AID, phosphorylated AID. The recombinant PKA subunit, which is known to autophosphorylate itself, can also be detected in samples where the enzyme was added (P-PKA). The nature of the additional faint band between P-AID and P-PKA is unclear, because it was observed only when high amounts of immunoprecipitate were used as substrate in the assay and was not visible by Coomassie. Immunoblot analysis with anti-AID antibodies documents that equivalent amounts of substrate were used in the assay (Lower). (c Upper) Forskolin treatment selectively induces phosphorylation of AID in vivo. FLAG precipitates were prepared from total cell extracts of 32P-labeled 293T cells transfected with the indicated vectors and treated with forskolin/3-isobutyl-1-methylxantine (IBMX) for 1 h before harvesting. The PKA inhibitor H89 was added together with 32P-orthophosphate at time 0 (i.e., 40 h after transfection). The apparent residual phosphate labeling in the AID-DM transfectants may reflect the presence of additional, minor, PKA-independent phosphorylation sites or background from the immunoprecipitation reaction. Immunoblot analysis with anti-AID antibodies on the same eluates is shown in Lower.(d Upper) PKA phosphorylates AID in vivo. Autoradiography of FLAG precipitates prepared from 32P-labeled 293T cells cotransfected with plasmids expressing HA-PKACA and 3XFLAG-AID (WT or DM), in the presence or absence of H89, added 30 min beforeγ-32P. (d Lower) Immunoblot analysis with anti-AID antibodies controls for equal loading.
To examine whether AID is phosphorylated by PKA in vivo, we performed metabolic labeling on 293T cells transfected with 3XFLAG-AID and either induced to activate endogenous PKA by treatment with the stimulator of adenylate cyclase Forskolin or cotransfected with plasmids expressing the free, active PKACA. Although very low phosphorylation levels are observed in unstimulated 293T cells (Fig. 2c, lane 2), phosphorylation was significantly induced after a 1 h exposure to forskolin plus the phosphodiesterase inhibitor 3-isobutyl-1-methylxantine (IBMX) (lane 3). Forskolin-induced phosphorylation of AID was completely blocked by pretreatment with the selective PKA inhibitor H89 (26) (lane 4), consistent with the involvement of PKA. Notably, the AID-DM protein could not be phosphorylated in this assay (lane 6), confirming that S38 and T27 are the critical phosphorylation targets in vivo. The direct involvement of PKA in this posttranslational modification event was further demonstrated by monitoring AID phosphorylation levels in 293T cells labeled with 32P-orthophosphate after cotransfection with HA-PKACA plasmids. Analogous to what was observed after pharmacologic activation of endogenous PKA, expression of exogenous active PKACA led to a dramatic induction of AID phosphorylation that was significantly attenuated by addition of H89 and was barely detectable in AID-DM (Fig. 2d, lanes 3, 4, and 6). The presence of phosphorylated PKA (P-PKA) could also be detected in the cotransfected samples, albeit at very low levels, most likely due to the high stringency of the immunoprecipitation (Fig. 7, which is published as supporting information on the PNAS web site). Overall, these data demonstrate that PKA phosphorylates AID in vivo at specific residues.
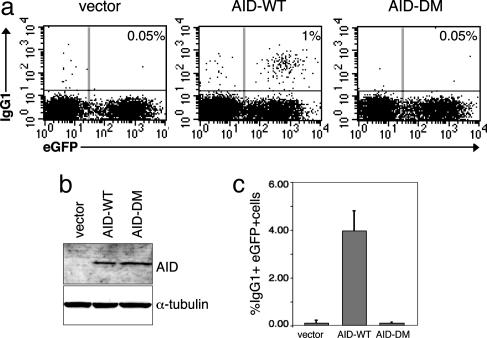
AID Phosphorylation Is Required for CSR. To examine whether PKA-mediated phosphorylation is required for AID activity, we tested whether the phosphorylation-defective AID-DM was able to restore CSR in B cells from AID-/- mice, which lack completely both CSR and SHM (6). To this end, AID-/- B cells, infected with retroviral vectors expressing either AID-WT or AID-DM and an eGFP indicator, were stimulated with LPS plus IL-4 to undergo CSR to IgG1. Although expressed at comparable levels, the AID-DM protein was unable to rescue CSR (IgG1+ cells in the virally transduced population, identified by eGFP expression: 0.1% = 0.05% of the total, comparable to cells transduced with vector alone), whereas a distinct IgG1+ population (≈5%, corresponding to 1% of the total) was detected in AID-WT transduced cells (Fig. 3). Although the low reconstitution efficiency of our assay may not allow an assessment of a possible residual activity in AID-DM, a study published while this manuscript was under revision tested individual mutations at S38 and T27 and showed that CSR is impaired, yet not completely abrogated, in both mutants (27). Thus, in human and mouse, integrity of these sites is required for full AID function in CSR (see Discussion). Although we cannot discard the possibility that other mechanisms, such as alterations in protein folding or loss of enzymatic activity, contribute to impaired AID function in the mutant, these results are consistent with a role for PKA-mediated phosphorylation in the regulation of AID activity.
Fig. 3.
Mutation of the phosphorylation sites abrogates AID activity on CSR. (a) Representative flow-cytometric profiles of AID-/- murine B cells, stimulated with LPS plus IL-4 and transduced with the indicated vectors. Cells were stained with anti-IgG1-APC, and numbers indicate the percentage of cells switched to IgG1 in the total population (upper right quadrant). The results of three independent experiments (mean IgG1+ eGFP+ cells ± standard deviation) are summarized in c. Note that although considerably lower than that reported in previous studies (most likely due to the vector used), the reconstitution efficiency was still 20-fold above background and, therefore, sufficient to demonstrate the difference in the activity of the AID mutant. (b) Immunoblot analysis of AID expression in whole-cell extracts from the AID-/- B cells shown in a. α-tubulin was used as control for loading.
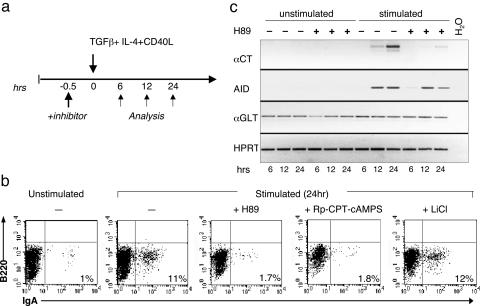
To further corroborate this notion, we tested whether pharmacologic inhibition of PKA prevents CSR in the CH12F3 cell line, an IgM+ B cell lymphoma line that can be induced to express AID and to switch specifically to IgA by treatment with TGFβ, IL-4, and CD40L (28) (Fig. 4a). Two potent and selective inhibitors of PKA, acting with distinct modalities, were used as follows: H89, an ATP analog, and Rp-8-CPT-cAMPs, a cell-permeable, cAMP antagonist (29). FACS analysis for surface IgA expression showed that ≈12% of untreated cells had switched at 24 h of stimulation, as expected (Fig. 4b, left two graphs); in contrast, <2% of cells had switched to IgA in the presence of either H89 or Rp-8-CPT-cAMPs (Fig. 4b Middle and Middle Right). This effect does not seem to be due to drug-related toxicity, because IgA expression was determined on live cells; furthermore, the two compounds induced different levels of cell death, which did not correlate with the extent of reduction in the percentage of IgA+ cells (Fig. 8, which is published as supporting information on the PNAS web site). Inhibitors of other kinases, including GSK3 (LiCl, 20 mM) (Fig. 4b Right) and PI3K (Wortmannin, 125 nM, data not shown), did not affect CSR, providing further support for the specific involvement of PKA in regulating this process. We also monitored the expression in these cells of Iα-Cμ circle transcripts (αCT), a sensitive and early molecular marker of CSR that is produced from circular DNA looped out after CSR (30). RT-PCR analysis at different time points after induction of CSR showed the appearance of αCT specifically in stimulated cells but not in stimulated cells pretreated with H89 (Fig. 4c). This effect cannot be ascribed to a general inhibition of transcription from the Iα promoter, because α-germline transcripts (α-GLT) were detectable at comparable levels in all samples (Fig. 4c); moreover, both AID RNA (Fig. 4c Center Top) and protein (data not shown) expression were induced efficiently in the presence of H89, consistent with the hypothesis that the block in CSR involves a posttranslational modification of AID. Collectively, these data suggest that inhibition of PKA impairs CSR by preventing AID phosphorylation.
Fig. 4.
Pharmacologic inhibition of PKA impairs CSR in B cells. (a) Treatment schedule for induction of CSR in CH12F3 cells. (b) FACS analysis of CH12F3 cells unstimulated (Left) or stimulated for 24 h with TGFβ, IL-4 and CD40L (remaining four graphs), in the absence or presence of two PKA specific inhibitors (H89 and Rp-8-CPT-cAMPS) (Middle and Middle Right) and the GSK3 kinase inhibitor LiCl (Right). The percentage of B220+IgA+ cells (gated on live cells) is indicated. Data are representative of several independent experiments. (c) Expression of Iα-Cμ circle transcripts (αCT), AID, and αGLT in CH12F3 cells treated for 6, 12, and 24 h as indicated in the scheme. Hypoxanthine phosphoribosyltransferase (HPRT) transcripts were amplified as an internal control. Data are representative of three independent experiments that gave analogous results.
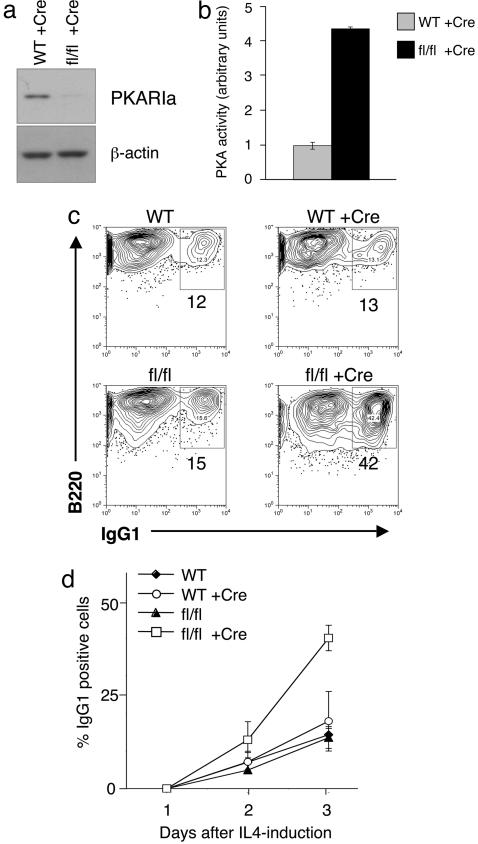
Constitutive PKA Activity Enhances CSR. We then examined whether PKA has a regulatory role on AID function in CSR by testing whether constitutive PKA signaling was associated with enhanced CSR. To this end, we analyzed B cells from mice (R1alphafl/fl) in which the PKA R1alpha gene can be conditionally inactivated via Cre-mediated deletion, leading to constitutive PKA activity (Fig. 9a, which is published as supporting information on the PNAS web site). Purified splenic B cells from R1alphafl/fl mice and WT littermates were first transduced with a permeable TAT-Cre protein or medium as control and then cultured in the presence of LPS +/- IL-4. PCR analysis confirmed efficient Cre-mediated deletion of the floxed allele (Fig. 9b), whereas immunoblot with specific antibodies documented the absence of PKAR1A protein expression (Fig. 5a), that, in turn, was associated with increased PKA activity, as determined by a kinase assays (Fig. 5b). The percentage of surface IgG1+ cells, measured by FACS analysis 3 days after the initiation of IL-4 treatment, was ≈13% in cells from WT or RIalphafl/fl mice that had not been transduced with TAT-Cre (Fig. 5 c and d). In contrast, 42% of the transduced RIalphafl/fl B cells were IgG1+ (Fig. 5 c and d). This enhancement was not due to an increase in the number of cell divisions, because activation of PKA in deleted R1alphafl/fl cells is actually associated with a reduction in cell proliferation (data not shown). These findings demonstrate that constitutive PKA activation promotes enhanced CSR, consistent with the notion that PKA-mediated phosphorylation of AID regulates its activity.
Fig. 5.
Enhanced CSR in conditional RIalphafl/fl mice. (a) Immunoblot analysis of PKAR1A expression in whole-cell extracts from purified splenic B cells of RIalphafl/fl and WT littermates 48 h after TAT-Cre transduction. Actin, loading control. (b) PKA kinase activity in the extracts from a measured by an in vitro assay and normalized to protein levels; the activity of WT B cells was arbitrarily set as 1. (c) FACS analysis of purified B cells from WT and R1alphafl/fl mice, untransduced or transduced with TAT-Cre and stimulated by LPS+IL-4. Numbers indicate the percentage of surface IgG1+ B220+ cells. One representative experiment of three giving comparable results is shown. (d) Time-dependent increase in IgG1+ cells after IL-4 treatment. Values represent the mean from three independent experiments, and error bars represent the standard deviation from the mean.
Discussion
Interaction Between PKA and AID. The PKA serine/threonine kinase is one of the most common downstream effectors of the cAMP signaling pathway (23, 31). In unstimulated conditions, PKA exists predominantly in the cytoplasm as an inactive tetrameric holoenzyme, consisting of a regulatory subunit dimer that serves as a receptor for cAMP, and two catalytic subunits that dissociate upon binding of cAMP, becoming catalytically active and ready to phosphorylate specific serine and threonine residues on substrate proteins (23, 31). We have found that the regulatory subunit of the PKA holoenzyme coimmunoprecipitates with AID from the cytoplasm of B cells and that both catalytic and regulatory subunits can interact with AID in transient transfection experiments on total cell extracts. However, the catalytic subunit was present at considerably lower levels in the cytoplasm of B cells, where it could be detected only after large-scale immunoprecipitation (Fig. 1c), and did not represent a major component in the AID-associated complex (Fig. 1a). Based on this observation, we cannot conclude on whether cytoplasmic AID interacts directly and independently with the regulatory and catalytic subunit or whether the interaction with the catalytic subunit is indirect within the PKA holoenzyme. Given the stronger interaction between PKAR1A and AID, one possible scenario is that PKAR1A serves as a direct binding “scaffold” to retain AID in the cytoplasmic compartment until specific signals are activated. At the same time, AID would be in close proximity to the PKA catalytic subunit and, therefore, readily available as a substrate for phosphorylation, once the hetero-tetrameric enzyme becomes dissociated. It remains to be determined whether AID is phosphorylated in the cytoplasm before shuttling to the nucleus, or directly in the nucleus after translocation of the two polypeptides. Indeed, both cytoplasmic and nuclear proteins have been identified as substrates for PKA (23, 32). We were not able to document AID-PKACA binding in immunoprecipitates from B cell nuclear extracts by Western blot analysis (data not shown); however, this result may be due to the very low amount of AID that can be isolated from this compartment and to the unstable nature of substrate-kinase physical interactions. Thus, additional studies will be required to elucidate the kinetic and subcellular location of the phosphorylation reaction.
Role of Phosphorylation in AID Function. Previous studies have reported that the activity of AID on in vitro transcribed SHM substrates depends on its interaction with replication protein A and that this interaction requires AID phosphorylation (18). Our results identify PKA as the critical effector of AID phosphorylation in vivo. Our results also identify S38 as the major target residue for PKA-mediated phosphorylation in the human AID sequence, with T27 representing a minor site. Notably, phosphorylation of AID on S38 was confirmed directly by mass spectrometric analysis of the endogenous murine protein in a recent study (27). The critical role for PKA-mediated phosphorylation of AID at S38 is supported by the observation that this residue is embedded in a canonical PKA recognition motif conserved in vertebrates proficient for both SHM and CSR (Fig. 2a). Indeed, the ex vivo complementation assay in AID-/- B cells (Fig. 3 and ref. 27) provides experimental evidence for an essential role of these two sites in conferring full biologic activity to the human AID sequence, at least in CSR. However, recent studies by Barreto et al. (33) have shown that teleost AID, where S38 is substituted by a glycine, is still capable of mediating CSR when expressed in AID-deficient B cells. One simple explanation for this apparent discrepancy may be that phosphorylation evolved later as a mechanism to allow tight regulation of AID activity in species undergoing CSR, a function lacking in Japanese puffer fish and zebrafish (34). Alternatively, it is possible that other residues mimic S38 phosphorylation in these organisms.
Biological Significance of PKA-Mediated Regulation of AID Function. The results herein suggest a role for PKA in modulating AID function and, therefore, in the control of antibody responses. Multiple signals activate PKA, including hormones and cytokines that act through cAMP, as well as mechanisms that act in a cAMP-independent fashion (35, 36). Given the multitude of PKA-activating factors, further work is necessary to identify those that may be critical for PKA-mediated regulation of AID. However, it is worth noting that the finding of AID as a target of PKA links AID regulation to other critical pathways that modulate B cell responses within the germinal center. In fact, PKA activation has been implicated in the control of B cell receptor (BCR) signaling and in the function of NF-κB (23, 35), a key transcriptional mediator of signals from the BCR and from T cells and cytokines. Thus, the present results suggest that, via regulating AID, PKA has a key role in integrating the signals from antigens and T cells that control AID-mediated affinity maturation and antibody class diversification in germinal center B cells.
Materials and Methods
Reagents. The cAMP analogue Rp-8-Br-cAMP was purchased from BioLog Life Sciences. Forskolin, 3-isobutyl-1-methylxantine, wortmannin, and LiCl were from Sigma, and H89 was obtained from Calbiochem.
Expression Vectors. The double-tagged pBABE-puro-AID-FH retroviral vector was constructed by introducing a Pfu-PCR-amplified human AID ORF into the BamHI-EcoRI sites of pBABE-puro, N-terminal, and in-frame to a tandem FLAG-HA sequence. The pCMV-3XFLAG-AID vector was constructed by subcloning the AID coding sequence into p3XFLAG-CMV-10 (Sigma), and mutants containing single or combined substitutions of T27 and S38 to alanine were generated by the QuikChange Site-Directed Mutagenesis kit (Stratagene). The HA-tagged constructs were obtained by cloning the human PKAR1α and PKACA ORF in pCMV-HA (Clontech). For the complementation assay, retroviral constructs were generated by subcloning WT and double-mutant AID coding sequences into a pMSCV-based EGFP-expressing retroviral vector. All constructs were verified by enzymatic digestion and sequencing.
Purification of AID-Interacting Proteins and Peptide Sequencing. The epitope tagging strategy to isolate AID-containing complexes is described in detail as Supporting Text, which is published as supporting information on the PNAS web site.
Transient Transfection, Western Blot, and Immunoprecipitation Assays. Transient transfections in 293T cells were performed by calcium phosphate precipitation. Equimolar amounts of control empty vectors containing the tag only were cotransfected in each experiment to monitor for nonspecific interactions between the tags. The protocol for immunoprecipitation/Western blot analysis and the antibodies against AID, HA, β-actin, and α-tubulin were described in ref. 8. Anti-PKAR1α (BD Transduction Laboratories) and anti-PKAαcat (C-20, Santa Cruz Biotechnology) antibodies were also used. The SuperSignal West Dura Extended Duration Substrate (Pierce) was applied for detection of endogenous proteins in coimmunoprecipitation arrays.
In Vitro PKA Phosphorylation Assay. PKA phosphorylation of AID was assessed in vitro by a PKA assay kit (Upstate Biotechnology) by using purified PKA (25 ng) and, as substrate, semipurified AID proteins obtained from total cell extracts of 293T cells, transfected with vectors expressing WT and mutant 3XFLAG-AID and immunoprecipitated with FLAG/M2 in high-salt buffer [50 mM Tris·Cl, pH 7.9/500 mM NaCl/1 mM EDTA/1% Triton X-100/10% glycerol/protease inhibitors (Sigma)], followed by 3XFLAG-peptide elution (Sigma). Similarly purified extracts from untransfected 293T cells served as a control. The reaction was stopped after 20 min at 30°C by adding equal volumes of 2× SDS sample buffer, and AID phosphorylation was analyzed by autoradiography after SDS/PAGE. Band intensities were quantified by using the nih image software.
Metabolic Labeling. 293T cells were transiently transfected with pCMV-3XFLAG-AID WT or DM (with or without pCMV-HA-PKACA), and labeled after 36-40 h in phosphate-free RPMI 1640 medium with 32P-orthophosphate (0.3 mCi/ml; 1 Ci = 37 GBq) for 4 h. Endogenous PKA was induced by treatment with forskolin (50 μM) and 3-isobutyl-1-methylxantine (100 μM) 1 h before harvesting. Where indicated, the H89 inhibitor (10 μM) was added 30 min before 32P-orthophosphate. Total cell extracts were immunoprecipitated with FLAG/M2 agarose and analyzed as described above.
Induction of CSR in CH12F3 Cells. CH12F3 cells were cultured and stimulated to undergo CSR as reported in ref. 28. Cells were stained with phycoerythrin-conjugated anti-B220 and FITC-conjugated anti-IgA antibodies and analyzed as described, gating for live cells by forward scatter versus side scatter (28). All antibodies were from BD-Pharmingen. Data were acquired on a FACSCalibur (Becton Dickinson) and analyzed with the cellquest software. Total RNA was extracted by TRIzol (Invitrogen); cDNA synthesis and RT-PCR amplification of Iα-Cμ circle transcripts, αGLT, AID and hypoxanthine phosphoribosyltransferase were performed as described in ref. 30.
AID Knockout and RIalpha Conditional Knockout Mice. The AID-/- mice were obtained from T. Honjo (Kyoto University, Kyoto) and maintained on a C57BL/6 background. The generation of RIalpha conditional knockout mice is described in Fig. 9.
Ex Vivo CSR Assay. Splenic B cells, purified as described in ref. 33, were cultured in RPMI medium 1640 containing 10% FBS, β-ME, and LPS (20 μg/ml) (Sigma) with or without IL-4 (10 ng/ml) (Pharmingen). For the complementation assay, AID-/- cells were transduced after 24 h with retroviral vectors expressing AID WT or -DM, and analyzed after 3 days as reported in ref. 33. Infection efficiency was consistently 20-25%, as determined by eGFP expression. In the RIalphafl/fl conditional deletion experiment, B220+ B cells were transduced with a permeable TAT-Cre protein (or control vehicle) immediately after purification and cultured as indicated above, except that IL-4 was added 48 h after transduction. Cells were collected 3 days later, stained with anti-B220-PE and anti-IgG1-APC, and analyzed on a LSR II (BD Biosciences) by using the flowjo software (TreeStar, Ashland, OR).
PKA Kinase Assay. The activity of PKA in cells from RIalphafl/fl mice was measured before and after Cre-mediated recombination by a PKA assay kit (Upstate Biotechnology) by using whole-protein extracts and Kemptide as substrate.
Supplementary Material
Acknowledgments
We thank W. Zhang for assistance in mass spectrometric analysis; W. Gu for advice in the purification of the AID complex; T. Honjo for the CH12F3 cell line and the AID-/- mice; K. Rajewsky for the permeable TAT-Cre protein; F. A. Alt for sharing unpublished data; J. Manis for the pMSCV vector; and U. Klein, D. Dominguez-Sola, and M. Li for suggestions. L.P. is a Special Fellow of the Leukemia and Lymphoma Society. Y.K. is a Senior Fellow of the Charles H. Revson Foundation. This work was supported by a National Institute of Health grant and a Specialized Center of Research grant from the Leukemia and Lymphoma Society (to R.D.-F.). H.G. and Y.K. are supported by The Irene Diamond Fund.
Author contributions: L.P. and R.D.-F. designed research; L.P. performed research; L.P. analyzed data; L.P. and R.D.-F. wrote the paper; Y.K. performed research and analyzed data on the PKA fl/fl mice; and H.G. designed the experimental plan involving the PKA fl/fl mice.
Conflict of interest statement: No conflicts declared.
Abbreviations: AID, activation-induced cytidine deaminase; AID-FH, C-terminal double-tagged AID-FLAG-HA; CSR, class switch recombination; HA, hemagglutinin; PKAR1A, protein kinase A regulatory subunit Iα; PKACA, PKA catalytic subunit α; SHM, somatic hypermutation.
References
- 1.Rajewsky, K. (1996) Nature 381, 751-758. [DOI] [PubMed] [Google Scholar]
- 2.Papavasiliou, F. N. & Schatz, D. G. (2002) Cell 109, Suppl., S35-S44. [DOI] [PubMed] [Google Scholar]
- 3.Manis, J. P., Tian, M. & Alt, F. W. (2002) Trends Immunol. 23, 31-39. [DOI] [PubMed] [Google Scholar]
- 4.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20, 165-196. [DOI] [PubMed] [Google Scholar]
- 5.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102, 565-575. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274, 18470-18476. [DOI] [PubMed] [Google Scholar]
- 8.Pasqualucci, L., Guglielmino, R., Houldsworth, J., Mohr, J., Aoufouchi, S., Polakiewicz, R., Chaganti, R. S. & Dalla-Favera, R. (2004) Blood 104, 3318-3325. [DOI] [PubMed] [Google Scholar]
- 9.Doi, T., Kinoshita, K., Ikegawa, M., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100, 2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honjo, T., Nagaoka, H., Shinkura, R. & Muramatsu, M. (2005) Nat. Immunol. 6, 655-661. [DOI] [PubMed] [Google Scholar]
- 11.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 12.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4, 452-456. [DOI] [PubMed] [Google Scholar]
- 13.Pham, P., Bransteitter, R., Petruska, J. & Goodman, M. F. (2003) Nature 424, 103-107. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422, 726-730. [DOI] [PubMed] [Google Scholar]
- 15.Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. (2003) Proc. Natl. Acad. Sci. USA 100, 4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu, K., Huang, F. T. & Lieber, M. R. (2004) J. Biol. Chem. 279, 6496-6500. [DOI] [PubMed] [Google Scholar]
- 17.Shinkura, R., Ito, S., Begum, N. A., Nagaoka, H., Muramatsu, M., Kinoshita, K., Sakakibara, Y., Hijikata, H. & Honjo, T. (2004) Nat. Immunol. 5, 707-712. [DOI] [PubMed] [Google Scholar]
- 18.Barreto, V., Reina-San-Martin, B., Ramiro, A. R., McBride, K. M. & Nussenzweig, M. C. (2003) Mol. Cell 12, 501-508. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri, J., Khuong, C. & Alt, F. W. (2004) Nature 430, 992-998. [DOI] [PubMed] [Google Scholar]
- 20.Pasqualucci, L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature 412, 341-346. [DOI] [PubMed] [Google Scholar]
- 21.Küppers, R. & Dalla-Favera, R. (2001) Oncogene 20, 5580-5594. [DOI] [PubMed] [Google Scholar]
- 22.Nakatani, Y. & Ogryzko, V. (2003) Methods Enzymol. 370, 430-444. [DOI] [PubMed] [Google Scholar]
- 23.Skalhegg, B. S. & Tasken, K. (2000) Front Biosci. 5, D678-D693. [DOI] [PubMed] [Google Scholar]
- 24.Doskeland, S. O., Maronde, E. & Gjertsen, B. T. (1993) Biochim. Biophys. Acta. 1178, 249-258. [DOI] [PubMed] [Google Scholar]
- 25.Amieux, P. S. & McKnight, G. S. (2002) Ann. N.Y. Acad. Sci. 968, 75-95. [DOI] [PubMed] [Google Scholar]
- 26.Chijiwa, T., Mishima, A., Hagiwara, M., Sano, M., Hayashi, K., Inoue, T., Naito, K., Toshioka, T. & Hidaka, H. (1990) J. Biol. Chem. 265, 5267-5272. [PubMed] [Google Scholar]
- 27.Basu, U., Chaudhuri, J., Alpert, C., Dutt, S., Ranganath, S., Li, G., Schrum, J. P., Manis, J. P. & Alt, F. W. (2005) Nature 438, 508-511. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura, M., Kondo, S., Sugai, M., Nazarea, M., Imamura, S. & Honjo, T. (1996) Int. Immunol. 8, 193-201. [DOI] [PubMed] [Google Scholar]
- 29.Christensen, A. E., Selheim, F., de Rooij, J., Dremier, S., Schwede, F., Dao, K. K., Martinez, A., Maenhaut, C., Bos, J. L., Genieser, H. G. & Doskeland, S. O. (2003) J. Biol. Chem. 278, 35394-35402. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita, K., Harigai, M., Fagarasan, S., Muramatsu, M. & Honjo, T. (2001) Proc. Natl. Acad. Sci. USA 98, 12620-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasken, K. & Aandahl, E. M. (2004) Physiol. Rev. 84, 137-167. [DOI] [PubMed] [Google Scholar]
- 32.Harada, H., Becknell, B., Wilm, M., Mann, M., Huang, L. J., Taylor, S. S., Scott, J. D. & Korsmeyer, S. J. (1999) Mol. Cell 3, 413-422. [DOI] [PubMed] [Google Scholar]
- 33.Barreto, V. M., Pan-Hammarstrom, Q., Zhao, Y., Hammarstrom, L., Misulovin, Z. & Nussenzweig, M. C. (2005) J. Exp. Med. 202, 733-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stavnezer, J. & Amemiya, C. T. (2004) Semin. Immunol. 16, 257-275. [DOI] [PubMed] [Google Scholar]
- 35.Zhong, H., SuYang, H., Erdjument-Bromage, H., Tempst, P. & Ghosh, S. (1997) Cell 89, 413-424. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, L., Duan, C. J., Binkley, C., Li, G., Uhler, M. D., Logsdon, C. D. & Simeone, D. M. (2004) Mol. Cell. Biol. 24, 2169-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.