Abstract
Recently, a multiple gene cassette with mutagenic translation synthesis activity was identified and shown to be under LexA regulation in several proteobacteria species. In this work, we have traced down instances of this multiple gene cassette across the bacteria domain. Phylogenetic analyses show that this cassette has undergone several reorganizations since its inception in the actinobacteria, and that it has dispersed across the bacterial domain through a combination of vertical inheritance, lateral gene transfer and duplication. In addition, our analyses show that LexA regulation of this multiple gene cassette is persistent in all the phyla in which it has been detected, and suggest that this regulation is prompted by the combined activity of two of its constituent genes: a polymerase V homolog and an alpha subunit of the DNA polymerase III.
INTRODUCTION
Adaptive mutation in bacteria has become a field of increasing interest in the last years. Also termed stationary-phase or stress-induced mutation (1), adaptive mutation concerns the mechanisms by which bacteria increase their mutation rate when placed under non-lethal selective conditions, thereby spontaneously generating mutations that relieve the selective pressure. Adaptive mutation is of special interest in microbiology because it has a broad influence on the mechanisms of evolution, and particularly on those taking place in closed and stressful media such as the hosts of bacterial pathogens. It is therefore of importance to the study of pathogen evolution inside a host, and more specifically to the analysis of antibiotic resistance generation and of bacterial co-evolution with the host immune response.
A major mechanism in adaptive mutation is translesion synthesis (TLS). Mediated by error-prone or lesion bypass polymerases, TLS allows the cell to replicate past a variety of DNA lesions and distortions, thereby promoting survival under endogenous or environmental insults, but also drastically increasing the stationary-phase mutation rate (1). Members of the recently discovered Y family of DNA polymerases, such as Escherichia coli polymerases IV (encoded by the dinB gene) and V (encoded by the umuD and umuC genes), have been shown to be error prone and poorly processive, lying at the core of TLS in most organisms (2). Indeed, Y family polymerases are present in all live domains and have been linked to a range of mutagenic activities: from adaptive mutagenesis in bacteria (3) to genetic instability in human cancer (4).
Well before their true nature was established, both E.coli dinB and umuDC products have already been linked to the SOS response (5). First described in E.coli, the SOS system is a global response mechanism against DNA damage that in E.coli regulates up to 40 genes involved in DNA repair and cell survival (6,7). Induced by single-stranded fragments of DNA generated by either DNA damage-mediated replication inhibition or enzymatic processing of broken DNA ends (8), the SOS response is governed by the LexA and RecA proteins, both of which are also members of this regulatory network. In normal conditions, LexA binds to operator sites of regulated genes and effectively represses their expression. Conversely, in the advent of DNA damage single-stranded DNA (ssDNA) fragments bind and activate the RecA protein, which acquires coprotease activity and promotes the autocatalytic cleavage of LexA (9). Cleaved LexA is unable to bind DNA, leading to the unrepressed expression of SOS genes. Once DNA damage has been addressed, ssDNA concentration falls and both RecA and LexA regain their usual conformation, repressing again the system. The LexA protein is widespread among bacteria and several distinct LexA-binding sites have been reported in different phylogenetic groups. E.coli LexA, for instance, binds a 16 bp consensus sequence (CTGTN8ACAG) (5) that is also found in most gamma and beta proteobacteria (10). Similarly, Gram-positive bacteria LexA has been shown to bind a palindromic motif with consensus GAACN4GTTY (11), which is very similar to that observed in cyanobacteria and green non-sulfur bacteria (12,13). Additional LexA-binding sites have been defined in the alpha proteobacteria (GTTCN7GTTC) (14,15), the Xanthomonadales (TTAN6TACTA) (16), Fibrobacter succinogenes (TGCNCN4GTGCA) (17) and the delta proteobacteria Myxococcus xanthus (CTRHAMRYBYGTTCAGS) (18) and Bdellovibrio bacteriovorus (TTACN3GTAA) (19).
Recently, a multiple gene cassette encoding two error-prone polymerases was described in Pseudomonas putida and homologs of this same cassette were identified in several gamma, beta and alpha proteobacteria species (20). The original gene cassette in P.putida encoded a copy of the LexA repressor (PP3116) recognizing a cyanobacteria-like LexA-binding sequence, a protein annotated in P.putida as SulA (PP3117), a dinB homolog (PP3118) and an alpha subunit of the DNA polymerase III (PP3119). This cassette was shown to be a DNA-damage inducible operon, self-regulated by its own encoded LexA protein, and homologs of this full P.putida gene cassette were identified in the genomes of Pseudomonas syringae, Pseudomonas fluorescens and all the sequenced Xanthomonadaceae (20). In addition, three-gene cassette derivatives lacking the lexA gene were also found in the close Pseudomonas aeruginosa, the beta proteobacterium Ralstonia solanacearum and many alpha proteobacteria species such as Agrobacterium tumefaciens (20). Interestingly, all the reported cassette instances harbored also an adapted LexA-binding site on the promoter region of their first gene, and both DNA-damage inducibility and LexA binding were experimentally confirmed for cassette homologs in A.tumefaciens, P.aeruginosa, Sinorhizobium meliloti and Xanthomonas campestris (20). Given the aforementioned divergence in LexA-binding sequences among these bacterial groups, the persistence of LexA regulation in all theses species hinted at a positive pressure towards LexA control of this multiple gene cassette.
Later work on Caulobacter crescentus (21) confirmed that its three-gene cassette homolog was also a DNA-damage inducible operon, negatively regulated by C.crescentus LexA protein, and named its constituents as imuA, imuB and dnaE2 (PP3117, PP3118 and PP3119 homologs, respectively). Furthermore, experimental assays demonstrated that the imuA-imuB-dnaE2 cassette is responsible for most DNA-damage induced mutations in C.crescentus, and is required for error-prone processing of DNA lesions in this species (21). In addition, phylogenetic analyses revealed that the cassette dnaE2 gene was related to Mycobacterium tuberculosis dnaE2 gene, whose product had been shown previously to mediate SOS mutagenesis in this species. Further experiments confirmed that the imuB and dnaE2 gene products of C.crescentus cooperated in lesion bypass, with a mutational signature different from that observed previously in the E.coli umuDC system. The fact that C.crescentus and all the sequenced alpha proteobacteria, in which the presence of the imuA-imuB-dnaE2 cassette is ubiquitous, lack an umuDC TLS system suggested that this gene cassette might be playing a TLS role in these species. This line of reasoning was strengthened with the identification of split cassette homologues in other species lacking an umuDC system, such as the Planctomycetes Rhodopirellula baltica or the Actinomycetales M.tuberculosis and Propionibacterium acnes, although the first gene of these split cassette instances in the actinobacteria, which we will here term imuA′, could not be shown to be an homolog of its proteobacteria equivalent imuA.
More recent work has also identified a DNA-damage inducible, LexA-regulated imuA-imuB-dnaE2-like cassette in the delta proteobacterium B.bacteriovorus (19), and has shown that its imuA gene, although annotated as and sharing partial homology with recA, does not trans-complement a recA defective E.coli strain. To further elucidate the nature and evolution of this multiple gene cassette, here we have conducted an extensive database search of imuA, imuA′, imuB and dnaE2 homologs across the bacteria domain, and we have made use of robust phylogenetic methods to infer the evolutionary history of this gene cassette, revealing its dispersal through duplications, vertical inheritance and lateral gene transfer (LGT), and its continued regulation by and influence on the SOS system.
MATERIALS AND METHODS
Gene, protein and genomic sequences
Protein and DNA sequences for lexA, imuA′, imuA, imuB and dnaE2 annotated homologs and their respective promoters were downloaded from NCBI GenBank, TIGR Comprehensive Microbial Resource and JGI Integrated Microbial Genomes database resources after identification of homologs by the on-site database textual or BLAST (22) search tools. Additional lexA, imuA′, imuA, imuB and dnaE2 DNA sequences from homologs in unfinished microbial genomes were downloaded from either JGI Integrated Microbial Genomes or TIGR Unfinished Microbial Resource after their identification through these resources own BLAST services, and DNA sequences were translated into their corresponding amino acid sequences with the EditSeq v5.0 program (DNAStar). Complete and incomplete genome assemblies for the organisms harboring imuA′, imuA, imuB and dnaE2 homologs were downloaded also from the aforementioned resources.
Identification of LexA regulatory motifs
LexA regulatory motifs in the promoter of lexA, imuA′, imuA, imuB and dnaE2 genes were searched using RCGScanner v2.1 (10), a consensus-building software for the prediction of regulatory motifs, in those species with known or inferred LexA-binding sequences. Taking advantage of the fact that lexA is a self-regulated gene, undescribed LexA regulatory motifs were identified by first looking for putative dyad motifs in the promoter region of lexA using custom MsWord macro routines to detect palindromic and direct repeat motifs with varying number of mismatches and spacer lengths. Candidate motifs were then sought in the promoter regions of recA and imuA′/imuA or dnaE2 genes with EditSeq's Find function allowing for ambiguity and, if found, the significance of these findings was independently corroborated using the motif discovery tool MEME (23) through its web-based interface at the San Diego Supercomputing Center.
Phylogenetic analyses
Alignments of protein sequences were carried out using a combined procedure to improve alignment quality. Protein sequences were first aligned through CLUSTALW v1.83 (24) using Gonnet matrices and default [10], 25 and 5 gap-opening penalties for the multiple alignment stage, thus generating three different alignments. These three slightly different alignments, together with a local alignment generated by T-COFFEE Lalign method, were integrated as libraries into T-COFFEE v1.37 (25) for optimization. T-COFFEE is an alignment tool for the optimization of parametric consistency in multiple alignments that draws on different alignments to reduce the impact of initial errors in greedy progressive alignment methods. The optimized alignment was then visually inspected with BioEdit v5.0.9 (26) and submitted to Gblocks v0.91b (27) with the half-gaps setting and otherwise default parameters to select conserved positions and discard poorly aligned and phylogenetically unreliable information.
Phylogenetic analyses of the T-COFFEE optimized alignments were carried out using both MrBayes v3.1.1 for Bayesian (28) and PHYML v2.4.1 (29) for maximum-likelihood (ML) inference of tree topologies. In both cases, a mixed four-category gamma distributed rate plus proportion of invariable sites model [invgamma] was applied and its parameters were estimated independently by each program. Four independent MrBayes Metropolis-Coupled Markov Chain Monte Carlo runs were carried out with four independent chains for 106 generations, and 1000 bootstrap replicates were used for ML inference with PHYML. The resulting phylogenetic trees were plotted with TreeView v1.6.6 (30) and enhanced for presentation using CorelDraw Graphic Suite v12 (Corel Corporation).
RESULTS AND DISCUSSION
Identification of additional instances of the imuA-imuB-dnaE2 cassette
Using BLAST and PSI-BLAST searches with the products of each of the cassette genes as queries, homologs of this multiple gene cassette were located in most bacterial phyla, ranging from the actinobacteria to the proteobacteria. Cast in the light of the accepted branching order of bacteria, as established by 16S RNA (31), RecA (32) and protein signature phylogenies (33), these findings reveal either a progressive accretion or fragmentation of the imuA-imuB-dnaE2 cassette in the actinobacteria, punctuated by several genetic reorganizations that gave rise to a range of different cassette configurations.
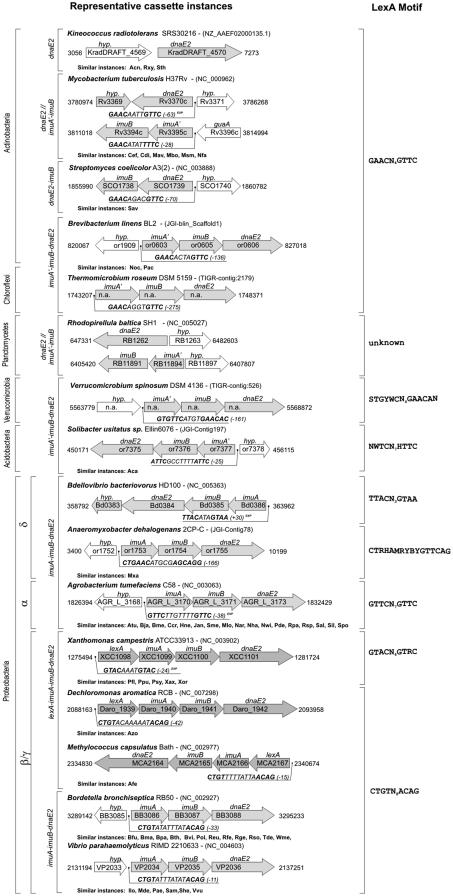
As it can be seen in Figure 1, the simplest instance of an imuA-imuB-dnaE2 cassette constituent can be traced back to some actinobacteria, such as Kineococcus radiotolerans, Symbiobacterium thermophilum or Actinomyces naeslundii, in which only the dnaE2 gene can be found. Close actinobacteria, like the Streptomycineae, show evidence of a two-gene cassette consisting of the dnaE2 and a dinB homolog which we here ascribe to imuB. On the other hand, several actinobacteria present split cassette homologs in which the dnaE2 gene is isolated while two additional genes, the RecA homolog here labeled as imuA′ and imuB, make up a two-gene cassette. This is the case of the previously reported M.tuberculosis, but also of all the sequenced Mycobacteriaceae, Corynebacterineae and of Nocardia farcinica. Finally, instances of an imuA′-imuB-dnaE2 cassette can also be found within the actinobacteria. P.acnes, Nocardioides sp. and Brevibacterium linens all possess a three-gene cassette, and this is very likely the layout of the imuA′-imuB-dnaE2 cassette that emerged from the actinobacteria into subsequent phyla.
Figure 1.
Schematic representation of representative cassette configurations and similar instances found in complete and incomplete genome sequences. All positions are relative to genome/contig start position. n.a. and (BLAST) indicate non-annotated cassette instances of incomplete genomes located through BLAST. EXP denotes experimentally verified LexA-binding motifs. Bacterial name abbreviations for similar instances are as follows: Aca—A.capsulatum, Acn—A.naeslundii, Afe—A.ferrooxidans, Atu—A.tumefaciens, Azo—Azoarcus sp., Bfu—Burkholderia fungorum Bja—Bradyrhizobium japonicum, Bma—Burkholderia mallei, Bme—Brucella melitensis, Bpa—Bordetella parapertussis, Bth—Burkholderia thailandensis, Bvi—Burkholderia vietnamiensis, Ccr—C.crescentus, Cdi—Corynebacterium diphtheriae, Cef—Corynebacterium efficiens, Hne—Hyphomonas neptunium, Ilo—I.loihiensis, Jan—Jannaschia sp., Mav—Mycobacterium avium, Mbo—Mycobacterium bovis, Mde—M.degradans, Mlo—Mesorhizobium loti, Msm—Mycobacterium smegmatis, Mxa—M.xanthus, Nar—Novosphingobium aromaticivorans, Nfa—N.farcinica, Nha—Nitrobacter hamburgensis, Noc—Nocardioides sp., Nwi—Nitrobacter winogradskyi, Pac—P.acnes, Pae—P.aeruginosa, Pde—Paracoccus denitrificans, Pfl—P.fluorescens, Pol—Polaromonas sp., Ppu—P.putida, Psy—P.syringae, Reu—Ralstonia eutropha, Rfe—Rhodoferax ferrireducens, Rge—Rubrivivax gelatinosus, Rpa—Rhodopseudomonas palustris, Rsp—Rhodobacter sphaeroides Rso—R.solanacearum, Rxy—R.xylanophilus, Sal—Sphingopyxis alaskensis, Sam—Shewanella amazonensis, Sav—Streptomyces avermitilis, She—Shewanella sp., Sil—Silicibacter sp., Sme—S.meliloti, Spo—Silicibacter pomeroyi, Sth—S.thermophilum, Tde—T.denitrificans, Vvu—Vibrio vulnificus, Wme—Wautersia metallidurans, Xax—Xanthomonas axonopodis and Xor—Xanthomonas oryzae. Additional information for each cassette instance is provided in Supplementary Table S1, which is included as Supplementary Data.
Leaving the actinobacteria, an imuA′-imuB-dnaE2 like cassette can be detected in several bacterial phyla preceding the proteobacteria, which is all the more remarkable due to the scant representation most of these phyla share in the sequenced microbial genomes databases. An imuA′-imuB-dnaE2 like cassette is found in the Chloroflexi Thermomicrobium roseum, the Planctomycetes R.baltica, the Verrucomicrobia Verrucomicrobium spinosum and the two partially sequenced acidobacteria species (Acidobacterium capsulatum and Solibacter usitatus). In all these instances, the imuA′-imuB-dnaE2 cassette presents the same configuration and apparently reflects a pattern of vertical transmission from a common ancestor with the actinobacteria that ultimately leads to the delta proteobacteria, where imuA-imuB-dnaE2 cassette instances can be identified in B.bacteriovorus and Anaeromyxobacter dehalogenans. Somewhere along this line, however, the imuA′ gene must have undergone drastic changes, involving either extensive mutation or, most probably, a partial or complete substitution by recombination that ultimately led to the imuA gene observed in the proteobacteria. A reliable phylogeny of the imuA′ and imuA genes cannot be reconstructed, since alignments generated with both sequences present almost no conserved positions, backing up the previous suggestion (21) that imuA and imuA′ are not homologs, but could be functional analogs. Moreover, separate phylogenies of imuA and imuA′ become only consistent when restricting the phylogeny to well-defined groups such as the actinobacteria or the proteobacteria (data not shown), making it impossible to precisely define the point at which the aforementioned recombination event took place.
The imuA-imuB-dnaE2 configuration found in the delta proteobacteria is also maintained in the alpha proteobacteria, where cassette instances can be found in all completely sequenced genomes and even in some plasmids (e.g. A.tumefaciens). Similar three-gene cassettes are also found in several gamma and beta proteobacteria species (e.g. Shewanella oneidensis, Vibrio parahaemolyticus), although cassette prevalence is not so regular in these bacterial classes. Neither imuA-imuB-dnaE2 cassettes, nor any of their constituent genes, can be found, for instance, in the Enterobacteriaceae and the Pasteurellaceae, although both are richly sampled families in terms of available genome sequences. In addition, both gamma and beta proteobacteria present several instances of lexA-imuA-imuB-dnaE2 cassettes. These include the previously described four-gene cassettes of P.putida, P.fluorescens, P.syringae and the Xanthomonadaceae (20), together with additional instances in Acidithiobacillus ferrooxidans, Methylococcus capsulatus and the beta proteobacteria Dechloromonas aromatica, Thiobacillus denitrificans and Azoarcus sp.
LexA regulation of imuA-imuB-dnaE2 cassettes
As mentioned above, a striking characteristic of the previously identified imuA-imuB-dnaE2 and lexA-imuA-imuB-dnaE2 cassettes in the alpha, gamma and beta proteobacteria was their persistent regulation by their host (or their own encoded) LexA protein in spite of the drastic differences between the regulatory motifs of these LexA proteins (10,15,20). Together with the existence of lexA-imuA-imuB-dnaE2 cassettes, in which a lexA gene seems to be specifically set to control the adjacent imuA-imuB-dnaE2 genes, and the complete absence of cassette instances in those phyla and classes lacking a lexA gene (e.g. Epsilon Proteobacteria), this suggested that LexA regulation of imuA-imuB-dnaE2-like cassettes ought to be markedly beneficial for their bacterial hosts. Having established that the presence of imuA-imuB-dnaE2 cassettes was not limited to the proteobacteria, and given that a new cassette instance in B.bacteriovorus (19) had also been shown to be regulated by LexA with yet another markedly divergent LexA-binding motif (TTACN3GTAA), we set about to examine LexA regulation in all the here-identified cassette instances.
Interestingly, even though no significant occurrences of the Gram-positive LexA-binding motif (11) can be located upstream of dnaE2 genes in those species (K.radiotolerans, S.thermophilum and A.naeslundii) presenting only this constituent of the imuA′-imuB-dnaE2 cassette, high-scoring and well-placed Gram-positive LexA-binding sites can already be found preceding the dnaE2 gene in the dnaE2-imuB cassettes of the Streptomycineae. As shown in Figure 1, a similar pattern of LexA regulation is conserved also in those actinobacteria presenting split cassette instances, with an imuA′-imuB tandem and an isolated dnaE2 gene. In these cases, of which M.tuberculosis is a well-known representative, apparent LexA regulation does not only concern the isolated dnaE2 gene [which has been experimentally confirmed, (34)], but extends also to the imuA′-imuB tandem. Even though in some cases, like N.farcinica, evidence of LexA regulation can be found only in the imuA′-imuB tandem, the systematic regulation of at least one of the cassette components in all these species suggests that it is their concerted activity that creates a positive pressure towards LexA regulation. In this context, it should be stressed that a combined activity of the imuB and dnaE2 genes of C.crescentus has already been singled out as the source of most TLS mutagenic activity in this species (21), and that unregulated TLS activity mediated by dinB has been shown previously to yield mutator phenotypes in E.coli (35), resulting in lowered adaptative fitness.
As expected, LexA regulation is maintained in most actinobacteria presenting full imuA′-imuB-dnaE2 cassettes. The sole exception concerns Rubrobacter xylanophilus, but this bacterium does not present a Gram-positive LexA-binding motif upstream of its lexA gene and its lexA gene sequence shows substantial divergence from that of other actinobacteria, suggesting that it may have evolved separately to recognize a new LexA-binding motif. Outside the actinobacteria, a Gram-positive LexA-binding motif can be identified upstream of both T.roseum lexA and imuA′ genes, suggesting that, in addition to the Dehalococcoidetes (13), this is also the LexA-binding motif of the Thermomicrobiales. Interestingly, a derivative of the Gram-positive LexA-binding motif with reminiscences of the one seen in E.coli (STGYWCAWNYGAACAN) can also be found upstream of V.spinosum lexA, recA and imuA′ genes, suggesting that this is the LexA-binding sequence in this species and that the imuA′-imuB-dnaE2 cassette is regulated by V.spinosum LexA. A similar situation is found also in the acidobacteria, where NWTCN7HTTC direct repeat motifs (close to the one associated with the alpha proteobacteria) (15) can be located upstream of the lexA and imuA′ genes in both species. This is all the more remarkable given that A.capsulatum presents two instances of the imuA′-imuB-dnaE2, and the aforementioned LexA-binding motif is present upstream of both imuA′ genes.
LexA regulation is also preserved in the delta proteobacteria. Besides the imuA-imuB-dnaE2 cassette already shown to be LexA regulated and DNA damage inducible in B.bacteriovorus (19), derivatives of the M.xanthus LexA-binding sequence (18) can also be found upstream of A.dehalogenans lexA, recA and imuA genes. Interestingly, in M.xanthus, where only the dnaE2 gene can be found, there is no evidence of a LexA-binding sequence upstream of it, suggesting again that it is the combined presence of imuB and dnaE2 in a bacterial genome that prompts LexA regulation. As mentioned earlier, the presence of imuA-imuB-dnaE2 cassettes among the alpha proteobacteria is pervasive, with some species harboring up to three copies of this cassette (e.g. A.tumefaciens) and frequent evidence of plasmid dissemination. Consistent with this widespread distribution, LexA regulation and DNA-damage induction of the imuA-imuB-dnaE2 cassettes have been reported in A.tumefaciens, C.crescentus and S.meliloti (20,21), and the corresponding LexA-binding motifs have been found upstream of imuA in other alpha proteobacteria species (15).
Concerning the gamma and beta proteobacteria, LexA regulation has been reported (10) or is apparent in all the identified instances of the imuA-imuB-dnaE2 cassette. It is interesting to note, however, that the previously reported (20) LexA-binding sequence of lexA-imuA-imuB-dnaE2 cassettes (GTACN4GTGC) is not present in all the here-identified four-gene cassette instances. A.ferrooxidans, T.denitrificans, D.aromatica and Azoarcus all present an E.coli-like LexA-binding sequence upstream of their cassette lexA gene which, in contrast to the Pseudomonadaceae and the Xanthomonadaceae, is the sole lexA gene in their genome.
Reorganization and lateral gene transfer of the imuA-imuB-dnaE2 cassette
Even though the overall distribution of the imuA-imuB-dnaE2 cassette is in approximate agreement with the established phylogeny of bacteria, its broad dispersal is punctuated by noticeable absences (e.g. cyanobacteria, epsilon proteobacteria) and seems to be intimately linked to the LexA network, as evidenced by its persistent regulation, the presence of four-gene cassettes including a lexA gene and the absence of cassettes in all classes and phyla lacking a lexA gene. Moreover, the identification of several imuA-imuB-dnaE2 cassette duplication events (e.g. A.capsulatum, A.tumefaciens) and its recurring existence in plasmids (e.g. R.solanacearum, S.meliloti) had suggested previously that the evolution of this cassette might have also been shaped by other factors, such as LGT. Therefore, to ascertain whether there existed relevant discrepancies in the particular phylogeny of the imuA-imuB-dnaE2 cassette, a rigorous phylogenetic analysis of this multiple gene cassette was conducted. Owing to the aforementioned lack of homology between imuA and imuA′, these sequences were discarded for the analysis. Similarly, optimized alignments of imuB sequences following our astringent protocols yielded too few conserved positions (below 30) for a sound phylogenetic analysis over the whole bacteria domain. Therefore, and given that imuB genes were also absent in some of the identified cassette instances (e.g. K.radiotolerans), the phylogenetic analysis was carried out using the dnaE2 gene product sequence, which was both the most phylogenetically informative of the cassette gene sequences (623 conserved positions) and also the only one providing full coverage among the identified cassette instances. Furthermore, to validate and reinforce the phylogenetic approach, the analysis was also conducted on the dnaE1 gene product sequence in all the species studied. The dnaE1 gene, corresponding to the normal (non-cassette) alpha subunit of the DNA polymerase III, is similar in length to the dnaE2 gene and ought to be subjected to similar structural constraints, thus providing a reliable contrast to the evolution of the dnaE2 gene and facilitating the identification of phylogenetic artifacts, such as those due to fast-clocked evolution.
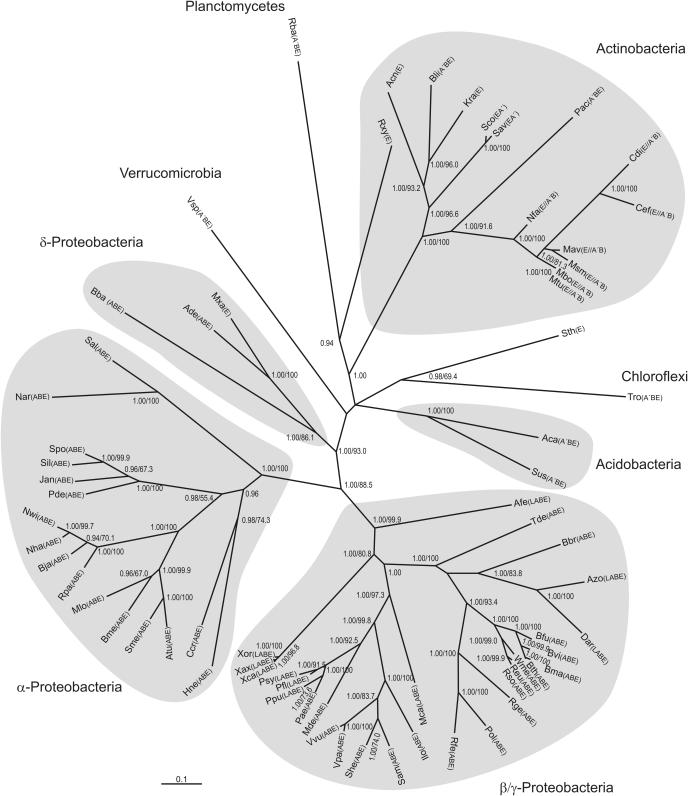
As it can be seen in Figure 2, the DnaE1 sequence produces a tree that is in broad agreement with the established phylogenies (31–33). A natural cluster groups all the actinobacteria except those (R.xylanophilus and S.thermophilum) that have already been suggested to be part of to-date undefined groups of actinobacteria (36–37). Both these species are positioned at the exit of the actinobacteria clade, intermingling with the intermediate phyla (Planctomycetes, green non-sulfur bacteria, acidobacteria and Verrucomicrobia) that then lead to the proteobacteria, in which the natural branching order (delta, alpha, beta and gamma) can be observed. In agreement with previous phylogenies (38,39), the Xanthomonadaceae branch earlier than gamma and beta proteobacteria, suggesting that they constitute a separate group of early gamma/beta proteobacteria.
Figure 2.
Unrooted DnaE1 sequence tree. Shaded areas correspond to established phylogenetic groups. Branch support values below 0.85 or 50% for Bayesian or ML inference phylogenetic analyses, respectively, are not shown. Cassette configurations are shown using constituent genes abbreviations: L—lexA, A′—imuA′, A—imuA, B—imuB, E—dnaE2. Name abbreviations not used in Figure 1 are as follows: Bba—B.bacteriovorus, Bbr—Bordetella bronchiseptica, Dar—D.aromatica, Kra—K.radiotolerans, Mca—M.capsulatus, Sco—Streptomyces coelicolor, Sus—S.usitatus sp., Tro—T.roseum, Vpa—V.parahaemolyticus and Vsp—V.spinosum. Additional information for each dnaE1 locus is provided in Supplementary Table S1, which is included as Supplementary Data.
A similar story is told by the DnaE2 sequence tree up to the branching point of beta and gamma proteobacteria (Figure 3). A main cluster encompasses most of the actinobacteria, displaying a branching order in which an imuA′-imuB-dnaE2 cassette configuration leads to split cassette instances and thus suggesting that the later originated following a genetic reorganization of the former. In this context, the persistent regulation of one or both members of the split cassettes suggests again a positive pressure towards LexA regulation. A separate group with long-branch lengths takes in those species purporting only a dnaE2 gene (S.thermophilum, R.xylanophilus, K.radiotolerans and T.roseum). The Streptomycetaceae, which display a unique dnaE2-imuB cassette configuration, also fall in this last group. Even though LGT events could be invoked to explain this clustering, its origin seems to be a long-branch attraction artifact due to the substantial divergence in these sequences, a fact that would be in agreement with a period of extensive genetic reorganization that led from three-gene imuA′-imuB-dnaE2 cassettes to the isolated dnaE2 and tandem dnaE2-imuB configurations seen in these species.
Figure 3.
Unrooted DnaE2 sequence tree. Shaded areas correspond to established phylogenetic groups. Branch support values below 0.85 or 50% for Bayesian or ML inference phylogenetic analyses, respectively, are not shown. Cassette configurations are shown using constituent genes abbreviations: L—lexA, A′—imuA′, A—imuA, B—imuB, E—dnaE2. P indicates that the cassette is located in a plasmid. Name abbreviations are as in Figure 2. Additional information for each dnaE2 locus is provided in Supplementary Table S1, which is included as Supplementary Data.
In contrast, the consistent placement of the Planctomycetes R.baltica deep within the proteobacteria cluster is in clear disagreement with both the established and the DnaE1 phylogenies. This, together with the fact that R.baltica possesses a split cassette with imuA and imuB genes (RB11894 and RB11891) that present high identity with P.putida imuA and imuB (instead of actinobacteria imuA′ and imuB), strongly suggests that R.baltica acquired its imuA-imuB-dnaE2 cassette through LGT from an ancestor of gamma and beta proteobacteria and later reorganized it into a split cassette with an imuA-imuB tandem and an isolated dnaE2 gene. In this respect, it should be noted that the clustering together of all imuA-imuB-dnaE2 cassette instances that are located in plasmids in the alpha proteobacteria strongly suggest that there has been extensive plasmid dissemination of the imuA-imuB-dnaE2 cassette in this bacterial class. In the light of this, and given the close placement of R.baltica in the DnaE2 tree (between alpha and gamma/beta proteobacteria), plasmid dissemination could certainly be the mechanism for the observed LGT event.
Duplication of the lexA-imuA-imuB-dnaE2 cassette
Another major source of disagreement between the DnaE2 and DnaE1 trees concerns the branching point of gamma and beta proteobacteria. While the DnaE1 tree supports a view that is in close agreement with the established phylogenies, the DnaE2 tree generates three separate clusters with low support values. Two of these clusters correspond, roughly, to the beta and gamma proteobacteria classes, while the third groups together nearly all instances of the lexA-imuA-imuB-dnaE2 cassette instances, intermingling beta and gamma proteobacteria species. Owing to the limited resolution of the DnaE2 tree at this level, it is difficult to infer an accurate account on the evolutionary history of the cassette at this point. LGT of a primordial four-gene cassette could certainly be invoked to explain the observed data, but repeated LGT events and ad hoc deletions would be still required in the most plausible LGT scenario, which cannot be supported solely on the available phylogenetic data. In contrast, several facts point to an ancestral duplication of the lexA-imuA-imuB-dnaE2 cassette, which may have then led through vertical inheritance and several deletions to the observed distribution of three- and four-gene cassettes among gamma proteobacteria. The consistent clustering of Xanthomonadaceae and Pseudomonadaceae, for instance, is reminiscent of a vertical inheritance pattern with missing intermediate gamma proteobacteria groups. Moreover, the fact that it is only in these two families that the lexA gene of the lexA-imuA-imuB-dnaE2 cassette presents a markedly divergent LexA-binding motif (GTACN4GTGC) not found in the other four-gene cassettes strongly suggests a common origin. Interestingly, lexA duplication has already been suggested in the Xanthomonadaceae (39), thus giving further support to the duplication hypothesis. In fact, a phylogeny of the LexA protein (data not shown) also clusters together both Xanthomonadaceae LexA proteins and one of the Pseudomonadaceae LexA proteins in a separate branch, while the remaining Pseudomonadaceae LexA protein clusters naturally with Microbulbifer degradans in the gamma proteobacteria cluster, thus suggesting again an ancestral duplication of lexA, and its attached cassette, at the root of the gamma and beta proteobacteria.
Taken together, thus, the results from the DnaE1, DnaE2 and LexA sequence trees support a scenario involving the duplication of the full lexA-imuA-imuB-dnaE2 cassette. The proposed scenario, outlined in Figure 4, places the formation of the original lexA-imuA-imuB-dnaE2 cassette in an ancestor of the gamma and beta proteobacteria, following a genomic reorganization that brought together the extant imuA-imuB-dnaE2 cassette and lexA gene already seen in earlier proteobacteria. Drawing from the available data in the DnaE1 tree, which places most lexA-imuA-imuB-dnaE2 cassette harboring species at the root of gamma and beta proteobacteria, this original lexA gene had already evolved or was on the way to evolve recognition of an E.coli-like motif. Shortly after, or during the own reorganization event, this lexA-imuA-imuB-dnaE2 cassette underwent a duplication, resulting in two copies of this four-gene cassette that for the sake of clarity we will here label as lexA1-imuA1-imuB1-dnaE21 and lexA2-imuA2-imuB2-dnaE22. Following duplication, the two cassette copies started to diverge, leading in their lexA gene to the full emergence of distinct LexA-binding motifs: the conventional E.coli one (CTGTN8ACAG) in lexA1-imuA1-imuB1-dnaE21 cassettes and that observed in the lexA2-imuA2-imuB2-dnaE22 cassette instances of the Xanthomonadaceae and the Pseudomonadaceae (GTACN4GTGC). Further evolution in the Xanthomonadaceae would have then led to their own lexA1 binding sequence [TTAN6TACTA, (16)], following a deletion of the rest of the lexA1-imuA1-imuB1-dnaE21 cassette. Based on the DnaE1 tree, it seems clear that a major deletion of the lexA2-imuA2-imuB2-dnaE22 cassette took place after the split between the Pseudomonadaceae and the rest of gamma proteobacteria, since there is no evidence of either the lexA2 gene or the rest of its accompanying cassette in the Vibrionaceae, the Pasteurellaceae, the Alteromonadaceae or the Enterobacteriaceae. A similar deletion must have taken place early in the evolutionary history of the beta proteobacteria, since there is again no evidence of any component of the lexA2-imuA2-imuB2-dnaE22 cassette in this bacterial subclass.
Figure 4.
Reconstruction of the proposed duplication hypothesis on a DnaE1 sequence tree of the gamma and beta proteobacteria classes, rooted using A.tumefaciens DnaE1 (Atu) as an out-group. Branch support values below 0.85 or 50% for Bayesian or ML inference phylogenetic analyses, respectively, are not shown. Cassette configurations and instances are shown on the right. Deletion and reorganization events are indicated by different symbols (see bottom left of Figure 4 for symbol explanation). Name abbreviations that are not used in Figure 2 are as follows: Eca—Erwinia carotovora, Hdu—Haemophilus ducreyi, Hin—Haemophilus influenzae, Msu—Mannheimia succiniciproducens, Plu—Photorhabdus luminescens laumondii, Pmu—Pasteurella multocida, Stm—Salmonella enterica serovar Typhimurium, Vvu—Vibrio vulnificus, Xfa—Xylella fastidiosa and Ype—Yersinia pestis. Additional information for each dnaE1 locus and cassette configurations is provided in Supplementary Table S1, which is included as Supplementary Data.
Reorganization of duplicated lexA-imuA-imuB-dnaE2 cassettes in the gamma and beta proteobacteria
The duplication scenario outlined in Figure 4 raises some interesting points. On the one hand, a repeated feature in both gamma and beta proteobacteria concerns the genetic reorganization of the lexA1-imuA1-imuB1-dnaE21 cassette, involving the excision of the lexA1 gene from the cassette. This seems to have occurred early in the evolutionary history of the beta proteobacteria, but it must have also taken place independently in the gamma proteobacteria and the Xanthomonadaceae. Such a recurring pattern of convergent evolution points to some form of positive pressure towards the split of the four-gene cassette, but it is difficult to speculate on its nature. An appealing possibility concerns the need for more efficient regulation of the constituents of the imuA1-imuB1-dnaE21 cassette. Even though operon organization is a straight and effective means of regulation, the inclusion in the operon of its own governing gene (lexA1) reduces the ability to fine-tune the expression of downstream genes. It can be thus speculated that the observed convergent split of the lexA1-imuA1-imuB1-dnaE21 may be due to a requirement to improve repression of the mutagenic imuA1-imuB1-dnaE21 cassette in normal conditions, or to boost its expression under adverse circumstances, without influencing negatively on the behavior of the whole LexA regulon. In this context, it is interesting to note a remarkable coincidence. The split of the lexA1-imuA1-imuB1-dnaE21 in the gamma proteobacteria is concurrent with the first instance of a sulA gene in this bacterial class (Figure 4). The sulA gene (40), encoding a cell-division inhibitor and under mandatory regulation by LexA in E.coli and Salmonella typhimurium, is absent from all other bacterial classes and phyla. Even though sulA is not directly associated with lexA in these two species, its first detectable homologs in Idiomarina loihiensis, the Shewanellaceae or the Pseudomonadaceae are in a lexA-sulA tandem operon configuration (20) and its transition into the E.coli configuration can still be traced through the analysis of its genomic surroundings in the Vibrionaceae (data not shown). It is thus reasonable to conjecture that the entrance of sulA in the gamma proteobacteria may have played a role in the split of the lexA1-imuA1-imuB1-dnaE21 cassette, especially since the imuA1 gene shares a degree of homology with E.coli sulA which has been found significant enough to incorrectly annotate this gene as sulA in some bacterial genomes (e.g. P.putida). This line of reasoning is further supported by what can be observed in the Xanthomonadaceae, where the split or partial deletion of the lexA1-imuA1-imuB1-dnaE21 gave way to a unique genetic reorganization involving lexA and recA, another gene sharing residual homology with imuA1, leading to the lexA1-recA-recX operon observed in this family (16).
Some other questions stemming from this scenario are also of substantial interest. For instance, the complete and final loss of the split imuA1-imuB1-dnaE21 cassette in the Enterobacteriaceae and other families (such as the Pasteurellaceae) calls for some explanation, as it appears to have occurred independently. Even though more data are required to shed more light on the matter, it seems relevant to note that the complete loss of the imuA1-imuB1-dnaE21 is strongly, though not univocally, linked to the emergence and prevalence of an umuDC operon in these families (10), which may have offered alternative means for adaptive mutagenesis, thus rendering the imuA1-imuB1-dnaE21 functionally redundant.
To summarize, the extensive analysis of the evolutionary history of the imuA-imuB-dnaE2 undertaken in this work has led to the identification of LGT and duplication events along its evolution, and has substantiated the claim that its mutagenic activity arises from a concerted activity of imuB and dnaE genes that calls for explicit LexA regulation, in spite of the marked changes in the regulatory sequence of this protein and some drastic turns in the evolutionary history of the imuA-imuB-dnaE2 cassette. Nevertheless, the scenarios suggested to explain the evolution of the imuA-imuB-dnaE2 cassette do also leave alluring open questions, such as the ultimate origin of the initial actinobacteria imuA′-imuB-dnaE2 cassette, or the influence of two cohabiting lexA copies in the evolution of the SOS regulatory network of gamma proteobacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are deeply grateful to Dr José Castresana for his enlightening discussions of bacterial phylogeny and for his critical reviewing of this manuscript, and to the reviewers of this paper for their insightful contributions. This work was funded by Grant BFM2004-02768/BMC from the Ministerio de Educación y Ciencia (MEC) de España and by the Consejo Superior de Investigaciones Científicas (CSIC). Dra. S. Campoy is recipient of a post-doctoral contract from INIA-IRTA. Funding to pay the Open Access publication charges for this article was provided by Grant BFM2004-02768/BMC from the Ministerio de Educación y Ciencia (MEC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kivisaar M. Stationary phase mutagenesis: mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ. Microbiol. 2003;5:814–827. doi: 10.1046/j.1462-2920.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- 2.Napolitano R., Janel-Bintz R., Wagner J., Fuchs R.P. All three SOS-inducible DNA polymerases (PolII, PolIV and PolV) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Layton J.C., Foster P.L. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bavoux C., Hoffmann J.S., Cazaux C. Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase kappa. Biochimie. 2005;87:637–646. doi: 10.1016/j.biochi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Walker G.C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández de Henestrosa A.R., Ogi T., Aoyagi S., Chafin D., Hayes J.J., Ohmori H., Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 7.Courcelle J., Khodursky A., Peter B., Brown P.O., Hanawalt P.C. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sassanfar M., Roberts J.W. Nature of SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 9.Little J.W. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 10.Erill I., Escribano M., Campoy S., Barbé J. In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon. Bioinformatics. 2003;19:2225–2236. doi: 10.1093/bioinformatics/btg303. [DOI] [PubMed] [Google Scholar]
- 11.Winterling K.W., Chafin D., Hayes J.J., Sun J., Levine A.S., Yasbin R.E., Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazón G., Lucena J.M., Campoy S., Fernández de Henestrosa A.R., Candau P., Barbé J. LexA-binding sequences in Gram-positive and cyanobacteria are closely related. Mol. Genet. Genomics. 2003;271:40–49. doi: 10.1007/s00438-003-0952-x. [DOI] [PubMed] [Google Scholar]
- 13.Fernández de Henestrosa A.R., Cuñé J., Erill I., Magnuson J.K., Barbé J. A green nonsulfur bacterium, Dehalococcoides ethenogenes, with the LexA binding sequence found in gram-positive organisms. J. Bacteriol. 2002;184:6073–6080. doi: 10.1128/JB.184.21.6073-6080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández de Henestrosa A.R., Rivera E., Tapias A., Barbé J. Identification of the Rhodobacter sphaeroides SOS box. Mol. Microbiol. 1998;28:991–1003. doi: 10.1046/j.1365-2958.1998.00860.x. [DOI] [PubMed] [Google Scholar]
- 15.Erill I., Jara M., Salvador N., Escribano M., Campoy S., Barbé J. Differences in LexA regulon structure among proteobacteria through in vivo assisted comparative genomics. Nucleic Acids Res. 2004;32:6617–6626. doi: 10.1093/nar/gkh996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campoy S., Mazón G., Fernández de Henestrosa A.R., Llagostera M., Brant-Monteiro P., Barbé J. A new regulatory DNA motif of the gamma subclass proteobacteria: identification of the LexA protein binding site of the plant pathogen Xylella fastidiosa. Microbiology. 2002;148:3583–3597. doi: 10.1099/00221287-148-11-3583. [DOI] [PubMed] [Google Scholar]
- 17.Mazón G., Erill I., Campoy S., Cortes P., Forano E., Barbe J. Reconstruction of the evolutionary history of the LexA-binding sequence. Microbiology. 2004;150:3783–3795. doi: 10.1099/mic.0.27315-0. [DOI] [PubMed] [Google Scholar]
- 18.Campoy S., Fontes M., Padmanabhan S., Cortes P., Llagostera M., Barbé J. LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus. Mol. Microbiol. 2003;49:769–781. doi: 10.1046/j.1365-2958.2003.03592.x. [DOI] [PubMed] [Google Scholar]
- 19.Campoy S., Salvador N., Cortes P., Erill I., Barbe J. Expression of canonical SOS genes is not under LexA repression in Bdellovibrio bacteriovorus. J. Bacteriol. 2005;187:5367–5375. doi: 10.1128/JB.187.15.5367-5375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abella M., Erill I., Jara M., Mazón G., Campoy S., Barbé J. Widespread distribution of a lexA-regulated DNA damage-inducible multiple gene cassette in the proteobacteria phylum. Mol. Microbiol. 2004;54:212–222. doi: 10.1111/j.1365-2958.2004.04260.x. [DOI] [PubMed] [Google Scholar]
- 21.Galhardo R.S., Rocha R.P., Marques M.V., Menck C.F.M. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005;33:2603–2614. doi: 10.1093/nar/gki551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 24.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notredame C., Higgins D., Heringa J. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 26.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 27.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist F., Huelsenbeck J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Page R.D. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 31.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domains: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisen J.A. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J. Mol. Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta R.S. The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 2000;26:111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- 34.Boshoff H.I., Reed M.B., Barry C.E., III, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., Sofuni T., Nohmi T., Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl Acad. Sci. USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kausar J., Ohyama Y., Terato H., Ide H., Yamamoto O. 16S rRNA gene sequence of Rubrobacter radiotolerans and its phylogenetic alignment with members of the genus Arthrobacter, gram-positive bacteria, and members of the family Deinococcaceae. Int. J. Syst. Bacteriol. 1997;47:684–686. doi: 10.1099/00207713-47-3-684. [DOI] [PubMed] [Google Scholar]
- 37.Ueda K., Yamashita A., Ishikawa J., Shimada M., Watsuji T.O., Morimura K., Ikeda H., Hattori M., Beppu T. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 2004;32:4937–4944. doi: 10.1093/nar/gkh830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig W., Klenk H.P. Overview: a phylogenetic backbone and taxonomic frame work for prokaryotic systematics. In: Boone D.R., Castelholtz R.W., Garrity G.M., editors. Bergey's Manual of Systematic Bacteriology, 2nd edn. NY: Springer-Verlag; 2001. pp. 49–65. [Google Scholar]
- 39.Martins-Pinheiro M., Galhardo R.S., Lage C., Lima-Bessa K.M., Aires K.A., Menck C.F.M. Different patterns of evolution for duplicated DNA repair genes in bacteria of the Xanthomonadales group. BMC Evol. Biol. 2004;4:29–40. doi: 10.1186/1471-2148-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi E., Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.