Abstract
RNA polymerase (pol) III transcription decreases when primary cultures of rat neonatal cardiomyocytes are exposed to low oxygen tension. Previous studies in fibroblasts have shown that the pol III-specific transcription factor IIIB (TFIIIB) is bound and regulated by the proto-oncogene product c-Myc, the mitogen-activated protein kinase ERK and the retinoblastoma tumour suppressor protein, RB. The principal function of TFIIIB is to recruit pol III to its cognate gene template, an activity that is known to be inhibited by RB and stimulated by ERK. We demonstrate by chromatin immunoprecipitation (ChIP) that c-Myc also stimulates pol III recruitment by TFIIIB. However, hypoxic conditions cause TFIIIB dissociation from c-Myc and ERK, at the same time as increasing its interaction with RB. Consistent with this, ChIP assays indicate that the occupancy of tRNA genes by pol III is significantly reduced, whereas promoter binding by TFIIIB is undiminished. The data suggest that hypoxia can inhibit pol III transcription by altering the interactions between TFIIIB and its regulators and thus compromising its ability to recruit the polymerase. These effects are independent of cell cycle changes.
INTRODUCTION
In most eukaryotic organisms, cellular oxygen concentrations are precisely regulated in order to maintain an adequate substrate supply for oxidative phosphorylation and other essential metabolic reactions. A decrease in oxygen tension (hypoxia) is a common feature of several pathological situations, such as tumourigenesis, ischemia and venous diseases. Mammalian cells can adapt to oxygen deprivation by inducing protective mechanisms, which include expression of specific gene products and cell cycle arrest (1–6). Another well-characterized consequence of hypoxic stress is a pronounced decrease in the rate of oxygen consumption and of energy turnover (7–9). This correlates with a substantial and rapid drop in the rate of protein biosynthesis, involving changes at the level of both translation and transcription (8,10–14).
RNA polymerase (pol) III plays a key role in protein synthesis by catalysing the production of small, untranslated RNA molecules, such as tRNA and 5S rRNA, which are involved in fundamental metabolic processes (15). Transcription factor IIIB (TFIIIB) and TFIIIC are two transcription factor complexes that are required for transcription of most pol III templates (15–17). In the majority of cases, TFIIIC is responsible for promoter recognition by binding directly to DNA. TFIIIC then recruits TFIIIB by protein–protein interactions and directs pol III to the transcription start site (15–17). The synthesis of tRNA and 5S rRNA by pol III is cell cycle regulated in higher organisms (18). In addition, overexpression of pol III products is a general feature of transformed cells (19–21). These observations can be explained by the fact that TFIIIB is strongly regulated by the tumour suppressor proteins RB and p53, as well as the proto-oncogene product c-Myc and the extracellular signal-regulated kinase ERK (22–26). Both RB and p53 repress pol III transcription by binding to TFIIIB and sequestering it in an inactive complex (22,26–30). In contrast, c-Myc and ERK can bind to and activate TFIIIB, causing a potent induction of pol III transcription (24,25).
Since hypoxia causes a down-regulation of RNA and protein synthesis (7), we investigated its effect on pol III transcription. Primary cultures of rat neonatal cardiomyocytes were utilized, since they have been widely used as a model to study the effect of hypoxia. When such cells were incubated in 1% oxygen, a decrease in protein synthesis was found to accompany an inhibition of pol III transcription. Under these conditions, hypoxia did not raise levels of p53, a known regulator of pol III. Indeed, a much lower oxygen concentration was found previously to be required to stabilize p53 levels in cardiomyocytes (31). The amount of c-Myc is also not altered in the low oxygen environment. However, co-immunoprecipitation revealed that the ability of c-Myc to bind TFIIIB is compromised. Furthermore, both the activity of ERK and its interaction with TFIIIB decrease during hypoxia. In contrast, binding of TFIIIB to its repressor RB increases when cardiomyocytes are exposed to low oxygen, an effect that correlates with dephosphorylation of RB. Chromatin immunoprecipitation (ChIP) shows that the association of pol III with tRNA genes is decreased in hypoxic cardiomyocytes, although TFIIIB and TFIIIIC remain bound. The data suggest that hypoxia inhibits pol III transcription under these conditions by reducing the recruitment of polymerase to promoters, most likely as a consequence of altered interactions between TFIIIB and its positive and negative regulators.
MATERIALS AND METHODS
Cell culture
Myocytes were dissociated from the ventricles of neonatal Sprague–Dawley rat hearts by a previously described adaptation of the method of Iwaki et al. (32,33). To deplete the myocytes of fibroblasts, the cells were pre-plated in DMEM/Medium-199 (4:1, v/v) supplemented with 10% (v/v) horse serum (HS), 5% (v/v) heat-inactivated foetal calf serum (FCS), penicillin (100 U/ml) and streptomycin (100 µg/ml). The (non-adherent) myocytes were then plated into 6-well culture dishes precoated with 1% (w/v) gelatin at a density of 1.4 × 103 cells/mm2. After 18 h, the medium was replaced with the same medium containing 4% HS and no FCS. Experiments were carried out in the same medium but containing 10% FCS. For hypoxia, cultures were incubated in a hypoxic incubator (Wolf Laboratories, UK) with a gas mixture containing 1% O2 and 5% CO2, balanced with nitrogen. Prior to placing the cells in the incubator, the culture medium was replaced with fresh medium that had been equilibrated overnight with the same gas mixture. Cells under normoxic conditions were treated in the same way, but using a gas mix of 20% O2, 5% CO2.
HIF-1α tetracycline (TetOn)-inducible SAOS2 cells were cultured using the method of Bardos et al. (34). Wild-type and c-myc-null fibroblasts were cultured as described previously (35). Preparation and use of c-MycER-transduced fibroblasts has been described (36).
Western immunoblotting analysis
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) then scraped in RIPA buffer [50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM EDTA, 10 mM NaF, 10 mM sodium phosphate, 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml soybean trypsin inhibitor and 10 µg/ml benzamidine]. Buffers used for hypoxic samples were de-oxygenated before use. Samples were centrifuged at 10 000 g for 15 min at 4°C. Western immunoblot analysis was performed as described by White et al. (37).
Antibodies used were N-15 against TFIIIC63, D-2 against ERK, 9E10 against c-Myc, C-15 against RB and 58C9 against TBP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The p53, phospho-RB (Ser-795) and phospho-ERK (Thr-202/Tyr-204) antibodies were from New England Biolabs. Peptide antisera 1900 against RPC155 subunit of pol III, Ab2 against TFIIIC220, 4286 against TFIIIC110 and 1898 against TFIIIC90 subunits of TFIIIC, and 2663 against Bdp1, and 128 against Brf1 subunits of TFIIIB have been characterized previously (29,38–41). Antiserum against BN51 was generously provided by Michael Ittman (42).
Protein synthesis measurement
Rates of protein synthesis were assayed in cardiomyocytes by measuring the incorporation of [35S]methionine/cysteine into acid-insoluble material as described previously (43). Approximately 10 µCi of radioisotope (>1000 Ci/mmol) was used per well of the culture dish.
In vitro transcription assay
Whole-cell extracts were prepared for transcription assays using the freeze-thaw procedure as previously described (37). Pol III transcription assays were carried out as in White et al. (44), except that transcriptions were for 60 min at 30°C.
Immunoprecipitations
Cells were washed twice in ice-cold PBS and scraped into IP buffer [50 mM HEPES pH 7.5, 5 mM EDTA, 10 mM NaF, 150 mM NaCl, 25% glycerol, 0.5% Triton X-100, 0.5 mM PMSF, 0.5 µg/ml leupeptin, 0.7 µg/ml pepstatin, 0.5 µg/ml aprotinin, 40 µg/ml bestatin, 1 mM sodium vanadate and 50 mM β-glycerophosphate]. After 15 min on ice, the extracts were passed three times through a 26G needle and insoluble material was removed by centrifugation at 14 000 g for 15 min prior to immunoprecipitation. Extracts (500 µg) were incubated on an orbital shaker with 30 µl of protein A–Sepharose beads carrying an equivalent amount of prebound IgG. Samples were then pelleted, supernatants removed and the beads washed three times with 300 µl Tris-buffered saline. The bound material was analysed by western blotting.
Northern blotting
Total cellular RNA was extracted using TRI reagent (Sigma), according to the manufacturer's instructions. Agarose gel electrophoresis, northern transfer and hybridization were carried out as described previously (22). The B2 gene probe was a 240 bp EcoRI–PstI fragment from pTB14 (44). The acidic ribosomal phosphoprotein P0 (ARPP P0) probe was a 1 kb EcoRI–HindIII fragment from mouse cDNA (45). The 18S rRNA probe was a 80 bp fragment from a highly conserved region of the gene (Ambion, Inc.).
RT–PCR analysis
RNA was extracted as above. Reverse transcription reactions were performed for 1 h at 42°C using 3 µg of RNA, 200 ng of Random Hexamers (Promega) and 400 U of Superscript II Reverse Transcriptase (Life Technologies) in a total volume of 40 µl of 1× First Strand Buffer (Life Technologies) containing 10 mM DTT and 0.5 mM concentration of each dNTP. PCRs were carried out using a Techgene thermal controller (Wolf Laboratories, UK). cDNA (2 µl) was amplified with 20 pmol of either tRNALeu primers (5′-GTCAGGATGGCCGAGTGGTCTAAGGCGCC-3′ and 5′-CCACGCCTCCATACGGAGACCAGAAGACCC-3′) to give an 88 bp product, tRNATyr primers (5′-CCTTCGATAGCTCAGCTGGTAGAGCGGAGG-3′ and 5′-CGGAATTGAACCAGCGACCTAAGGATGTCC-3′) to give an 84 bp product, 5S rRNA primers (5′-GGCCATACCACCCTGAACGC-3′ and 5′-CAGCACCCGGTATTCCCAGG-3′) to give a 107 bp product or ARPP P0 primers (5′-GCACTGGAAGTCCAACTACTTC-3′ and 5′-TGAGGTCCTCCTTGGTGAACAC-3′) to give a 265 bp product. Amplification reactions contained 0.5 U of Taq DNA polymerase (Promega) in a total volume of 1× Taq DNA polymerase buffer (Promega) containing 1.5 mM MgCl2 and 0.2 mM concentration of each dNTP and 1.8 µCi of [α-32P]dCTP (10 mCi/ml, 3000 Ci/mmol). PCR was performed under the following cycling parameters: (i) ARPP P0, 95°C for 2 min, 18 cycles of 95°C for 1 min, 58°C for 30 s and 72°C for 1 min; 72°C for 3 min; (ii) tRNALeu, 95°C for 3 min, 25 cycles of 95°C for 30 s, 68°C for 30 s and 72°C for 30 s; 72°C for 5 min; (iii) tRNATyr, 95°C for 3 min, 25 cycles of 95°C for 1 min, 62°C for 30 s and 72°C for 30 s; 72°C for 5 min; (iv) 5S rRNA, 95°C for 3 min, 18 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 1 min; 72°C for 5 min. Reaction products were resolved on a 7% polyacrylamide gel containing 7M urea and 0.5× TBE. Radioactivity was visualized by autoradiography and quantified by phosphorimaging.
Polymerase assay
Random polymerase assays were performed as before (46). Each mixture contained 5 µg of poly(dA–dT) template and 10 µg of whole-cell extract. Pol III activity was calculated by subtracting the polymerization obtained in the presence of 200 µg/ml α-amanitin (due to pol I) from that obtained in the presence of 1 µg/ml α-amanitin (due to pol III plus pol I).
Chromatin immunoprecipitation assay
ChIP was performed as in Gomez-Roman et al. (25). Immunoprecipited DNA was quantified by PCR performed using previously described primers and amplification procedures (25). Antibodies used were C-18 against TFIIB (Santa Cruz Biotechnologies), 128 against Brf1, 2663 against Bdp1, MTBP-6 against TBP, 4286 against TFIIIC110, Ab2 against TFIIIC220, 114 against pol III subunit RPC53 and 1900 against pol III subunit RPC155 (29,38–41). Serial dilutions of chromatin were used to establish that PCRs were within a linear range.
RESULTS
Pol III transcription decreases when cardiomyocytes are exposed to low oxygen
Exposure of many cell types to hypoxic conditions has been shown to decrease protein synthesis, although the mechanisms responsible have not been fully elucidated (10–14). This is also observed when cardiomyoctes are incubated in 1% oxygen (Figure 1A). Protein synthesis starts to decrease between 4 and 8 h following exposure of these cells to hypoxia. No significant difference in the level of apoptosis or necrosis was detected between cells incubated in the normoxic or hypoxic environment (data not shown). The effect of hypoxia on pol III transcription can clearly be seen by monitoring pol III transcripts derived from the middle repetitive gene family B2, which have a short half-life and so provide a reliable indication of transcriptional output. The abundance of B2 RNA was reduced by 40% when cardiomyocytes were incubated for 8 h in 1% O2 (Figure 1B and C). In contrast, levels of the pol I-transcribed 18S rRNA did not diminish under the same conditions (Figure 1B). This effect was confirmed using RT–PCR, in which levels of pol III transcripts B2, tRNALeu and tRNATyr were measured. Primers that hybridize specifically to the intron sequences of short-lived precursors of the tRNAs were used. Because introns are processed from primary transcripts and then degraded very rapidly, their levels in a cell reflect the rate of ongoing transcription (47). Levels of B2 and the primary transcripts tRNATyr and tRNALeu were significantly reduced after 8 h incubation in hypoxia (Figure 1D). In contrast, levels of mRNA encoding ARPP P0 did not diminish under the same conditions.
Figure 1.
Incubation of cardiomyocytes in low oxygen reduces pol III activity. (A) Cardiomyocytes were incubated in 20% (N) or 1% O2 (H) for 2, 4, 8, 16 or 24 h and [35S]methionine added to the medium 30 min prior to harvesting. Samples were processed to measure incorporation of label into trichloroacetic acid-precipitable material. Data are expressed as d.p.m./µg protein ± SD, where n = 3. (B) Northern blot analysis of total RNA (20 µg) extracted from cardiomyocytes cultured in 20% (N) or 1% O2 (H) for 4, 8 or 16 h. The upper panel shows the blot probed with a B2 gene, and the lower panel shows the same blot that has been stripped and reprobed with the rDNA gene. (C) Results from (B) were quantified and normalized against the rRNA signal. The graph shows the mean and SDs from three independent experiments; values obtained for the 4 h normoxic sample were set to 100 and other values calculated as a percentage of this. (D) cDNAs were generated by reverse transcription of RNA from cells treated as in (B), as indicated. These cDNAs were PCR amplified using primers specific for the indicated template.
The levels of pol III, TFIIIB and TFIIIC remain relatively constant in hypoxia-treated cardiomyocytes
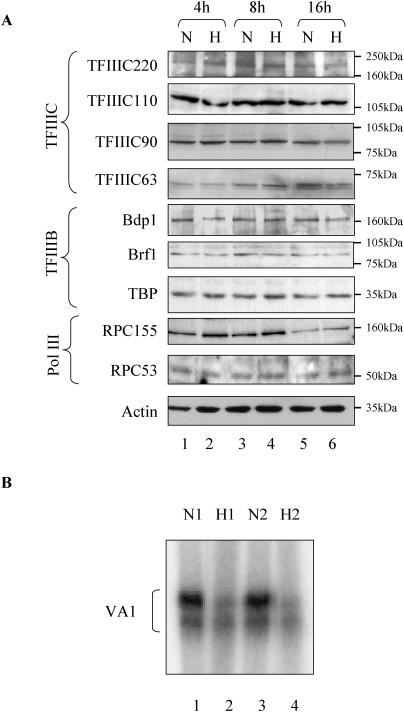
To investigate whether the reduction in pol III activity under hypoxic conditions is due to lower levels of pol III transcription factors, western blots were carried out with extracts from cells grown in 1% oxygen for various times. Up to 16 h in hypoxia produced little or no change in levels of the Bdp1, TBP and Brf1 subunits of TFIIIB, or the 220, 110, 90 or 63 kDa subunits of TFIIIC (Figure 2A). Levels of the RPC155 and RPC53 subunits of pol III also remain unchanged in response to hypoxia, as does that of actin (Figure 2A). Although antibodies are not available to monitor every subunit of TFIIIC and pol III in rat cells, the available data provide no evidence for a change in the abundance of pol III-specific transcription factors or pol III itself when cardiomyocytes become hypoxic.
Figure 2.
Down-regulation of pol III activity in hypoxia is not due to a decrease in the abundance of TFIIIB, TFIIIC or pol III subunits. (A) Cardiomyocytes cultured in 20% (N) or 1% O2 (H) for 4, 8 or 16 h were resolved on an SDS–7.8% polyacrylamide gel and then analysed by western immunoblotting with antibodies specific to the 63, 90, 110 and 220 kDa subunits of TFIIIC, the TBP, Brf1 and Bdp1 subunits of TFIIIB, and the 53 and 155 kDa subunits of pol III, as indicated. (B) Whole-cell extract was prepared from cardiomyocytes cultured for 16 h in 1% (H1 and H2) or 20% (N1 and N2) O2. Template (500 ng) VA1 was used for in vitro transcription assay using 15 µg of these extracts.
We assayed the ability of pol III to catalyse randomly initiated RNA synthesis independently of transcription factors, using a poly(dA–dT) template. No significant difference in the level of random polymerization by pol III was observed between extracts prepared from cells incubated in normoxic or hypoxic conditions (data not shown). Nevertheless, pol III transcriptional activity was clearly diminished in these extracts. Thus, extracts prepared from cardiomyocytes treated for 16 h in hypoxia were found to transcribe a pol III template significantly less actively than extracts prepared from cells grown in normoxic conditions, reflecting the situation in vivo (Figure 2B).
Pol III recruitment to tRNA promoters is reduced under hypoxic conditions
Although hypoxia caused little or no variation in the abundance of specific pol III transcription factors nor in the catalytic activity of the pol III enzyme, we investigated whether occupancy at pol III promoters was affected. ChIP experiments were carried out on cardiomyocytes incubated for 16 h under hypoxic or normoxic conditions. Formaldehyde cross-linked soluble chromatin was prepared from these cells and, following normalization for DNA content, these were immunoprecipitated (IP) with antibodies against TFIIIB (Bdp1, Brf1 and TBP) as well as TFIIIC (220 and 110 subunits) and pol III (subunit RPC53). PCR analysis of precipitated DNA showed that TFIIIC, Bdp1 and TBP occupancy of tRNALeu genes are essentially unaffected when cardiomyocytes are incubated in 1% oxygen (Figure 3). However, pol III occupancy at tRNA genes is decreased by 60% compared with normoxic cells, suggesting a defect in polymerase recruitment when hypoxic stress is applied. In contrast, Brf1 crosslinking is elevated under hypoxic conditions. Since Brf1 is known to bind directly to pol III, this apparent anomaly may be explained if the accessibility of Brf1 to antibody increases when pol III occupancy falls. The observed drop in pol III interaction with its templates offers an explanation for the decrease in transcription of these genes under low oxygen.
Figure 3.
Culture of cardiomyocytes in hypoxia compromises the promoter occupancy by pol III. (A) ChIP assay showing levels of Bdp1, Brf1, TBP, TFIIIC (TFIIIC 110 and 220 subunits), pol III (RPC53 subunit) and TFIIB associated with the tRNALeu gene in cardiomyocytes cultured for 16 h in normoxic (N) or hypoxic (H) conditions. (B) Quantification of ChIP assays. PCR products from three independent ChIP experiments were quantified for tRNA genes; after normalization to input, the mean and SDs are shown with the normoxic value in each case set to 100 and the hypoxic value expressed relative to this (*significantly different from control, P < 0.05).
Hypoxia alters the interactions of TFIIIB with its regulators RB, c-Myc and ERK
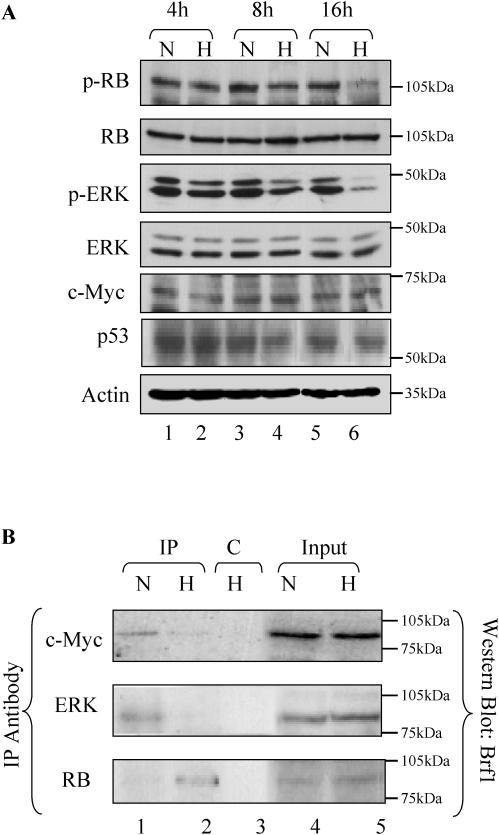
Several regulatory factors have been shown to influence the function of the pol III machinery (19,21,48). For example, TFIIIB is a specific target for repression by RB and p53 (22,26–30,49). Conversely, both c-Myc and ERK can bind TFIIIB and activate pol III transcription (24,25). Western blot analysis detected little or no change in the levels of RB, p53, c-Myc or ERK when cardiomyocytes were exposed for up to 16 h in 1% oxygen (Figure 4A). Since pol III transcription is sensitive to the phosphorylation status of RB, this was also measured in these cells; it is the underphosphorylated form of RB that binds and represses TFIIIB (50). Western blotting with a phospho-specific antibody revealed a decrease in RB phosphorylation at Ser795 within 8 h of incubation in 1% oxygen (Figure 4A). This correlates with the down-regulation of pol III transcription observed in these cells. ERK activation in response to mitogens stimulates pol III transcription via phosphorylation of TFIIIB (24). Since ERK must be phosphorylated before it can activate TFIIIB, the phosphorylation status of the kinase was measured. Although hypoxia can cause a rapid, acute activation of ERK in some cell types (51,52), incubating cardiomyocytes for 8 h in low oxygen resulted in a decrease in phospho-ERK and phosphorylation was almost abolished by 16 h (Figure 4A).
Figure 4.
The phosphorylation status of RB and ERK decreases when cardiomyocytes are cultured in 1% O2. (A) Cardiomyocytes cultured in 20% (N) or 1% O2 (H) for 4, 8 or 16 h were resolved on an SDS–7.8% polyacrylamide gel and then analysed by western immunoblotting with antibodies specific to RB phosphorylated at Ser-795 (p-RB), RB, ERK phosphorylated at Thr-202 and Tyr-204 (p-ERK), ERK, c-Myc, p53 and actin, as indicated. (B) Cell extracts (500 µg) were prepared from rat neonatal cardiomyocytes cultured for 16 h in normoxic (lanes 1 and 4) or hypoxic (lanes 2, 3 and 5) conditions. These were IP with anti-c-Myc (lanes 1 and 2, upper panel), anti-ERK (lanes 1 and 2, middle panel), anti-RB (lanes 1 and 2, lower panel) or anti-4E-BP1 (lane 3, all panels). Precipitates were resolved by SDS–PAGE and then analysed by western blotting with anti-Brf1 antibody. 10% input samples are shown in lanes 4 and 5.
ERK, c-Myc and RB all regulate pol III transcription by interacting with TFIIIB (24,25,27–29). We therefore tested whether hypoxia alters the association of these proteins using co-immunoprecipitation. Extracts were prepared from cardiomyocytes exposed for 16 h in normoxic or hypoxic conditions and c-Myc, RB or ERK immunoprecipitates were probed by western using a specific antibody against the Brf1 subunit of TFIIIB. Both ERK and c-Myc were found to associate with Brf1 in cells grown at normal oxygen concentrations, but this interaction is substantially reduced in low oxygen (Figure 4B, upper two panels). In contrast, the amount of RB that associates with Brf1 is greatly increased in hypoxia (Figure 4B, lower panel). These co-precipitations reflect specific interactions, since Brf1 is not co-immunoprecipitated in either a normoxic or hypoxic environment with a control antiserum against the translational regulator 4E-BP1 (Figure 4B, lane 3, and data not shown). Whereas all previous studies have used immortalized cell lines, these data confirm that TFIIIB also binds to c-Myc, RB and ERK in primary cells. They also reveal that the constellation of regulators associated with TFIIIB is dynamic, and changes dramatically in response to hypoxia.
HIF-1α can associate with c-Myc and regulate pol III transcription
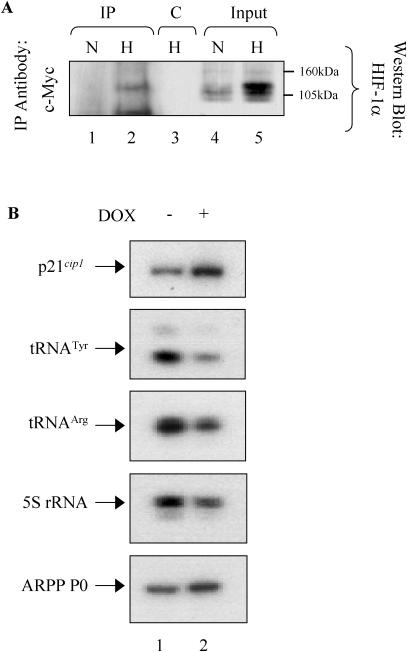
The altered interaction of TFIIIB with ERK and RB correlates with the observed change in phosphorylation status of these proteins in 1% oxygen (Figure 4A). In contrast, phosphorylation has not been shown to influence the binding of c-Myc to TFIIIB. However, recent work has suggested that c-Myc function is compromised under low oxygen conditions through the action of HIF-1α, which displaces c-Myc from some of its targets (53,54). These studies reported a weak and probably indirect association of HIF-1α with c-Myc in hypoxic human cell lines, so we investigated if this could also be detected in primary rat cardiomyocytes. To this end, we immunoprecipitated c-Myc from cardiomyocytes incubated for 16 h in normoxia or hypoxia, and tested by western blotting for binding of HIF-1α. Treatment with 1% oxygen resulted in induction of HIF-1α (Figure 5A, lanes 4 and 5) and its appearance in anti-c-Myc immunocomplexes (Figure 5A, lane 2). This is consistent with a model in which HIF-1α interacts with c-Myc in hypoxic cardiomyocytes and interferes with its ability to activate pol III transcription. To begin to test this hypothesis, we used a cell system in which HIF-1α can be induced with doxycycline under normoxic conditions (34); these cells are negative for both RB and p53, thus excluding two known mediators of pol III regulation. Since it has been shown that HIF-1α induces p21cip1 gene expression by blocking c-Myc function (53), we used this gene as a positive control. Indeed, addition of doxycycline to these cells did enhance p21cip1 mRNA levels, as reported previously (Figure 5B). Furthermore, this was accompanied by a specific decrease in tRNA and 5S rRNA, suggesting that expression of HIF-1α can inhibit synthesis of pol III transcripts, possibly through an effect on c-Myc. As above, the tRNA primers used in this experiment are specific for the short-lived primary transcripts and therefore provide a measure of ongoing transcription.
Figure 5.
HIF-1α can bind c-Myc and regulate pol III transcription. (A) Cell extracts (500 µg) were prepared from rat neonatal cardiomyocytes cultured for 16 h in normoxic (lanes 1 and 4) or hypoxic (lanes 2, 3 and 5) conditions. These were IP with anti-c-Myc (lanes 1 and 2) or anti-4E-BP1 (lane 3) antibodies. Precipitates were resolved by SDS–PAGE and then analysed by western blotting with anti-HIF-1α antibody. Input samples (10%) are shown in lanes 4 and 5. (B) Saos-2 HIF-1α TetOn-inducible cells were treated with or without doxycycline (DOX; 2 µg/ml) for 18 h. cDNAs were generated by reverse transcription and these were amplified by PCR using primers specific for p21cip1, tRNATyr, tRNAArg, 5S rRNA and ARPP P0, as indicated.
Pol III recruitment is stimulated by c-Myc
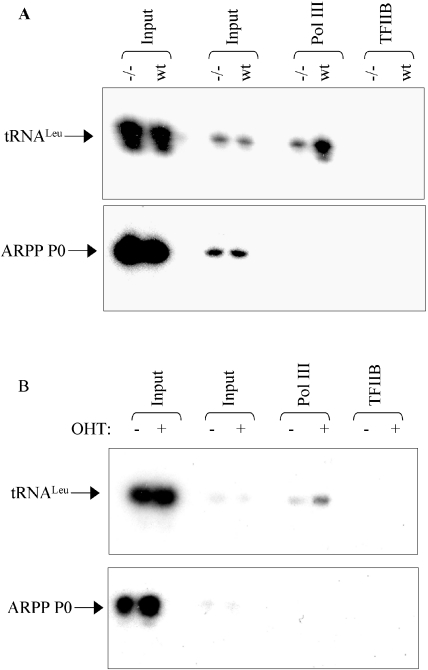
The ChIP assays above showed a specific reduction in template occupancy by pol III in 1% oxygen. Although ERK and RB have both been demonstrated to influence pol III recruitment by TFIIIB, this has yet to be shown for c-Myc. To test this, we used ChIP to compare pol III occupancy in matched wild-type and c-Myc-knockout Rat1 cells. This revealed that the interaction of pol III with tRNA genes is markedly diminished in the cells from which c-Myc has been selectively deleted (Figure 6A). The association is specific, as the pol II transcription factor TFIIB which was used a control was not detected at the tRNA genes and the pol III antibody did not immunoprecipitate DNA from within the coding region of the ARPP P0 gene. As an independent test, we also used a murine fibroblast cell line carrying a c-MycER construct, which can be rapidly induced by the addition of hydroxy-tamoxifen (OHT) for 4 h. These cells were starved of serum, to silence expression of endogenous c-Myc, and then harvested for ChIP analysis either with or without OHT treatment. Pol III occupancy of tRNA genes increased substantially and specifically within 4 h of c-MycER induction (Figure 6B). We conclude that c-Myc can promote the recruitment of pol III to its targets, consistent with its established ability to stimulate pol III transcription. The diminished interaction between c-Myc and TFIIIB is therefore likely to contribute to the observed reduction in polymerase occupancy under low oxygen conditions.
Figure 6.
Pol III recruitment to tRNA genes is stimulated by c-Myc. (A) ChIP assay showing levels of pol III (RPC155 subunit) and TFIIB associated with tRNALeu and ARPP P0 genes in Myc+/+ (wt) and Myc−/− (−/−) cells. (B) ChIP assay showing levels of pol III (RPC155 subunit) and TFIIB associated with tRNALeu and ARPP P0 genes in MycER-transduced murine fibroblasts treated with 4-OHT for 0 h (uninduced) or 4 h (induced). Quantitative PCR was performed with equivalent DNA input amounts determined by PCR on 1:10 and 1:100 diluted input chromatin (upper panel).
DISCUSSION
In primary cultures of rat cardiomyocytes, low oxygen conditions reduce the transcriptional output of pol III, as well as its association with target genes. Although not all subunits were examined, we detected no change in the level or catalytic activity of pol III. We also found little or no change in the expression of TFIIIB and TFIIIC or in their occupancy of tRNA genes. However, the interaction of TFIIIB with regulatory factors was altered quite markedly, with increased binding to its inhibitor RB and decreased binding to its activators c-Myc and ERK. TFIIIB is responsible for recruiting pol III to its templates (16,17). Previous studies have shown that ERK stimulates the interaction between TFIIIB and pol III (24), whereas RB has the opposite effect (29). Here we have demonstrated that c-Myc can also stimulate the ability of TFIIIB to recruit pol III to its target genes. The dissociation of TFIIIB from c-Myc and ERK, as well as its binding to RB, can therefore explain the reduced occupancy of tRNA genes by pol III in low oxygen and hence the diminished rates of transcription.
Our data suggest a model in which the dissociation of c-Myc from TFIIIB might be mediated by HIF-1α. It has been reported that the stabilization of HIF-1α under hypoxic conditions antagonizes c-Myc function and displaces it from several promoters, including that of the p21cip1 gene (53,54). We confirmed the effect of HIF-1α on p21cip1 expression using cells in which doxycycline was added to specifically induce HIF-1α activity (34); such a system allows one to distinguish the effects of HIF-1α from other responses to hypoxic conditions. Induction of HIF-1α also resulted in a significant decrease in pol III transcript levels. Thus, inactivation of c-Myc by HIF-1α might contribute to the decrease in pol III transcription at low oxygen tension, although much further work will be required to test this rigorously. Although we have yet to establish the mechanism(s) responsible for dissociating c-Myc from TFIIIB, the binding of ERK and RB to TFIIIB is known to be regulated by phosphorylation (24,50). Interaction of ERK with TFIIIB requires that the former is phosphorylated and activated by MEK (24); western blotting showed a reversal of ERK activation in low oxygen, thereby explaining its release from TFIIIB. Conversely, it is the hypophosphorylated form of RB that binds and regulates TFIIIB (50) and it is this form that predominates under hypoxic conditions. Oxygen deprivation therefore leads to an overall exchange of TFIIIB partners from activators to a potent repressor.
We had anticipated that p53 might contribute to the regulation of pol III transcription under these conditions, as hypoxia is known to be one of the stresses that can stabilize and activate p53 (55). However, studies of the p53 response to low oxygen generally use chemical mimetics of anoxia or oxygen concentrations of 0.1% or less (31,55,56). We found that the milder conditions of 1% oxygen used here were insufficient to provoke a p53 response in cardiomyocytes, even though other molecular changes were clearly observed. This is consistent with a previous study, using a colon carcinoma cell line, which found that p53 induction begins at 0.5% oxygen, but is not apparent in 1% oxygen, even after 48 h incubation (56).
Assays of protein–protein interactions, in vitro and in vivo, indicated that RB and ERK influence the binding of TFIIIB to both TFIIIC and pol III; ERK stimulates these steps, whereas RB blocks them (24,29). Since TFIIIB is recruited to tRNA genes by TFIIIC, we had predicted that its promoter occupancy would respond to RB and ERK. This is supported by ChIP analyses which confirmed that RB-bound TFIIIB is not associated with tRNA genes in cycling HeLa cells (57). We were therefore surprised to find that release of ERK and increased binding of RB does not dissociate TFIIIB from promoters in hypoxic cardiomyocytes, although we do observe the predicted change in pol III occupancy. Although differences in cell type might be invoked, we suggest that a key difference between this and previous studies is that the cardiomyocytes used here have withdrawn permanently from the cell cycle. In proliferating cells, we would expect pol III transcription complexes to be displaced from promoters by DNA replication (58); their stability may therefore be enhanced considerably in non-replicating, post-mitotic cardiomyocytes. Perhaps in the absence of replication, mammalian TFIIIB can remain promoter-bound even if its contacts with TFIIIC have been weakened; this is certainly the situation in Saccharomyces, where TFIIIC is required to recruit TFIIIB, but not for its subsequent retention (59). In contrast, the recruitment of pol III by TFIIIB must occur for every round of transcription and may accordingly be much more immediately responsive to changes in ERK and/or RB activity. This may explain why it is only pol III occupancy that decreases in hypoxic cells; even a stable preinitiation complex must recruit polymerase to allow multiple rounds of transcription. A failure of pol III recruitment is clearly sufficient to give the observed decrease in transcriptional output. However, the complexes remain poised on tRNA gene promoters, ready to resume activity if oxygen levels rise.
The ability to reduce macromolecular synthesis is an important component of the protective response to hypoxia. We have demonstrated that oxygen deprivation causes a clear reduction in gene expression by pol III. This is likely to provide a substantial saving in energy consumption, since pol III is responsible for around 10% of all nuclear transcription. When aerobic metabolism is curtailed, such energy conservation may make a significant contribution to a cell's chances of survival.
Acknowledgments
We thank Margaret Ashcroft for helpful discussions and for access to the HIF-1α TetOn-inducible cells, Carla Grandori for c-MycER-transduced fibroblasts, Sonia Rocha for RNA samples, Arnie Berk for antisera against TFIIIC220 and Michael Ittmann for antisera against RPC53. This work has been supported by project grants from The Wellcome Trust (068710 and 062625) and Chest, Heart and Stroke Scotland (02/A62). Funding to pay the Open Access publication charges for this article was provided by University of Glasgow.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gardner L.B., Li Q., Park M.S., Flanagan W.M., Semenza G.L., Dang C.V. Hypoxia inhibits G1/S transition through regulation of p27 expression. J. Biol. Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 2.Goda N., Ryan H.E., Khadivi B., McNulty W., Rickert R.C., Johnson R.S. Hypoxia-inducible factor 1α is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris A.L. Hypoxia—a key regulatory factor in tumour growth. Nature Rev. Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 4.Kumar G.K., Klein J.B. Analysis of expression and posttranslational modification of proteins during hypoxia. J. Appl. Physiol. 2004;96:1178–1186. doi: 10.1152/japplphysiol.00818.2003. [DOI] [PubMed] [Google Scholar]
- 5.Denko N.C., Fontana L.A., Hudson K.M., Sutphin P.D., Raychaudhuri S., Altman R., Giaccia A.J. Investigating hypoxic tumour physiology through gene expression patterns. Oncogene. 2003;22:5907–5914. doi: 10.1038/sj.onc.1206703. [DOI] [PubMed] [Google Scholar]
- 6.Roy S., Khanna S., Bickerstaff A.A., Subramanian S.V., Atalay M., Bierl M., Pendyala S., Levy D., Sharma N., Venojarvi M., et al. Oxygen sensing by primary cardiac fibroblasts: A key role of p21(Waf1/Cip1/Sdi1) Circ. Res. 2003;92:264–271. doi: 10.1161/01.res.0000056770.30922.e6. [DOI] [PubMed] [Google Scholar]
- 7.Casey T.M., Pakay J.L., Guppy M., Arthur P.G. Hypoxia causes downregulation of protein and RNA synthesis in noncontracting mammalian cardiomyocytes. Circ. Res. 2002;90:777–783. doi: 10.1161/01.res.0000015592.95986.03. [DOI] [PubMed] [Google Scholar]
- 8.Koumenis C., Naczki C., Koritzinsky M., Rastani S., Diehl A., Sonenberg N., Koromilas A., Wouters B.G. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2 alpha. Mol. Cell. Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur P.G., Giles J.J., Wakeford C.M. Protein synthesis during oxygen conformance and severe hypoxia in the mouse muscle cell line C2C12. Biochim. Biophys. Acta. 2000;1475:83–89. doi: 10.1016/s0304-4165(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 10.Hochachka P.W., Buck L.T., Doll C.J., Land S.C. Unifying theory of hypoxia tolerance: molecular/metabolic defence and rescue mechanisms for surviving oxygen lack. Proc. Natl Acad. Sci. USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettersen E.O., Juul N.O., Ronning O.W. Regulation of protein-metabolism of human-cells during and after acute-hypoxia. Cancer Res. 1986;46:4346–4351. [PubMed] [Google Scholar]
- 12.Clark L., Fillenwarth M.J., Shen W.H., Boyle D.W. Effect of hypoxia on translation initiation factors in the C2C12 skeletal muscle cell line over time. FASEB J. 2003;17:A1285. [Google Scholar]
- 13.Tinton S.A., Buc-Calderon P.M. Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 1999;446:55–59. doi: 10.1016/s0014-5793(99)00185-4. [DOI] [PubMed] [Google Scholar]
- 14.Kraggerud S.M., Sandvik J.A., Pettersen E.O. Regulation of protein-synthesis in human-cells exposed to extreme hypoxia. Anticancer Res. 1995;15:683–686. [PubMed] [Google Scholar]
- 15.White R.J. RNA Polymerase III Transcription 3rd edn. Austin: Landes Bioscience; 2002. [Google Scholar]
- 16.Geiduschek E.P., Kassavetis G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 17.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 18.White R.J., Gottlieb T.M., Downes C.S., Jackson S.P. Cell cycle regulation of RNA polymerase III transcription. Mol. Cell. Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White R.J. RNA polymerase III transcription—a battleground for tumour suppressors and oncogenes. Eur. J. Cancer. 2004;40:21–27. doi: 10.1016/j.ejca.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Felton-Edkins Z.A., Kenneth N.S., Brown T.R.P., Daly N.L., Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–184. [PubMed] [Google Scholar]
- 21.White R.J. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 22.Cairns C.A., White R.J. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White R.J., Trouche D., Martin K., Jackson S.P., Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 24.Felton-Edkins Z.A., Fairley J.A., Graham E.L., Johnston I.M., White R.J., Scott P.H. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 2003;22:2422–2432. doi: 10.1093/emboj/cdg240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 26.Chesnokov I., Chu W.M., Botchan M.R., Schmid C.W. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol. Cell. Biol. 1996;16:7084–7088. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larminie C.G.C., Cairns C.A., Mital R., Martin K., Kouzarides T., Jackson S.P., White R.J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu W.-M., Wang Z., Roeder R.G., Schmid C.W. RNA polymerase III trancription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J. Biol. Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 29.Sutcliffe J.E., Brown T.R.P., Allison S.J., Scott P.H., White R.J. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 2000;20:9192–9202. doi: 10.1128/mcb.20.24.9192-9202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crighton D., Woiwode A., Zhang C., Mandavia N., Morton J.P., Warnock L.J., Milner J., White R.J., Johnson D.L. p53 represses RNA polymerase III transcription by targetting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–2820. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long X.L., Boluyt M.O., Hipolito M.D., Lundberg M.S., Zheng J.S., O'Neill L., Cirielli C., Lakatta E.G., Crow M.T. p53 and the hypoxia-induced apoptosis of cultured neonatal rat cardiac myocytes. J. Clin. Invest. 1997;99:2635–2643. doi: 10.1172/JCI119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwaki K., Sukhatme V.P., Shubeita H.E., Chien K.R. Alpha-adrenergic and beta-adrenergic stimulation induces distinct patterns of immediate early gene-expression in neonatal rat myocardial-cells. fos/jun expression is associated with sarcomere assembly—egr-1 induction is primarily an alpha-1-mediated response. J. Biol. Chem. 1990;265:13809–1381. [PubMed] [Google Scholar]
- 33.Bogoyevitch M.A., Ketterman A.J., Sugden P.H. Cellular stresses differentially activate c-jun n-terminal protein-kinases and extracellular signal-regulated protein-kinases in cultured ventricular myocytes. J. Biol. Chem. 1995;270:29710–29717. doi: 10.1074/jbc.270.50.29710. [DOI] [PubMed] [Google Scholar]
- 34.Bardos J.I., Chau N.-M., Ashcroft M. Growth factor-mediated induction of HDM2 positively regulates hypoxia-inducible factor 1α expression. Mol. Cell. Biol. 2004;24:2905–2914. doi: 10.1128/MCB.24.7.2905-2914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateyak M.K., Obaya A.J., Adachi S., Sedivy J.M. Phenotypes of c-myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 36.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 37.White R.J., Gottlieb T.M., Downes C.S., Jackson S.P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alzuherri H., White R.J. Regulation of a TATA-binding protein-associated factor during cellular differentiation. J. Biol. Chem. 1998;273:17166–17171. doi: 10.1074/jbc.273.27.17166. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y., Igo M., Yalamanchili P., Berk A.J., Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felton-Edkins Z.A., White R.J. Multiple mechanisms contribute to the activation of RNA polymerase III transcription in cells transformed by papovaviruses. J. Biol. Chem. 2002;277:48182–48191. doi: 10.1074/jbc.M201333200. [DOI] [PubMed] [Google Scholar]
- 41.Fairley J.A., Scott P.H., White R.J. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003;22:5841–5850. doi: 10.1093/emboj/cdg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ittman M., Ali J., Greco A., Basilico C. The gene complementing a temperature-sensitive cell cycle mutant of BHK cells is the human homologue of the yeast RPC53 gene, which encodes a subunit of RNA polymerase C (III) Cell Growth Differ. 1993;4:503–511. [PubMed] [Google Scholar]
- 43.Welsh G.I., Proud C.G. Regulation of protein-synthesis in swiss 3t3 fibroblasts. Rapid activation of the guanine-nucleotide-exchange factor by insulin and growth-factors. Biochem. J. 1992;284:19–23. doi: 10.1042/bj2840019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White R.J., Stott D., Rigby P.W.J. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- 45.Hurford R.K., Cobrinik D., Lee M.-H., Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 46.Roeder R.G. Multiple forms of nucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. J. Biol. Chem. 1974;249:241–248. [PubMed] [Google Scholar]
- 47.Winter A.G., Sourvinos G., Allison S.J., Tosh K., Scott P.H., Spandidos D.A., White R.J. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumours. Proc. Natl Acad. Sci. USA. 2000;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White R.J. RNA polymerases I and III, growth control and cancer. Nature Rev. Mol. Cell. Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 49.Hirsch H.A., Gu L., Henry R.W. The retinoblastoma tumour suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell. Biol. 2000;20:9182–9191. doi: 10.1128/mcb.20.24.9182-9191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott P.H., Cairns C.A., Sutcliffe J.E., Alzuherri H., McLees A., Winter A.G., White R.J. Regulation of RNA polymerase III transcription during cell cycle entry. J. Biol. Chem. 2001;276:1005–1014. doi: 10.1074/jbc.M005417200. [DOI] [PubMed] [Google Scholar]
- 51.Minet E., Michel A.G., Roland I., Mottet D., Raes M., Remacle J., Michiels C. ERK activation upon hypoxia: involvement in HIF-1 activation. FEBS Lett. 2000;468:53–58. doi: 10.1016/s0014-5793(00)01181-9. [DOI] [PubMed] [Google Scholar]
- 52.Jin N.J., Hatton N., Swartz D.R., Xia X.L., Harrington M.A., Larsen S.H., Rhoads R.E. Hypoxia activates Jun-N-terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries. Am. J. Respir. Cell Mol. Biol. 2000;23:593–601. doi: 10.1165/ajrcmb.23.5.3921. [DOI] [PubMed] [Google Scholar]
- 53.Koshiji M., Kageyama Y., Pete E.A., Horikawa I., Barrett J.C., Huang L.E. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koshiji M., To K.K.-W., Hammer S., Kumamoto K., Harris A.L., Modrich P., Huang L.E. HIF-1α induces genetic instability by transcriptionally downregulaing MutSα expression. Mol. Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 55.Graeber T.G., Peterson J.F., Tsai M., Monica K., Fornace A.J., Giaccia A.J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achison M., Hupp T.R. Hypoxia attenuates the p53 response to cellular damage. Oncogene. 2003;22:3431–3440. doi: 10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch H.A., Jawdekar G.W., Lee K.-A., Gu L., Henry R.W. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumour suppressor protein. Mol. Cell. Biol. 2004;24:5989–5999. doi: 10.1128/MCB.24.13.5989-5999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolffe A.P., Brown D.D. DNA-replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell. 1986;47:217–227. doi: 10.1016/0092-8674(86)90444-7. [DOI] [PubMed] [Google Scholar]
- 59.Kassavetis G.A., Braun B.R., Nguyen L.H., Geiduschek E.P. S.cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]