Abstract
Monocyte-to-macrophage differentiation with the cytokine granulocyte–macrophage colony-stimulating factor induces expression of the cyclic nucleotide phosphodiesterase PDE1B2. However, what role PDE1B2 plays in macrophage biology has not been elucidated. We have addressed this question by inhibiting PDE1B2 induction by using RNA interference. Using a retrovirus-based system, we created HL-60 stable cell lines that express a short-hairpin RNA targeting PDE1B2. HL-60 cells treated with phorbol-12-myristate-13-acetate differentiate to a macrophage-like phenotype and up-regulate PDE1B2. However, expression of PDE1B2 short hairpin RNA effectively suppresses PDE1B2 mRNA, protein, and activity up-regulation. Using the HL-60 PDE1B2 knockdown cells and agonists for either adenylyl or guanylyl cyclase, it was found that PDE1B2 predominantly regulates cGMP and plays a lesser role in cAMP regulation in response to cyclase agonists. Furthermore, in intact HL-60 cells, PDE1B2 activity can be regulated by changes in Ca+2 levels. Inhibiting PDE1B2 up-regulation does not prevent HL-60 cell differentiation, because several markers of macrophage differentiation are unaffected. However, suppression of PDE1B2 expression alters some aspects of the macrophage-like phenotype, because cell spreading, phagocytic ability, and CD11b expression are augmented. The cAMP analog 8-Bromo-cAMP reverses the changes caused by PDE1B2 knockdown. Also, PDE1B2 knockdown cells have lower basal levels of cAMP and alterations in the phosphorylation state of several probable PKA substrate proteins. Thus, the effects of PDE1B2 on differentiation may ultimately be mediated through decreased cAMP. In conclusion, PDE1B2 regulates a subset of phenotypic changes that occur upon phorbol-12-myristate-13-acetate-induced differentiation and likely also plays a role in differentiated macrophages by regulating agonist-stimulated cGMP levels.
Keywords: cyclic AMP, cyclic GMP, phosphodiesterase
Monocytes are peripheral immune cells that have the potential to differentiate into a variety of cell types depending on the combination of cytokines and stimuli to which they are exposed. The differentiated cell types range from professional antigen-presenting dendritic cells to osteoclasts to various macrophage populations found throughout the body (1–3). Different combinations of cytokines regulate signaling pathways that define the phenotype of the differentiated cell (4, 5). The characteristics that define cellular phenotype include not only expression of cell surface markers and morphology but also functional immune capabilities such as cytokine release, antigen presentation, phagocytic ability, and microbiocidal activity.
Cyclic nucleotide phosphodiesterases (PDEs) are enzymes that regulate the amplitude and duration of cAMP and cGMP signaling by controlling cyclic nucleotide (cNT) degradation. Eleven different PDE gene families have been described and characterized based on their sequences and regulatory properties (6–8). Recent work has described changes in the expression of several PDE isoforms in monocytes and monocyte-derived cells upon cytokine stimulation (9–11). Thus, each monocyte-derived cell type has a unique system for regulating cyclic nucleotide levels. For instance, differentiation of a monocyte into a peritoneal-like macrophage with the cytokine macrophage colony-stimulating factor (M-CSF) induces PDE2A expression, whereas differentiation to an alveolar-like macrophage with granulocyte–macrophage colony-stimulating factor (GM-CSF) up-regulates PDE1B2 instead of PDE2A (11). However, addition of IL-4 in the presence of GM-CSF changes the differentiation to a dendritic cell and suppresses PDE1B2 up-regulation (10, 12). Although it is assumed that these changes in PDE activity have important consequences in the cell, the function of most of the individual PDEs up-regulated with differentiation has not yet been addressed.
It could be predicted that PDE induction would play some role in regulating monocyte differentiation, because earlier results have shown that changes in cyclic nucleotides can affect the differentiation process. For example, with primary monocytes in culture, it has been shown that increases in either cAMP or cGMP can promote the development of an intermediate cell that has a phenotype with characteristics of both a dendritic cell and a macrophage (13). However, final differentiation into either the mature macrophage or dendritic cell is inhibited by high levels of either of these cyclic nucleotides. Similarly, monocyte differentiation in the presence of adenylyl cyclase agonists such as PGE2 (14–16) or the guanylyl cyclase agonist atrial natriuretic factor (ANF) (17) can polarize dendritic cells toward promoting Th1 or Th2 responses, respectively. Likewise, generation of osteoclasts from bone marrow precursors is potentiated by treatments that increase either cAMP or cGMP such as PGE2 (18, 19) and nitric oxide (20). Finally, differentiation of monocyte cell lines, such as the HL-60 (21–23) and U937 (24, 25) lines, can be initiated by increased cAMP or cGMP levels.
Because we have earlier found that PDE1B2 is highly induced during the course of monocyte-to-macrophage differentiation with GM-CSF, we wished to determine whether PDE1B2 plays a role in the differentiation process. PDE1B2 is a Ca+2/calmodulin-stimulated PDE that hydrolyzes both cAMP and cGMP but has a preference for cGMP as substrate in vitro (12, 26). The study of PDE1B2 function in these cells has been precluded by the lack of a cell-permeant PDE1B2 selective inhibitor. Therefore, we have taken a genetic approach to studying PDE1B2 and used retroviral infection to create stable HL-60 cell lines expressing a short-hairpin RNA (shRNA) that inhibits PDE1B2 expression. We chose HL-60 cells as a model to study the involvement of PDE1B2 in the differentiation process, because they can be differentiated to a macrophage-like phenotype with phorbol-12-myristate-13-acetate (PMA) treatment (27, 28) and highly up-regulate PDE1B2 during the process (12). In comparing the PDE1B2 knockdown cells with control cells, we have found that PDE1B2 is a primary regulator of agonist-stimulated cGMP and has only modest effects on agonist-stimulated cAMP. Furthermore, we have found that PDE1B2 is not required for overall differentiation to a macrophage-like cell but does play a role in regulating a subset of characteristics that are acquired with PMA-induced differentiation. Our results suggest that induction of PDE1B2 plays a role in specifying the phenotype of differentiated macrophages.
Results
RNA Interference (RNAi) Suppresses PDE1B2 Induction. To try to determine how PDE1B2 affects macrophage function, RNAi was used to selectively inhibit expression of the enzyme. A major impediment to using RNAi in primary nondividing monocytes or macrophages is their resistance to transfection. This is also true for HL-60 cells, which do divide. HL-60 cells are promyelocytic leukemia cells that can be differentiated to a macrophage-like phenotype by treatment with PMA (27, 28) and highly up-regulate PDE1B2 in the process (12). To circumvent the problem of low transfection efficiency in these cells, we used a retrovirus to introduce a gene for expression of a shRNA into the cells and then used antibiotic resistance to select for stably expressing cells. Sequences containing either a shRNA targeting PDE1B or a shRNA with a scrambled sequence were inserted behind a U6 promoter into a retrovirus also encoding a puromycin resistance gene. The plasmids were transfected into packaging cells, and the subsequently produced virus was used to infect HL-60 cells. We performed two separate infections and puromycin-selected mixed populations of cells to ensure that any effects seen were not clonal artifacts due to positions of insertion. Data from one of these populations are shown in this report, and data from the second population are shown in Supporting Text, which is published as supporting information on the PNAS web site. The results for the two populations were virtually identical. The morphology and phenotype of cells infected with the scrambled shRNA virus highly resemble noninfected cells. Importantly, we have previously demonstrated that expression of PDE1B2 before differentiation is negligible (12); therefore, the expression of the PDE1B2 shRNA would not be expected to affect the phenotype of the dividing undifferentiated cells, and long-term compensatory changes due to the knockdown of PDE1B2 before PMA-induced differentiation are unlikely.
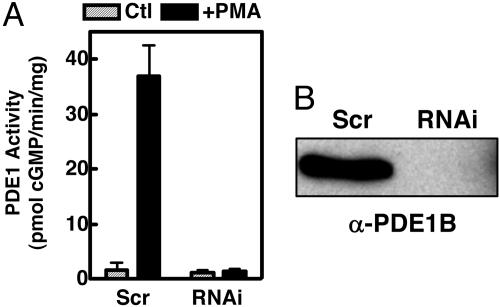
As shown in Fig. 1A, PMA treatment induces a large increase in PDE1 activity in the scrambled shRNA-expressing cells that is almost completely suppressed in the PDE1B2 shRNA-expressing cells. Also, PDE1B2 protein was not found in the PDE1B2 shRNA cells, as determined by immunoprecipitation and Western blotting (Fig. 1B). Using sensitive RT-PCR to detect PDE1B2 mRNA in the PMA-differentiated cells, we found that PDE1B2 mRNA levels were reduced to just barely above the limit of detection in the PDE1B2 shRNA cells (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the PDE1B2 shRNA sequence used successfully reduces PDE1B2 expression to nearly undetectable levels.
Fig. 1.
RNAi suppresses PDE1B2 induction. HL-60 cells were infected with a retrovirus encoding expression of a PDE1B2-targeting shRNA (RNAi) or a shRNA with a scrambled sequence (Scr), and stable cell lines were generated by puromycin selection. (A) PDE1 activity in untreated cells (Ctl) and cells differentiated for 3 days with PMA (+PMA) was determined. (B) The expression of PDE1B2 protein in PMA differentiated cells was detected by immunoprecipitation with the ACC-1 antibody and Western blotting with an α-PDE1B antibody. Activity values are means ± SEM of four separate differentiations, and the Western blot shown is representative of three separate differentiations.
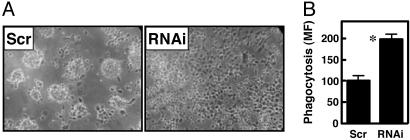
PDE1B2 Is Not Required for Differentiation but Affects the Final Phenotype of Differentiated HL-60 Cells. To explore the function of PDE1B2 in HL-60 cells, the cell lines expressing the PDE1B2 shRNA were compared with control cell lines expressing the scrambled shRNA. Multiple phenotypic changes occur when HL-60 cells are differentiated with PMA to a macrophage-like cell. Using the PDE1B2 knockdown cells, it was first determined whether PDE1B2 up-regulation is a trigger for differentiation and is required for initiation of this process. One of the most easily observable changes that occurs upon monocyte-to-macrophage differentiation is the transition from a nonadherent cell to one that is strongly adherent and spreads across the growth surface. In HL-60 cells, knockdown of PDE1B2 did not prevent the cells from becoming adherent after PMA treatment (Fig. 2A). This indicates that a major functional marker for differentiation of the cells is still occurring after PMA treatment, and therefore PDE1B2 is not required for this process. However, the morphology of PDE1B2 shRNA-expressing cells was altered. Scrambled shRNA cells tended to form clumps of cells that were more rounded, whereas the PDE1B2 shRNA cells were less clumped and more uniformly dispersed and spread to a greater degree. Thus, it can be inferred that PDE1B2 regulates genes involved in defining morphology or regulates the adherence and spreading processes directly.
Fig. 2.
PDE1B knockdown alters PMA differentiated HL-60 cell phenotype. HL-60 cells expressing either a scrambled shRNA (Scr) or a PDE1B-targeting shRNA (RNAi) were differentiated for 3 days with PMA and cell phenotype was analyzed. (A) Cell morphology was examined by using phase-contrast microscopy. PDE1B2 shRNA cells display increased spreading in comparison with cells infected with the scrambled shRNA. (B) Phagocytosis of 2-μm fluorescently labeled microspheres by differentiated HL-60 cells was determined after 4 h by FACS for the scrambled and PDE1B2 shRNA cells. Values shown are means ± SEM of four separate experiments. *, Significantly different from scrambled cells (P < 0.05).
Expression levels of many genes change with PMA-induced differentiation of HL-60 cells, as demonstrated by using cDNA microarray analysis (29, 30). Expression of cell-surface markers whose levels increase (CD11b and CD87) or decrease with differentiation (CD71) were measured by FACS to verify whether differentiation was occurring despite the loss of PDE1B2 and also to begin to characterize the phenotype of the differentiated cell. CD71 (transferrin receptor) is highly expressed on dividing cells but is down-regulated in differentiated cells (31, 32). As has been seen previously, CD71 expression was decreased with PMA treatment, and the expression levels of CD71 were equally low in the scrambled and PDE1B2 shRNA cells (Table 1). CD87 (urokinase plasminogen activator receptor) is a receptor involved in processes such as cell migration and adherence (33), whose expression is induced with PMA treatment. CD87 was found to be induced by PMA treatment, and its expression was not affected by PDE1B2 knockdown (Table 1). The only marker tested whose expression was altered by PDE1B2 knockdown was the integrin CD11b (MAC-1). CD11b expression is very low on undifferentiated cells, but its expression is highly induced with PMA treatment (34, 35). PDE1B2 shRNA cells up-regulate CD11b to a greater degree than scrambled shRNA cells. Although the differences are not large (23% and 67% increases over scrambled shRNA cells for the two different infections), they were found to be statistically significant (P < 0.05). Confirming that the effect is due to the PDE1B2 shRNA, the CD11b mean fluorescence for differentiated noninfected cells (438 ± 81) was not statistically different from values for the two scrambled shRNA lines (594 ± 84 and 498 ± 68). These results indicate that only a subset of genes altered during PMA induced differentiation are affected by PDE1B2 knockdown.
Table 1. Expression of CD11b, CD87, and CD71 in undifferentiated cells and cells differentiated for 3 days with PMA (+PMA) was measured by flow cytometry.
| Condition | CD11b | CD87 | CD71 |
|---|---|---|---|
| Scr | 8.6 ± 2.9 | 6.6 ± 1.6 | 145 ± 27 |
| Scr + PMA | 594 ± 84 | 51 ± 7.6 | 18 ± 1.4 |
| RNAi | 8.1 ± 1.5 | 5.9 ± 0.8 | 132 ± 22 |
| RNAi + PMA | 728 ± 118* | 57 ± 12 | 18 ± 1.2 |
Numbers listed are mean fluorescence values and are means ± SEM of four to six separate experiments.
Significantly different from scrambled cells (P < 0.05)
PMA differentiation induces changes in HL-60 cells that also make them functionally more macrophage-like. One of these changes is an increase in the phagocytic ability of the cells (36, 37). The phagocytic ability of HL-60 cells was measured with an assay that uses fluorescently labeled 2-μm microspheres opsonized with human serum as surrogates for opsonized pathogens. The microspheres were incubated with cells, and phagocytosis of the beads was measured by flow cytometry. An initial time course experiment was performed to ensure that time-dependent phagocytosis was being measured linearly. Both PDE1B2 and scrambled shRNA cells displayed a linear time-dependent uptake of the microspheres (Fig. 7, which is published as supporting information on the PNAS web site). However, the PDE1B2 shRNA cells had a nearly 2-fold greater phagocytic ability. This difference was seen with the PDE1B2 shRNA cells from both infections at the 4-h time point (Fig. 2B and Fig. 8, which is published as supporting information on the PNAS web site). Taken together, the results regarding adherence, cell surface marker expression and phagocytosis suggest that PDE1B2 is not required for differentiation of HL-60 cells, but that PDE1B2 affects the development of a subset of the macrophage-like qualities of the cells.
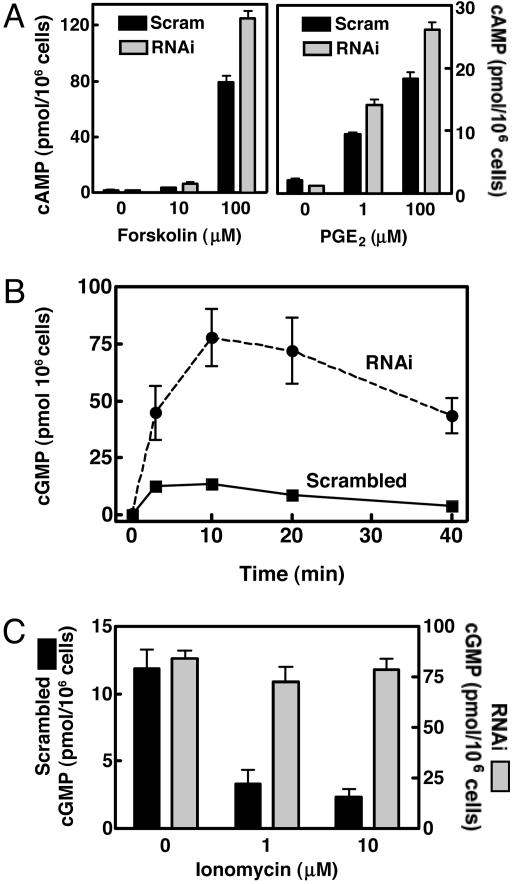
PDE1B2 Regulates Cyclic Nucleotide Levels in Differentiated HL-60 Cells. Monocytes and macrophages respond to a variety of stimuli by increasing cNT levels. To determine to what degree PDE1B2 regulates cAMP and cGMP in HL-60 cells, differentiated scrambled and PDE1B2 shRNA cells were treated with agonists that stimulate either adenylyl or guanylyl cyclase, and the accumulation of cAMP and cGMP was measured. A robust increase in cAMP was achieved with forskolin treatment, and only slightly higher cAMP levels were observed in the PDE1B2 shRNA cells compared with the scrambled shRNA cells (Fig. 3A). When cells were treated with the physiological agonist PGE2, an increase in cAMP also was seen, although the nonspecific PDE inhibitor 3-isobutyl-1-methylxanthine (IBMX) had to be added to observe a significant accumulation of cAMP (Fig. 3A). That IBMX had to be added to obtain an elevation of cAMP, even when PDE1B2 expression is decreased, suggests that PDE1B2 is not a major regulator of PGE2 elevated cAMP. Again, only slightly higher levels of cAMP accumulated in the PDE1B2 shRNA cells after treatment with PGE2. Thus, it appears that PDEs other than PDE1B2 are the primary regulators of cAMP in response to these adenylyl cyclase agonists.
Fig. 3.
PDE1B2 regulation of cAMP and cGMP in response to agonists. HL-60 cells expressing either scrambled or PDE1B2 shRNA were differentiated for 3 days with PMA. (A) Cells were treated with forskolin for 15 min. Cyclic nucleotides were then extracted by incubating the cells in ice-cold EtOH/HCl for 10 min. cAMP levels were determined by EIA. Cells were also treated with various concentrations PGE2 for 15 min after a 10-min pretreatment with 200 μM 3-isobutyl-1-methylxanthine. Values shown are means ± SEM from four separate determinations. (B) HL-60 cells were treated with 100 nM ANF, and the cGMP content of cells was determined at various time points by EIA. (C) Cells were treated with 100 nM ANF for 10 min after 10 min of pretreatment with varying concentrations of the calcium ionophore ionomycin. cGMP values for the cells expressing the scrambled shRNA are plotted on the left y axis, and the values from the cells expressing the PDE1B2 shRNA are plotted on the right y axis. All cGMP values are means ± SEM from three separate experiments.
Atrial natriuretic factor (ANF) is known to elevate cGMP in macrophages (38, 39) and has been shown to be produced by macrophages themselves (40, 41). In scrambled shRNA cells, ANF elicited an increase in cGMP production in a concentration- (Fig. 9, which is published as supporting information on the PNAS web site) and time-dependent manner (Fig. 3B). In this case, a marked increase in cGMP accumulation was seen in PDE1B2 shRNA cells over what was observed in scrambled shRNA cells (6- to 10-fold). 3-Isobutyl-1-methylxanthine was not needed to see the response. Furthermore, preincubating the scrambled shRNA cells with the Ca+2 ionophore ionomycin decreased the accumulation of cGMP (Fig. 3C). In contrast, ionomycin had no effect on cGMP accumulation in PDE1B2 shRNA cells. Because PDE1B2 is activated by Ca+2/CaM binding, Ca+2 entry facilitated by ionomycin would be expected to activate PDE1B2 and decrease cGMP accumulation only in the scrambled shRNA cells and not in the PDE1B2 shRNA cells, if PDE1B2 is a major regulator of cGMP in these cells. This experiment functionally confirms that PDE1B2 expression has been effectively suppressed and, more importantly, it demonstrates that a portion of PDE1B2 is active in the cells at basal Ca+2 levels, because substantially higher cGMP levels are seen with ANF treatment in the PDE1B2 shRNA cells. Also, a portion of the cellular PDE1B2 is inactive under basal conditions and can be activated by increasing intracellular Ca+2. Taken together, these experiments with the adenylyl and guanylyl cyclase agonists establish that PDE1B2 functions in the cell primarily to regulate elevated cGMP and to a lesser degree cAMP. This cyclic nucleotide selectivity is consistent with what is known about the in vitro kinetic properties of recombinant PDE1B2, which has a preference for cGMP to cAMP as substrate based on Km and Vmax values (12).
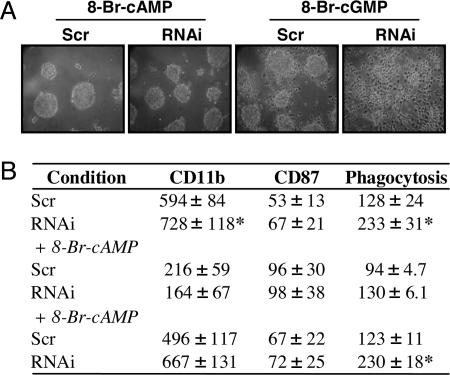
Cyclic Nucleotide Regulation of Differentiation. Given the effects of PDE1B2 on cAMP and cGMP regulation shown in Fig. 3, and that both cyclic nucleotides have been shown to modulate early steps of the differentiation of monocytes (13–17), we hypothesized that any modification of differentiation by PDE1B2 would most likely be mediated by alterations in cGMP. To test this hypothesis, we examined the effects of 8-bromo (8-Br)-cAMP-modified cNTs on differentiation of the scrambled and PDE1B2 shRNA cells. Because 8-Br-cAMP and 8-Br-cGMP are both cell-permeable cyclic nucleotide analogs that are more resistant to hydrolysis by PDE1B than the native cNTs (42–44), it was expected that they would mimic many of the long-term effects of elevated cAMP or cGMP in these cells. However, somewhat unexpectedly, 8-Br-cGMP had only a small effect on morphology, because cell spreading was only slightly inhibited. When HL-60 cells were differentiated with PMA in the presence of 8-Br-cAMP, cell spreading was greatly diminished, and the cells formed clumps of round cells (Fig. 4A). This was the case for both the scrambled and PDE1B2 shRNA cell lines, because the two had nearly identical morphology in the presence of 8-Br-cAMP, and the differences noted in Fig. 2 were abolished. Similarly, 8-Br-cAMP significantly inhibited CD11b up-regulation (Fig. 4B), whereas 8-Br cGMP did not. The expression of CD11b in the scrambled and PDE1B2 shRNA cells was reduced to nearly equal levels by 8-Br cAMP but not by 8-Br-cGMP. As another measure of function, 8-Br-cAMP slightly increased CD87 expression (Fig. 4B) with no effect by 8-Br-cGMP. The phagocytic ability of the differentiated PDE1B2 shRNA cells was also reduced by 8-Br-cAMP to the level of the scrambled cells (Fig. 4B) but was not affected by 8-Br-cGMP. Finally, the effects of 8-Br-cAMP on cell spreading and CD11b were also mimicked by the PKA activator N6-benzoyl cAMP (data not shown) and are thus likely mediated through PKA. The unexpected reversal of the PDE1B2 knockdown phenotype with 8-Br-cAMP and not 8-Br-cGMP suggests that the effects of PDE1B2 knockdown on differentiation are most likely mediated by a reduction in basal cAMP.
Fig. 4.
8-Br-cAMP prevents HL-60 cell spreading, decreases CD11b expression, and decreases phagocytic ability. HL-60 PDE1B2 shRNA (RNAi) and scrambled shRNA (Scr) cells were differentiated for 3 days with 100 nM PMA in the presence of 500 μM 8-Br-cAMP or 8-Br-cGMP. After 3 days, cellular morphology was assessed by using phase-contrast microscopy (A). (B) CD11b and CD87 expression was measured by FACS, and phagocytosis of fluorescent microspheres was determined for cells differentiated in the presence or absence of 500 μM 8-Br-cNTs. Numbers reported are mean fluorescence values. Pictures shown are representative of at least three separate differentiations under each condition, whereas FACS and phagocytosis values are means ± SEM from four separate experiments. *, Significantly different from scrambled shRNA cells (P < 0.05).
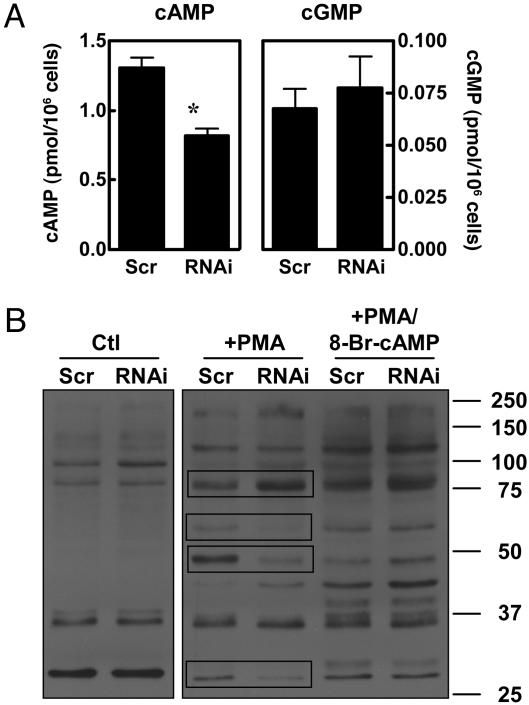
In the experiments shown in Fig. 3, cNT levels were determined under basal conditions after differentiation before agonist treatment. However, because the basal cyclic nucleotide values are very low compared with the values after agonist treatment, it is difficult to see the results at the scale of Fig. 3. Therefore, the basal values in the absence of agonist treatment are replotted and are shown in Fig. 5A. In Fig. 5A, it is apparent that the PDE1B2 knockdown cells have lower basal cAMP (47% reduction) but nearly equal cGMP levels (Fig. 5A). This seems to be a paradox, because knocking out a PDE would be expected to increase levels of a cyclic nucleotide. However, as was demonstrated in Fig. 3, the primary role of PDE1B2 appears to be regulation of cGMP and not cAMP. Hypothetically, reduction of PDE1B2 activity could bring about a decrease in cAMP indirectly either by activating a PDE or by decreasing the activity of an adenylyl cyclase. At present, it is not known which of these mechanisms is occurring in these cells.
Fig. 5.
PDE1B2 knockdown lowers basal cAMP and alters PKA substrate phosphorylation. HL-60 PDE1B2 shRNA (RNAi) and scrambled shRNA cells (Scr) were differentiated for 3 days with 100 nM PMA. Basal cAMP and cGMP levels were determined by EIA. Values shown are means ± SEM from eight separate differentiations. *, Significantly different from scrambled shRNA cells (P < 0.05). (B) Lysates from cells treated for 3 days with 100 nM PMA were Western-blotted by using an antibody that recognizes a phospho-Ser/Thr PKA substrate motif. Untreated cells (Ctl), PMA treated cells (+PMA), and PMA-treated cells with 8-Br-cNT added (+PMA/8-Br-cAMP) are compared. Protein bands that were consistently different between the two cell populations are boxed in and described in the text. The gel shown is representative of three experiments, and quantitation is graphed in Fig. 10.
Consistent with the changes in cAMP, we found a difference in the pattern of PKA substrate phosphorylation between the scrambled and PDE1B2 shRNA cells (Fig. 5B). This was determined by using an antibody that recognizes a phospho-Ser/Thr residue in a consensus motif for cAMP-dependent protein kinase (PKA). Although PKA is likely to be the favored kinase that phosphorylates substrates having this motif, it is likely that in some proteins, it can be phosphorylated by other kinases. Before differentiation, the pattern of substrate phosphorylation between the cell lines was nearly identical when compared by Western blot (Fig. 5B, Ctl). Consistent with the lower basal cAMP levels in the PDE1B2 shRNA cells after PMA treatment, a reduction in phosphorylation of several phosphorylated bands was found in the PDE1B shRNA cells in comparison to the scrambled shRNA cells (Fig. 5B, compare +PMA Scr vs. +PMA RNAi). For example, the protein bands shown in the boxes at ≈60, 48, and 27 kDa were less phosphorylated in the PDE1B2 shRNA cells. However, a few bands were more heavily phosphorylated in PDE1B2 shRNA cells, as exemplified by the band at ≈76 kDa (Fig. 5).
These data suggest that the absence of PDE1B2 activity may affect localized pools of cyclic nucleotide that can regulate different protein kinase targets. They also suggest that the overall decrease in cAMP may be due to indirect or compensatory mechanisms. Phosphorylation of each of the major substrates phosphorylated in Fig. 5B was increased by 8-Br-cAMP, and the differences in phosphorylation of these bands between the two cell types were abolished, just as the differences between their phenotypes were abolished with 8-Br-cAMP treatment. The differences in the bands at 48 and 76 kDa were the largest and most consistent and were quantitated, as was the total phosphorylation of PMA-treated cells in the absence or presence of 8-Br-cAMP. These values are shown in Fig. 10, which is published as supporting information on the PNAS web site.
Discussion
One characteristic difference between monocyte-derived cell types is their PDE expression profiles. We previously identified PDE1B2 as a PDE that is highly induced upon monocyte to macrophage differentiation with the cytokine GM-CSF. We have now used RNAi to prevent induction of PDE1B2 during differentiation of HL-60 cells to a macrophage-like phenotype to study its function. The generation of stable RNAi expressing cell lines in this case was an attractive experimental strategy, because the protein to be knocked down (PDE1B2) is not expressed under normal growth conditions, and therefore the expression of the PDE1B2-targeting shRNA presumably would not cause any developmental or long term changes. We found that our RNAi approach effectively suppressed the PDE1B2 mRNA, protein, and activity up-regulation that is normally triggered by PMA-induced differentiation.
Using the PDE1B2 knockdown cells, it was determined that PDE1B2 primarily regulates cGMP and not cAMP elevated by agonists, and that endogenous PDE1B2 activity is regulated in the cell by intracellular Ca+2 (Fig. 3). cAMP in immune cells and macrophages has been extensively studied and is accepted to be generally anti-inflammatory (45, 46). In contrast, significantly less is known regarding how cGMP affects macrophage function, and its effects are somewhat controversial. Our results conclusively show that PDE1B2 is a major regulator of ANF-stimulated cGMP in the macrophage cell type studied. This is consistent with an earlier finding of higher up-regulation of the ANF receptor GC-A in primary macrophages that up-regulate PDE1B2 upon differentiation from a monocyte (11). Our experiments with the Ca+2 ionophore ionomycin indicate that PDE1B2 is functionally activated in intact cells by changes in Ca+2 levels. Although some of the cellular PDE1B2 is active under basal conditions, a portion can be further activated by increases in Ca+2 triggering attenuation of cGMP accumulation. Thus, PDE1B2 may not only affect differentiation but also may modulate the effects of ANF or other cGMP agonists on macrophage immune functions, particularly those that involve changes in intracellular Ca+2.
PDE1B2 knockdown did not prevent most of the changes characteristic of PMA-induced HL-60 cell differentiation, because many markers of differentiation were unaffected, but a suppression of PDE1B2 expression did affect select aspects of the final cell phenotype. Somewhat unexpectedly, a significant increase in basal total cGMP after differentiation was not detected with PDE1B2 knockdown. There could be an increase in the level of a localized cGMP pool that was not detectable by measuring total cell cGMP that modulates aspects of the final phenotype. Because lower basal cAMP levels were found in PDE1B2 shRNA cells after differentiation, crosstalk with the cAMP pathway is also possible. The concept that PDE1B2 could regulate a specific localized cyclic nucleotide pool is supported by the observation that the phosphorylation state of several substrates having PKA consensus sites was differentially altered by PDE1B2 knockdown.
An inverse relationship between cGMP and cAMP has been noted before in macrophages. For example, in human peritoneal macrophages, ANF and sodium nitroprusside were found to elevate cGMP while decreasing cAMP (47), and in murine bone marrow-derived macrophages, ANF had the same cAMP-lowering effect (48). A plausible explanation for the reduced cAMP levels with PDE1B2 knockdown would be an activation of the cGMP-stimulated PDE2 by cGMP. However, PDE3 and PDE4 are the primary cAMP PDEs expressed in these cells (see Fig. 11, which is published as supporting information on the PNAS web site). PDE2 activity was not detected in HL-60 cell homogenates, as measured by either erythro-9-(2-hydroxy-3-nonyl)adenine inhibition of cGMP activity or cGMP stimulation of cAMP activity in the presence of a PDE3 inhibitor. It is therefore likely that the mechanism by which repression of PDE1B2 induction causes a decrease in basal cAMP is more indirect. Possible mechanisms would be indirect activation of another PDE or inhibition of an adenylyl cyclase. Hopefully, future work will define the mechanism by which PDE1B2 alters differentiation and cAMP.
The immune system has likely endowed monocytes with phenotypic plasticity so that a variety of specialized cell types could be generated to mount an effective customized immune response to different pathogenic stimuli. A highly tailored and localized immune response to specific insults would be more effective at eliminating invaders while minimizing damage to host tissue. We previously reported differences in cGMP PDE expression between primary cell types, because GM-CSF-differentiated macrophages highly expressed PDE1B2, whereas M-CSF-differentiated macrophages expressed lower PDE1B2 and higher PDE2 (11). The finding that PDE1B2 knockdown increases phagocytic ability and alters morphology and marker expression may indicate changes in phenotype that are analogous to differences between GM-CSF and M-CSF macrophage phenotypes (2). Finally, identification of how unique PDEs expressed in the different macrophage cell types affects their generation and function should provide therapeutic opportunities to selectively target one cell type involved in a specific pathology, such as how to control monocytes recruited to become macrophages in atherosclerotic lesions. Thus, in a broader context, the elucidation of how PDEs define the phenotype of monocyte-derived cell types could be useful in developing methods for treating inflammatory pathologies.
Methods
Preparation of PDE1B RNAi Constructs and Virus Production. Viruses for RNAi were prepared by using the pSiren retro vector from BD Biosciences (Palo Alto, CA) that contains a gene for puromycin resistance. Details of the methods used to prepare the viruses and the sequences of the PDE1B2-targeting and scrambled oligonucleotides are listed in Supporting Text. Briefly, oligonucleotides encoding a shRNA targeting PDE1B or a scrambled sequence were ligated into the pSiren retro plasmid according to the manufacturer's instructions. Phoenix-Ampho packaging cells were transfected with the pSiren plasmids using Lipofectamine 2000 (Invitrogen). The virus-containing media were collected and used to infect HL-60 cells. After 48 h of infection, the virus-containing media were removed, and the cells were resuspended in fresh media containing 1 μg/ml puromycin for selection of infected cells.
Analysis of PDE1B2 Activity, Protein, and mRNA. HL-60 cells were harvested, washed once in cold PBS, suspended in lysis buffer, ruptured by sonication, and PDE activities were measured in the cell homogenates as described previously (see Supporting Text for details), with the exception that cilostamide was used as a PDE3 inhibitor instead of milrinone (11, 49). PDE1B2 was immunoprecipitated from HL-60 cell cytosol by using the ACC-1 monoclonal antibody as described (49). To measure PDE1B2 mRNA, total RNA was isolated from 5 × 106 cells, and RT-PCR was performed as described (11). RT-PCR primer sequences used for amplification of the two cDNA targets are listed in Supporting Text.
cAMP/cGMP Enzyme Immunoassay (EIA). cAMP and cGMP content of HL-60 cells was measured by using EIA kits from American Qualex (San Clemente, CA). Cells were differentiated for 3 days with PMA, and the media from the adherent cells were removed and replaced with fresh RPMI 1640 media. Cells were treated with various agonists and, at the end of the designated treatment time, the cell media were removed. Cells were washed once with cold PBS and then incubated in a solution of ice-cold ethanol:1 M HCl in a ratio of 99:1 vol/vol for 10 min. The samples were then evaporated in a Savant (Holbrook, NY) Sc110 speed vac concentrator, and cNT content was subsequently determined by using the EIA kits according to the manufacturer's instructions. Representative cells from untreated wells were harvested by scraping, dispersed by vigorous pipetting to break up aggregates, and counted by using trypan blue and a hemocytometer to normalize values between experiments.
HL-60 Cell Differentiation and Analysis of Morphology and FACS. For HL-60 cell differentiation, cells were incubated at 6 × 105 cells/ml with 100 nM PMA in RPMI 1640 medium supplemented with penicillin/streptomycin/10% FBS and buffered with 10 mM Hepes. FACS was used to determine expression of cell surface marker proteins. For FACS analysis, cells were harvested by scraping, washed with PBS, stained with fluorescently labeled antibodies, then fixed in PBS containing 1% paraformaldehyde and analyzed by using a Becton Dickinson FACScan flow cytometer as described (11). Cell morphology was observed by using phase-contrast microscopy and a ×20 objective. Cells were differentiated in six-well plates for 3 days with PMA, and images of the adherent cells were obtained.
Phagocytosis of Fluorescent Microspheres. Cells were differentiated in 12-well plates for 3 days. The media were then removed and replaced with RPMI 1640 medium. Two-micrometer nile red microspheres were opsonized by incubation with human serum for 30 min at 37°C. Opsonized beads were added to HL-60 cells in a ratio of 10 beads per cell. The beads were incubated with the cells for various lengths of time at 37°C in a CO2-regulated tissue culture incubator. At the end of the incubation, the cells were washed three times with PBS, scraped into PBS, and analyzed by FACS.
Supplementary Material
Acknowledgments
A.T.B. is a trainee under University of Washington Cardiovascular Pathology Training Grant NIH HL-07312. This work was supported by National Institutes of Health Grants DK21723 and HL44948 (to J.A.B.).
Author contributions: A.T.B. and J.A.B. designed research; A.T.B. performed research; A.T.B. and J.A.B. analyzed data; and A.T.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: PDE, phosphodiesterase; GM-CSF, granulocyte–macrophage colony-stimulating factor; shRNA, short-hairpin RNA; PMA, phorbol-12-myristate-13-acetate; cNT, cyclic nucleotide; RNAi, RNA interference; ANF, atrial natriuretic factor; 8-Br, 8-bromo; EIA, enzyme immunoassay.
References
- 1.Takahashi, K., Naito, M. & Takeya, M. (1996) Pathol. Int. 46, 473-485. [DOI] [PubMed] [Google Scholar]
- 2.Akagawa, K. S. (2002) Int. J. Hematol. 76, 27-34. [DOI] [PubMed] [Google Scholar]
- 3.Stout, R. D. & Suttles, J. (2004) J. Leukocyte Biol. 76, 509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaves, D. R. & Gordon, S. (2002) Int. J. Hematol. 76, 6-15. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, S. & Gordon, S. (1998) J. Leukocyte Biol. 63, 153-168. [DOI] [PubMed] [Google Scholar]
- 6.Francis, S. H., Turko, I. V. & Corbin, J. D. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 1-52. [DOI] [PubMed] [Google Scholar]
- 7.Soderling, S. H. & Beavo, J. A. (2000) Curr. Opin. Cell Biol. 12, 174-179. [DOI] [PubMed] [Google Scholar]
- 8.Juilfs, D. M., Soderling, S., Burns, F. & Beavo, J. A. (1999) Rev. Physiol. Biochem. Pharmacol. 135, 67-104. [DOI] [PubMed] [Google Scholar]
- 9.Gantner, F., Kupferschmidt, R., Schudt, C., Wendel, A. & Hatzelmann, A. (1997) Br. J. Pharmacol. 121, 221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantner, F., Schudt, C., Wendel, A. & Hatzelmann, A. (1999) Pulm. Pharmacol. Ther. 12, 377-386. [DOI] [PubMed] [Google Scholar]
- 11.Bender, A. T., Ostenson, C. L., Giordano, D. & Beavo, J. A. (2003) Cell. Signalling 16, 365-374. [DOI] [PubMed] [Google Scholar]
- 12.Bender, A. T., Ostenson, C. L., Wang, E. H. & Beavo, J. A. (2005) Proc. Natl. Acad. Sci. USA 102, 497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giordano, D., Magaletti, D. M., Clark, E. A. & Beavo, J. A. (2003) J. Immunol. 171, 6421-6430. [DOI] [PubMed] [Google Scholar]
- 14.Morelli, A. E. & Thomson, A. W. (2003) Trends Immunol. 24, 108-111. [DOI] [PubMed] [Google Scholar]
- 15.Kalinski, P., Hilkens, C. M., Snijders, A., Snijdewint, F. G. & Kapsenberg, M. L. (1997) J. Immunol. 159, 28-35. [PubMed] [Google Scholar]
- 16.Lee, J. J., Takei, M., Hori, S., Inoue, Y., Harada, Y., Tanosaki, R., Kanda, Y., Kami, M., Makimoto, A., Mineishi, S., et al. (2002) Stem Cells 20, 448-459. [DOI] [PubMed] [Google Scholar]
- 17.Morita, R., Ukyo, N., Furuya, M., Uchiyama, T. & Hori, T. (2003) J. Immunol. 170, 5869-5875. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, Y., Mizoguchi, T., Take, I., Kurihara, S., Udagawa, N. & Takahashi, N. (2005) J. Biol. Chem. 280, 11395-11403. [DOI] [PubMed] [Google Scholar]
- 19.Wani, M. R., Fuller, K., Kim, N. S., Choi, Y. & Chambers, T. (1999) Endocrinology 140, 1927-1935. [DOI] [PubMed] [Google Scholar]
- 20.Holliday, L. S., Dean, A. D., Lin, R. H., Greenwald, J. E. & Gluck, S. L. (1997) Am. J. Physiol. 272, F283-F291. [DOI] [PubMed] [Google Scholar]
- 21.Boss, G. R. (1989) Proc. Natl. Acad. Sci. USA 86, 7174-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang, B. E., Ericsen, C. & Aarbakke, J. (1994) Pharmacol. Toxicol. 75, 108-112. [DOI] [PubMed] [Google Scholar]
- 23.Chaplinski, T. J. & Niedel, J. E. (1982) J. Clin. Invest. 70, 953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavison, R., Matzner, Y. & Fibach, E. (1988) Isr. J. Med. Sci. 24, 697-701. [PubMed] [Google Scholar]
- 25.Brodsky, A., Davio, C., Shayo, C., Lemos Legnazzi, B., Barbosa, M., Lardo, M., Morelli, A., Baldi, A., Sanchez Avalos, J. C. & Rivera, E. (1998) Eur. J. Pharmacol. 350, 121-127. [DOI] [PubMed] [Google Scholar]
- 26.Kakkar, R., Raju, R. V. & Sharma, R. K. (1999) Cell Mol. Life Sci. 55, 1164-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rovera, G., Santoli, D. & Damsky, C. (1979) Proc. Natl. Acad. Sci. USA 76, 2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, P. & Ralph, P. (1985) J. Leukocyte Biol. 37, 407-422. [DOI] [PubMed] [Google Scholar]
- 29.Trayner, I. D., Bustorff, T., Etches, A. E., Mufti, G. J., Foss, Y. & Farzaneh, F. (1998) Leukocyte Res. 22, 537-547. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, X., Ravatn, R., Lin, Y., Shih, W. C., Rabson, A., Strair, R., Huberman, E., Conney, A. & Chin, K. V. (2002) Nucleic Acids Res. 30, 4489-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou, F. F., Boyce, J., Zhang, Y. & Owen, W. F., Jr. (2000) Immunol. Cell Biol. 78, 205-213. [DOI] [PubMed] [Google Scholar]
- 32.Barker, K. A. & Newburger, P. E. (1990) Exp. Cell Res. 186, 1-5. [DOI] [PubMed] [Google Scholar]
- 33.Bene, M. C., Castoldi, G., Knapp, W., Rigolin, G. M., Escribano, L., Lemez, P., Ludwig, W. D., Matutes, E., Orfao, A., Lanza, F., et al. (2004) Leukemia 18, 394-400. [DOI] [PubMed] [Google Scholar]
- 34.Aihara, H., Asaoka, Y., Yoshida, K. & Nishizuka, Y. (1991) Proc. Natl. Acad. Sci. USA 88, 11062-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins, S. J. (1987) Blood 70, 1233-1244. [PubMed] [Google Scholar]
- 36.Stendahl, O., Dahlgren, C. & Hed, J. (1982) J. Cell Physiol. 112, 217-221. [DOI] [PubMed] [Google Scholar]
- 37.Bard, J., Hall, J. & Levitt, D. (1987) J. Gen. Microbiol. 133, 899-910. [DOI] [PubMed] [Google Scholar]
- 38.Kiemer, A. K. & Vollmar, A. M. (2001) Ann. Rheum. Dis. 60 Suppl. 3, iii68-iii70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattana, J. & Singhal, P. C. (1993) Am. J. Physiol. 265, C92-C98. [DOI] [PubMed] [Google Scholar]
- 40.Vollmar, A. M. & Schulz, R. (1994) J. Clin. Invest. 94, 539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmar, A. M., Colbatzky, F. & Schulz, R. (1992) Cell Tissue Res. 268, 397-399. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, R. B., Jr., & Miller, J. P. (1974) Life Sci. 14, 1019-1040. [DOI] [PubMed] [Google Scholar]
- 43.Hei, Y. J., MacDonell, K. L., McNeill, J. H. & Diamond, J. (1991) Mol. Pharmacol. 39, 233-238. [PubMed] [Google Scholar]
- 44.Francis, S. H., Noblett, B. D., Todd, B. W., Wells, J. N. & Corbin, J. D. (1988) Mol. Pharmacol. 34, 506-517. [PubMed] [Google Scholar]
- 45.Aronoff, D. M., Canetti, C., Serezani, C. H., Luo, M. & Peters-Golden, M. (2005) J. Immunol. 174, 595-599. [DOI] [PubMed] [Google Scholar]
- 46.Essayan, D. M. (2001) J. Allergy Clin. Immunol. 108, 671-680. [DOI] [PubMed] [Google Scholar]
- 47.Houdijk, A. P., Adolfs, M. J., Bonta, I. L. & De Jonge, H. R. (1990) Eur. J. Pharmacol. 179, 413-417. [DOI] [PubMed] [Google Scholar]
- 48.Kiemer, A. K., Lehner, M. D., Hartung, T. & Vollmar, A. M. (2002) Endocrinology 143, 846-852. [DOI] [PubMed] [Google Scholar]
- 49.Sonnenburg, W. K., Rybalkin, S. D., Bornfeldt, K. E., Kwak, K. S., Rybalkina, I. G. & Beavo, J. A. (1998) Methods 14, 3-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.