Abstract
We carried out in vitro selection experiments to systematically probe the effects of TATA-box flanking sequences on its interaction with the TATA-box binding protein (TBP). This study validates our previous hypothesis that the effect of the flanking sequences on TBP/TATA-box interactions is much more significant when the TATA box has a context-dependent DNA structure. Several interesting observations, with implications for protein–DNA interactions in general, came out of this study. (i) Selected sequences are selection-method specific and TATA-box dependent. (ii) The variability in binding stability as a function of the flanking sequences for (T-A)4 boxes is as large as the variability in binding stability as a function of the core TATA box itself. Thus, for (T-A)4 boxes the flanking sequences completely dominate and determine the binding interaction. (iii) Binding stabilities of all but one of the individual selected sequences of the (T-A)4form is significantly higher than that of their mononucleotide-based consensus sequence. (iv) Even though the (T-A)4 sequence is symmetric the flanking sequence pattern is asymmetric. We propose that the plasticity of (T-A)n sequences increases the number of conformationally distinct TATA boxes without the need to extent the TBP contact region beyond the eight-base-pair long TATA box.
INTRODUCTION
Protein–DNA interactions are at the heart of many cellular processes, including DNA replication, transcription, recombination and DNA packaging within the nucleosome. It is now clearly evident that there is more to protein–DNA interactions than simple readout of hydrogen bond donor and acceptor groups (‘direct readout’). In the current picture of protein–DNA interactions, sequence-dependent conformation and the ability to change it at low energetic cost (‘deformability’) can contribute extensively to sequence-specific recognition by regulatory proteins (‘indirect readout’). This latter type, also called structural recognition, is usually a subsidiary mechanism to direct sequence recognition. However, sequence-specific regulatory proteins that bind the DNA double helix through the minor groove must operate mainly by indirect readout since the symmetric positioning of donor and acceptor groups in this groove makes it difficult to differentiate an A.T base pair from an T.A one [and likewise for G.C and C.G base pairs (1)]. The binding of the TATA-binding protein (TBP) to its target sites, ‘TATA boxes’, is an exemplar of structure and flexibility-based specificity mechanism, since the binding is through the minor groove (2,3). Moreover, the TATA box consensus sequence is T-A-T-A-A/T-A-A/T-A/G (4) (http://www.epd.isb-sib.ch/promoter_elements). Thus, it is composed mainly of different combinations of T.A and A.T base pairs.
TBP is a universal transcription factor, required for initiation of transcription by all three eukaryotic RNA polymerases (5–7). Gene expression by RNA polymerase II (Pol II) is regulated mainly at the stage of the initiation of transcription. The formation of the TBP/TATA-box complex is the first step in the assembly of a set of transcription factors necessary for the initiation of transcription of genes that are transcribed by Pol II (8,9). The other transcription factors (except for TFIIB) do not contact the DNA in a sequence-specific manner (8,10). Thus, sequence-specific TBP/TATA-box interactions are central for the regulation of gene transcription.
We have previously looked at the signals for TBP/TATA-box interaction (11). In this study, we observed a novel mechanism of indirect readout, in which there is a differential effect of the identity of the DNA sequence in the region flanking the core 8 bp TATA box on the kinetics of dissociation of the TBP/TATA-box complex (11). Changing the sequences that flank the core TATA box did not change the stability of the complex formed with the major-late promoter (MLP) TATA box (TATAAAAG, compare wtMLP with fsMLP in Table 1). However, the half-life of the complex of TBP with the TATA-box variant TATATATG was significantly increased when the flanking sequences were changed from G-tracts to alternating (G-C) tracts (compare T5T7 with fsT5T7 in Table 1) (11). Similar observations on the effects of the TATA flanking sequences on TBP binding were made by Wolner and Gralla (12). We have previously suggested (11) that the MLP TATA box is not affected by changes in the adjoining sequences because of the dominant and invariable character of homopolymeric A-tract sequences (defined as An, n ≥ 4) (13), which are not easily changed by the nature of the sequences adjacent to them (14). (A-T)n runs, on the other hand, are polymorphic in structure (15), and their conformation is dependent on sequence context and crystal packing forces (16).
Table 1.
Selected TBP/TATA-box complexes studied individually
| Name | Sequence | Half-life Ba,b (min) | ‘B’ fraction | ‘A’ fraction | Reference |
|---|---|---|---|---|---|
| Dissociation kinetics | |||||
| MLPk93 | CCTCGGTATAAAAGGGCGCT | 249 (±23) | 0.81 (±5) | 0.19 (±5) | This work |
| MLPk62 | TTGGCGTATAAAAGCGCGCG | 213 (±4) | 0.79 (±5) | 0.21 (±5) | This work |
| MLPk52 | TGACGGTATAAAAGTGCCTA | 212 (±7) | 0.73 (±3) | 0.27 (±3) | This work |
| MLPk88 | TTCGTCTATAAAAGGGCGTG | 206 (±17) | 0.70 (±4) | 0.30 (±4) | This work |
| MLPkconc | TTTGGCGTATAAAAGTTTAGG | 232 (±12) | 0.75 (±4) | 0.25 (±4) | This work |
| wt MLP | CGGGCTATAAAAGGGGGTGG | 255 (±24) | 0.83 (±3) | 0.17 (±3) | d |
| fsMLPe | CGGACTATAAAAGCGCGTGC | 271 (±20) | 0.86 (±2) | 0.14 (±2) | d |
| E4k28 | TCCTAGTATATATACTGAGT | 333 (±14) | 0.93 (±2) | 0.07 (±2) | This work |
| E4k60 | TTGGGGTATATATAGTGTGG | 325 (±12) | 0.89 (±2) | 0.11 (±2) | This work |
| E4k56 | GGGTCTTATATATAGGGCGT | 254 (±11) | 0.89 (±2) | 0.11 (±2) | This work |
| E4k30 | TAGCGCTATATATATGGTCT | 243 (±6) | 0.89 (±3) | 0.11 (±3) | This work |
| E4k67 | GGTCGATATATATACGCCGT | 197 (±3) | 0.86 (±1) | 0.14 (±1) | This work |
| E4k55 | GGAAGCTATATATACACCCC | 192 (±27) | 0.76 (±4) | 0.24 (±4) | This work |
| E4k53 | TGAACCTATATATACGCAGC | 175 (±8) | 0.78 (±5) | 0.22 (±5) | This work |
| E4k36 | TGGTGCTATATATAGACTGG | 147 (±5) | 0.83 (±3) | 0.17 (±3) | This work |
| E4kcon mono | GGGGCTATATATACGGGGGGGG | 145 (±5) | 0.90 (±3) | 0.10 (±3) | This work |
| E4kcon high | CCCGCTATATATACGCGGGG | 182 (±8) | 0.88 (±1) | 0.12 (±1) | This work |
| (TA)4 | CGGGCTATATATAGGGGTGG | 163 (±6) | 0.74 (±4) | 0.26 (±4) | d |
| fs(TA)4e | CGGACTATATATACGCGTGC | 157 (±5) | 0.81 (±5) | 0.19 (±5) | This work |
| wt E4 | AGTCCTATATATACTCGCTC | 70 (±4) | 0.81 (±5) | 0.19 (±5) | This work |
| T5T7 | CGGGCTATATATGGGGGTGG | 78 (±6) | 0.87 (±2) | 0.13 (±2) | d |
| fsT5T7e | CGGACTATATATGCGCGTGC | 155 (±5) | 0.74 (±2) | 0.26 (±2) | d |
| Binding affinity | Kd (nM)b | ||||
| E4t10 | CCCTGCTATATATACCCTGG | 2.4 (±0.3) | This work | ||
| E4t16 | AGCCGCTATATATAACGGCA | 3.5 (±0.4) | This work | ||
| E4t45 | CCACCCTATATATAGGCTTG | 4.1 (±0.7) | This work | ||
| E4t6 | GTCCGATATATATATCACGC | 7.5 (±0.4) | This work | ||
| E4tconc | TCCGTTATATATAGGTTGGC | 3.3 (±0.3) | This work | ||
| wtE4 | AGTCCTATATATACTCGCTC | 19 (±2) | This work | ||
aEquation used is . A and B are the fraction of molecules dissociating with macroscopic rate constants k1 and k2, respectively. The half-life was determined from t1/2B = ln2/k2.
bNumbers in parentheses are the standard error of the mean. It includes the experimental error between the different independent experiments (5–9 experiments for each sequence) and the difference between the experimental points and the curve-fitting model.
cThese consensus sequences are based on linked higher-order sequence motifs, but also agree with a mononucleotide-based consensus sequence (see text for details).
dExperiments are from (11), reanalyzed here by the equation above.
efs stands for ‘flanking sequences’, and is the name given to these sequences by (11), where only one flanking-sequence variant was analyzed for each TATA box.
TATA flanking sequences can also affect the binding affinity of TBP to TATA boxes. Librizzi et al. (17) observed that changes in the sequence flanking the MLP TATA box slightly decrease the equilibrium association constant relative to that observed in the natural sequence context of the MLP TATA box. Librizzi et al. (17) attributed this observation to a difference in the rate of association of TBP to these TATA boxes. Wolner and Gralla (18) showed that sequences flanking the core TATA box influence the level of basal transcription as well as the response to activators. They have used two prototype adenovirus TATA boxes, MLP and E4. The first is a strong basal promoter and is only weakly responsive to activators, whereas E4 promoter is a weak basal promoter, but is highly responsive to protein activators [discussed in ref. (18)]. By swapping the blocks of sequences surrounding these two TATA boxes they showed that substituting the E4 flanking sequences into the MLP increased its response to activators, whereas substituting the MLP sequences into the E4 promoter had the opposite effect (18).
We have probed systematically the role of the sequences flanking the MLP and E4 TATA boxes, as prototype TATA boxes having A4 versus (T-A)4 tracts, respectively, on TBP/TATA-box interactions using in vitro evolution methods. We searched for sequences with optimal binding affinities (low equilibrium dissociation constant), as well as for those with optimal binding stabilities (low kinetic off rate). We show that not only do TATA flanking sequences influence the binding of TBP to the E4 TATA box to a much larger extent than their effect on the TBP/MLP TATA-box interaction, but the variability observed in TBP/E4 binding as a function of the flanking sequences is as large as that observed when the core TATA box itself is changed. Moreover, we show that the influence of the flanking sequences on TBP/E4 interaction is asymmetric and directional. Thus, one abutting side is more conserved than the other side. We ascribe this behavior to the multi-step binding mechanism of TBP to TATA boxes. We suggest here that structural modulation of certain TATA-boxes by their flanking sequences increases the number of different sequences that constitute a valid TATA box. Since TBP/TATA-box interaction is the first step in the assembly of the preinitiation complex, this enhances the fine-tuning of gene regulation attainable at this initial stage of transcription.
MATERIALS AND METHODS
DNA
TATA-box variants were chemically synthesized on an automated DNA synthesizer at the Keck Foundation Resource Laboratory (Yale University) or by Sigma Genosys (Israel) and purified using the standard protocols (19). For the in vitro selection experiments two DNA templates were used. MLP template, GTAGGTGTAGGCCACGTGACCGGGTGTTCCT-(N)10TATAAAAG(N)10CGCGTTCGTCCTCACTCTCTTCCGCATCG, derived from the sequence of the Adenovirus MLP positions −79 to 10, and E4 template, GGGTGTTTTTTGTGGACTTTAACCGTTACGTCA(N)10TATATATA(N)10ACTTGGCCCTTTTTACACTGTGACTGATTG, derived from the sequence of the Adenovirus E4 promoter positions −82 to 10, except that each template contained 20 random positions. Primers were designed to be complementary to templates in their 5′ and 3′ regions. DNA for binding affinity or dissociation kinetics experiments of individual selected sequences, or the various consensus sequences, was synthesized as hairpin constructs with 20 or 22 bp double-stranded stems and 5 cytosines in the loop (located at the 3′ side of the molecules, as written in Table 1). It was radioactively labeled as described previously (11). The competitor DNA molecules for dissociation kinetics experiments of individual selected sequences were 21 bp linear duplexes with either the wtMLP or the (T-A)4 sequence (Table 1), for probing sequences selected from the pool of MLP or E4 templates, respectively. fs(T-A)4 and wtE4 were challenged with competitor DNA (21 bp linear duplex) of the same DNA sequence as the hairpin constructs to which they were added as competitor, to concur with the binding stability study of T5T7, fsT5T7 and (T-A)4, all of which were studied by us previously (11). The rational for using a labeled hairpin DNA and linear duplex competitor DNA are discussed elsewhere (11).
Protein
The c-terminal domain of yeast TBP (yTBPc) was a kind gift from S. Juo (Yale University). The overexpression and purification of the protein were as described by Kim et al. (20). The fraction of yTBPc active for DNA binding was determined as described previously (11) and found to be 50%. We have used yeast TBP for the selection of sequences flanking a TATA box of a human pathogen to concur with our previous study (11). The crystal structures of TBP from different sources are all similar to each other (21–23), as well as the co-crystal structures of TBP/TATA-box complexes (20,24–28). The deletion of the N-terminal results in enhanced gel stability and it does not affect TBP/TATA-box interactions (29).
Preparation of double-stranded DNA oligonucleotides for in vitro selection experiments
The two DNA templates described above were made double-stranded using PCR. The two oligonucleotide primers for each template were labeled using [γ-32P]ATP (3000 Ci/mmol) and T4 polynucleotide kinase for 30 min at 37°C. Free radioactivity was separated from radioactive oligonucleotides using G-25 spin columns. An aliquot of 1 nmol of each primer was mixed with 20 pmol of the appropriate single-stranded template pool, in a PCR mixture that included 1 mM dNTPs, 4 mM MgCl2, 5 U of BIO-X-ACT™ Short DNA Polymerase (Bioline), OptiBuffer and Hi-Spec Additive supplied with the polymerase. This amount of template ensures that the pool of the first round of selection contained at least 10 double-stranded molecules from each possible sequence (of 420 different DNA sequences). Since a large DNA amount was used in the initial PCR, the PCR mixture was divided to 10 identical reactions.
For the initial selection experiment, the double-stranded DNA was generated and amplified by three cycles of PCR. After PCR the products were immediately loaded on a native gel (16%, acrylamide/bisacrylamide 39:1) and run at 350 V until the BPB dye migrated 24 cm in a 32 cm × 16 cm × 1.5 cm gel together with a double-stranded DNA size marker [21 bp, VW (30)]. The gels were visualized using a Fujii FLA 5000 phosphoimager. PCR products corresponding to the correct size were extracted from the gel using standard procedures (19), and the purified double-stranded oligonucleotides from each individual PCR were combined to one pool.
Selection for high binding affinity
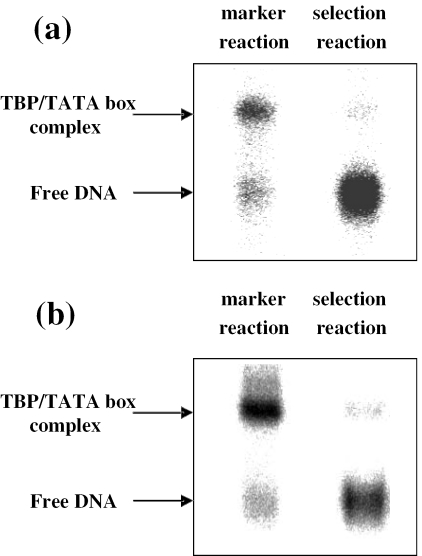
In the first kind of selection experiments, the oligonucleotides were selected based on high binding affinity to yTBPc. In these experiments an excess of double-stranded oligonucleotide pool was incubated with a limited amount of protein, and the partition of the selected from non-selected pool was by the EMSA. In the first round of selection, the double-stranded pool of oligonucleotides (40 nM) and yTBPc (0.4 nM active protein) were incubated for 60 min at 30°C in binding buffer (4 mM DTT, 10 mM Tris–HCl, pH 7, 30 mM KCl, 0.4% Brij58, 5 mM MgCl2 and 10% glycerol). After 60 min loading dye was added, and the reaction mixture was immediately loaded on a running native gel (8%, acrylamide/bisacrylamide ratio 39:1, 10% glycerol), which was run at 450 V and 30°C for 2 h in a buffer containing 1× TG (25 mM Tris–HCl, 190 mM Glycine, pH 8.3) and 5 mM MgAc. The wet gels were visualized using a Fujii FLA 5000 phosphoimager. The fraction of oligonucleotides that was bound to yTBPc was extracted from the gel using standard procedures (19). The conditions for TBP/TATA-box binding were changed as selection progressed, so that in each cycle only 1% of the DNA targets, those with the highest affinity for yTBPc, were in complex with it (Figure 1a). This was achieved by lowering continuously the concentration of yTBPc protein relative to DNA from 1/100 ratio of active yTBPc/DNA to 1/500 and by increasing the KCl concentration in the binding buffer from 30 to 150 mM between the first and the fifth (and last) rounds of selection. The selected and extracted oligonucleotides were amplified by PCR for 8 cycles, using 50 pmol of each 32P-labeled primer. The PCR products were purified and treated as discussed above and were then used in the next round of selection. To be certain of the exact position and boundaries of the selected band on the gel, despite the low fraction of DNA molecules bound to the protein, a ‘marker reaction’ was used to help in locating the band of the TBP/TATA-box complex on the gel. The DNA used for the marker reaction and the selection reaction was taken from the same double-stranded oligonucleotide pool. The composition of the marker reaction was similar to the composition of the selection reaction except that the amount of DNA was equimolar to the amount of the protein. Therefore, the amount of TBP/TATA-box complex was sufficiently high to be easily observed on the gel. However, in all cases a weak selected band was observed at the same location of the marker reaction band (Figure 1a). After extraction from the gel, the selected DNA pool was PCR amplified and gel purified (as discussed above) before being used in the next selection cycle. Five rounds of selection were carried out by the thermodynamic criteria. After each round the pool was cloned and several transformants were sequenced from each template. After the last round, 54 transformants were sequenced from each pool. The data contained multiple identical copies of several transformants. The list of unique selected sequences is given in the Supplementary Table S1. Thus, additional rounds of selection will not result in further enrichment of the population of the selected molecules. On the other hand, further selection may increase the PCR bias in the population.
Figure 1.
In vitro selection experiments. EMSA conducted for the separation of selected and non-selected DNA templates. (a) Selection for high binding affinity from a DNA pool containing the MLP TATA box and ten random flanking sequences on each side. (b) Selection for high complex stability from a DNA pool containing the E4 TATA box and 10 random flanking sequences on each side. The gels shown are those after the first selection cycle.
Selection for high complex stability
The second type of selection experiments was based on high kinetic stability. In the first round of selection, the double-stranded pool of oligonucleotides (20 nM) and yTBPc (20 nM active protein) were incubated for 60 min at 30°C in the binding buffer described above before adding a large molar excess of unlabeled 21 bp linear duplex competitor (2.62 µM, 131-fold excess of the cold competitor over active protein). The sequence of the competitor DNA molecules was either wt MLP or (T-A)4 (Table 1) for selection from the pool of MLP or E4, respectively. The reaction mixture, including the competitor DNA, was incubated for 4.5 h at 30°C. Loading dye was then added and the reaction mixture was immediately loaded on a running native gel (8%, acrylamide/bisacrylamide ratio 39:1, 10% glycerol), which was run at 450 V and 30°C for 2 h in 1× TG and 5 mM MgAc buffer. The conditions for TBP/TATA-box binding reaction were changed as selection progressed, so that only 1% of the most stable DNA targets were selected (Figure 1b). This was carried out by extending continuously the incubation time after the addition of the unlabeled DNA competitor from 4.5 to 7.5 h and by increasing the KCl concentration in the binding buffer from 30 to 150 mM between the first and fifth rounds of selection. Here too we have used a ‘marker reaction’ to help in locating the weakly observed band of the complex on the gel. The marker reaction was identical to the selection reaction except that the incubation was stopped after 1 h and no competitor DNA was added. The gels were visualized using a Fujii FLA 5000 phosphoimager. The fraction of oligonucleotides that was bound to TBP was extracted from the gel using standard procedures (19). It must be noted that even if the 21 bp competitor DNA is eluted together with the 88 bp selected DNA, it has no influence on the outcome of the selection as it cannot be amplified or cloned. The selected oligonucleotides were amplified by PCR and gel purified, as described above, and then used in the next round of selection. After each round the pool was cloned and several transformants were sequenced for each pool. After the fifth cycle, 51 transformants were sequenced from each template. Supplementary Table S1 gives the list of unique transformants. Here too multiple identical copies appeared for several transformants, and hence the selection was stopped after the fifth round.
Cloning and sequencing
The selection was started using 420 = 1.1 × 1012 different DNA sequences of each template. At each round we took a maximum of 1% of bound DNA. Thus, after five rounds of selection the final pool should contain ∼110 different DNA sequences. Hence, we stopped the selection after five rounds and cloned the resultant pool. We also cloned a sample from each previous round to check the progress of selection. Before cloning, the DNA pool was amplified by PCR, using 200 pmol of each unlabeled primers for 20 cycles. The PCR products were purified using QiAquick PCR Purification kit (Qiagen). The PCR amplified oligonucleotides were then ligated into the commercial pGEM-T Easy vector system (Promega) using the standard protocol of Promega. The ligated plasmids were transformed into XL/BLUE competent cells by the standard heat-shock procedure. The cells were grown first in Luria–Bertani (LB)/ampicillin medium at 37°C for 1.5 h and then on LB plates with ampicillin/IPTG/X-Gal at 37°C overnight. The recombinant plasmids were isolated using the Wizard Plus SV Minipreps DNA Purification System (Promega). These plasmids were analyzed by digestion with 20 U of the restriction enzyme EcoRI (New England Biolabs) for 1–2 h. The digestion products were run on a 1.6% agarose gel, and 51–54 plasmids that contained inserts were sequenced by the Macrogen Company (Korea).
Dissociation kinetics experiments
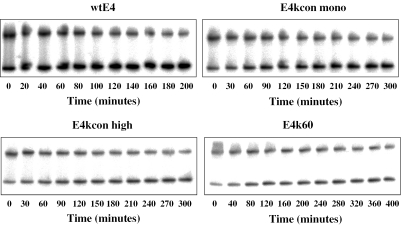
Dissociation kinetics experiments were carried out as determined previously (11). In short, individual selected sequences, containing 5–9 bp from each side flanking the TATA box (MLP or E4), were embedded in hairpin constructs described above. Radiolabeled hairpin duplexes (0.4 nM) and yTBPc (27 nM active protein) were incubated at 30°C in the same binding buffer used in the initial cycle of the thermodynamic or the kinetic selection experiments, before adding unlabeled competitor DNA [wtMLP or (T-A)4, 1.76 µM, 65-fold excess over active protein]. We used this buffer in order to concur with our previous study (11). Ten time points were taken for each sequence, they were adjusted according to the half-life derived from initial experiments, and ranged between 20 and 40 min each (Figure 2). Dried gels were quantified using a Fujii FLA 5000 phosphoimager, as described previously (11). F(t), the fraction of bound DNA at the different time points, was calculated from the equation: F(t) = (PSL-bg)complex(t)/[(PSL-bg)complex(t) + (PSL-bg)free(t)], where PSL is the photostimulated luminescence and bg is the background. ln[F(t)/F(0)] was plotted as a function of time (t) after the addition of the unlabeled competitor. The data were fitted to a two-phase first-order kinetic equation F(t)/F(0) = Ae−kA × t + Be−kB × t, where A and B are fractions of molecules dissociating with rate constants kA and kB, respectively. Half-life of complexes dissociating by the B process was calculated from: t1/2 B = ln2/kB
Figure 2.
Gels showing representative results for the dissociation kinetics of yTBPc (27 nM) from several E4-related TATA boxes embedded in hairpin constructs (0.4 nM). The number below each gel specifies the time after adding competitor DNA (1.76 mM).
Binding affinity measurements
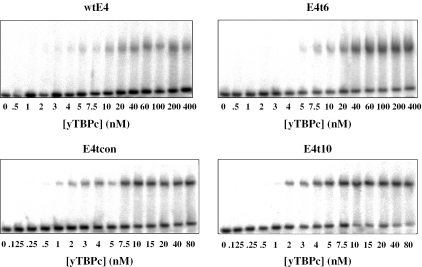
Individual selected sequences containing 5–7 bp from each side flanking the E4 TATA box were embedded in hairpin constructs, as described above. Radiolabeled hairpin duplexes (50 pM) and increasing amounts of yTBPc were incubated at 30°C for 2 h in the buffer used during the selection cycles, but with 100 mM KCl. Complexes were resolved from free DNA by electrophoresis on native gels (10%, 75:1 acrylamide:bisacrylamide, 10% glycerol), using the same buffer and running conditions as in the dissociation kinetics experiments.
Dried gels were quantified using a Fujii FLA 5000 phosphoimager. Equal-sized boxes were defined surrounding each band on the gel. We have extended the boxes from the band on the complex to the band of the free DNA, to account for dissociation of the complex during electrophoresis, as described previously (31). Association binding constants were calculated using non-linear least-squares methods of parameter estimation (SigmanPlot, Jandel Scientific, CA). Initially, we used equations describing a regular one binding site system, as described previously (31). However, the fit between the experimental points and the model was not always good. As can be noted from the gels of Figure 5, there is a plateau in the end of the titration. At the last few points of each gel, there is a small amount of free DNA that does not decrease upon further increase in protein concentration. Plotting the fraction bound as a function of 1/[yTBPc] revealed that 12–22% of the DNA molecules were inactive for binding, as the graph reached a plateau when 78–88% of the DNA molecules were bound (data not shown). Hence, we used the end points as additional adjustable parameters, as is customarily done in quantitative footprinting analyses of protein–DNA interactions [e.g. (32)]. This procedure yielded a much better fit between the experimental points and the curves of the fitted model (data not shown).
Figure 5.
Gels showing representative binding affinity measurements for yTBPc complexes with several E4-related TATA boxes embedded in hairpin constructs (50 pM).
Sequence analysis
A non-redundant dataset was created from each sequenced DNA pool by deleting duplicate sequences from each pool. The four DNA datasets were aligned using the web version of the program MEME (33). For the two E4 datasets we used the sequence on both strands in the alignment. From this non-redundant dataset we deleted sequences containing additional or alternative TATA boxes, on the same strand (shifted laterally) or on the complementary strand. It is this unambiguous dataset that was further analyzed. The information content of the sequences was calculated as described previously (34). The same procedure was applied to natural promoters containing the MLP and E4 TATA boxes, found in the Eukaryotic Promoter Database [EPD, Release 82 (35)]. The sequences from the EPD were checked to ensure that the TATA boxes were bone fide ones and were not in the transcribed region of the retrieved promoter. In addition, promoters with ambiguous TATA boxes, such as those containing a mixed E4 and MLP sequence (e.g. TATATATAAAAG), were deleted from the analysis, as well as sequences containing (T-A)n tracts with n ≥ 4. For sequences from the EPD we used MEME just to align the one strand given in the EPD.
To assess the significance of occurrence of sequence motifs larger than mononucleotides we calculated the Z-statistics of each motif, which is the deviation of observed frequency of occurrence of DNA tracts longer than mononucleotide from that expected based on the mononucleotides components of that DNA tract (36). This analysis was carried out for dinucleotides, trinucleotides and tetranucleotides. Equal occurrence of all bases at each position was assumed since in the starting pool each position contained an equal amount of each base. The same analysis was carried out on three sets of random sequences, containing 40, 42 and 47 sequences, and 10 random positions.
RESULTS
Analysis of the sequences selected in the in vitro selection experiments
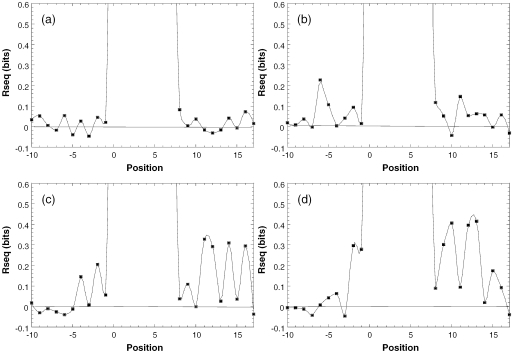
An unequivocal non-redundant dataset, from which we deleted sequences that contained additional and alternative TATA boxes of each sequence family, was analyzed by calculating the information content of the binding sites (34,37) (Table 2). Information content is a measure of the amount of specificity required for the recognition of DNA by proteins independent of the mechanism of recognition. The information content for each position was calculated and plotted as a function of the position of each base in the sequence (Figure 3). This calculation is based on mononucleotide frequencies and assumes independence of the occurrence of each mononucleotide. The consensus sequences are shown in Table 2. By definition, a consensus sequence lists the most frequent base at each position. In Table 2 we list as consensus only those bases at specific positions where the reduction in uncertainty (Rseq) in this position is larger by at least 1 SD than that expected for a sample of that size. We denote these bases by a small letter. Positions where the nucleotide frequency is >50% are denoted by a uppercase letter. The sequences deleted from the original non-redundant datasets were those containing alternative consensus-like TATA boxes. For the two E4 datasets we deleted only those sequences with alternative TATA boxes with half-life of 180 min and above (Hana Faiger and Tali E. Haran, unpublished data), i.e. sequences that formed complexes that were more stable that that formed with the consensus sequence (see below). However, deleting weaker alternative TATA boxes did not significantly change the pattern of selected sequences (data not shown).
Table 2.
Consensus sequences and information content of sequences studied here
| Sequence | mononucleotide consensus | Total Rseq (bits) | Total number of sequencesb | Sequence used in analysisc | |||
|---|---|---|---|---|---|---|---|
| 5′ Side | TATA box | 3′ side | total | ||||
| 1 3 5 7 9 11 13 15 17 19 21 23 25 27 | |||||||
| Consensus of selected sequences based on mononucleotide frequenciesa | |||||||
| MLP therm. | nknngnnnknTATAAAAGbnnnnnnndn | 0.1 (1) | 15.6 (1) | 0.2 (1) | 15.9 (3) | 47 | 42 |
| MLP kinetic | nnnnktnnsnTATAAAAGktnrgkgnkn | 0.5 (1) | 15.6 (1) | 0.5 (1) | 16.6 (2) | 45 | 42 |
| E4 therm. | nnnnnnsnGnTATATATAngnTGnCnbn | 0.3 (1) | 15.6 (1) | 1.4 (1) | 17.3 (2) | 51 | 47 |
| E4 kinetic | nnnnnggnGCTATATATAcgsgGGnggn | 0.6 (1) | 15.6 (1) | 1.9 (1) | 18.1 (3) | 42 | 40 |
| Consensus of selected sequences based on the mononucleotide frequencies observed in higher-order motifsd | |||||||
| MLP therm. | nnnnnnnnkkTATAAAAGgttnnnnnnn | ||||||
| MLP kinetic | nnswTTgGsrTATAAAAGktbrskgdbn | ||||||
| E4 therm. | nnnnntCCGyTATATATAssytsvsvcn | ||||||
| E4 kinetic | nnnnnsscgcTATATATAcgsgggsGGG | ||||||
| Consensus of natural promotersa,e | |||||||
| MLP eukaryote | vnnnggwggSTATAAAAGcvGvngbrcg | 0.85 (3) | 15.90 (3) | 0.75 (3) | 17.50 (5) | 185 | 176 |
| MLP human | rnnngGnGnSTATAAAAGcvGnngGnsg | 1.4 (2) | 15.5 (1) | 1.1 (2) | 18.0 (3) | 42 | 38 |
| E4 eukaryote | wynwnawcncTATATATASngngnnnnn | 0.3 (1) | 15.7 (1) | 0.4 (1) | 16.3 (2) | 70 | 59 |
aTATA boxes are underlined. Boldface letters in the flanking sequences indicate that the reduction in uncertainty for that position (Rseq) is larger than 1 SD from that expected for a sample of that size. Uppercase letters indicate that the frequency of that nucleotide is >50%. An ambiguous code is used whenever there are several nucleotides that are within 1 SD of the most frequent one, and is denoted by a uppercase letter when at least one nucleotide frequency is >50%. K = G or T; S = C or G; W = A or T; B = C, G or T; D = A, G or T; V = A, C or G.
bTotal number of sequences in the non-redundant data.
cUnequivocal TATA-box sequences only. Sequences were deleted if they contained additional and alternative TATA boxes in the flanking sequences.
dBased only on higher-order motifs (2, 3 or 4 bp long) that are statistically significant in the selected sequences (see Table 3 for details). Ambiguous codes are given as discussed in Footnote a. Uppercase letters indicate that this nucleotide is the only one observed in this position, in all three higher-order levels. Italicized letters indicate that the frequency of this base is >50% in all three levels.
eSequences were retrieved from the Eukaryotic Promoter Database [release 82 (35)].
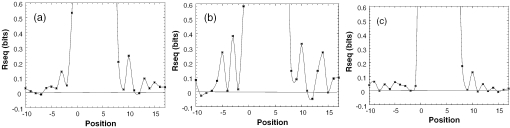
Figure 3.
Information content of in vitro selected sequences flanking TATA boxes. (a) Constructs containing MLP-like sequences selected for high binding affinity. (b) Constructs containing MLP-like sequences selected for high complex stability. (c) Constructs containing E4-like sequences selected for high binding affinity. (d) Constructs containing E4-like sequences selected for high complex stability. Rseq is the reduction in uncertainty at each position of the binding site. The Rseq of the core TATA box is close to 2. We rescaled the graphs at Rseq= 0.6 to show the pattern in the flanking sequences more clearly.
Given the off rates obtained in the individually studied sequences from the pool selected for optimal binding stability (see below and Table 1), we were probably selecting for an ensemble that is present later in the approach to equilibrium but not 100% there, in the initial round of selection. However, the half-life of TBP/TATA-box complexes decrease with increase in KCl concentration (data not shown) and hence, in later cycles (from the third cycle onwards), the complexes must have reached equilibrium at the selection stage. Even though less cycles of selection may have been carried out at true equilibrium, the selection has probably reached saturation, because each dataset derived from selecting for high binding affinity contained multiple identical sequences.
The analysis of sequences flanking MLP-like TATA boxes shows that no significant pattern emerged when we selected these sequences based on high binding affinity of TBP to these target sites (Table 2 and Figure 3a). When we selected for high complex stability (low kinetic off rate) a pattern emerges contributing ∼6% to the overall information content (3% from each side, Figure 3b). Since the second target site, E4 [=(T-A)4] is totally symmetric we could not establish the orientation of TBP on these sequences. The pattern for the sequences selected at the regions flanking E4-like TATA boxes was thus established by aligning these sequences (33) using both strands to maximize the alignment. Relative to the pattern observed for the selected MLP templates, the information content observed at the sequences flanking the E4 TATA box is more significant (Figure 3c and d). The total contribution to the information content is 11% for the thermodynamic selection and 16% for the kinetic selection (Table 2). More surprising is that both the thermodynamic as well as the kinetic selection show a significant asymmetry in the information content pattern in the two flanking sequences. One side has low information content, similar to that observed in the MLP targets, whereas the other side has significantly higher values. This could not arise simply from the alignment of the sequences (which was carried out without the constant sequence region of the primers). Have the sequences had a similar pattern at both sides [as expected from a totally symmetric sequence, and from a protein that is known to bind TATA boxes in both orientations (38)] then even after this alignment the pattern, and particularly the information content, should have remained similar at both ends. The standard error of the difference for the two sides in both thermodynamic and kinetic selections is 0.02, whereas the differences between the two sides are 1.1 and 1.3 for the thermodynamic and kinetic selections, respectively. This is about 55- and 65-fold greater than the standard error, which shows that the phenomenon is highly significant statistically. Possible sources for this behavior are discussed below.
To compare the results obtained by the in vitro selection experiment to sequence pattern observed in sequences flanking the TATA box of natural promoters, we calculated the information content for 179 eukaryotic promoters containing an unambiguous MLP TATA box and 59 eukaryotic promoters containing an unambiguous E4 TATA box found in the EPD (out of 4809 sequences). In addition, 38 human promoters containing an unambiguous MLP TATA box (out of 1871 sequences) were similarly analyzed. The current number of available human promoters containing a uniquely defined E4 TATA box (15 promoters) is not sufficient for a significant analysis of their information content. The information content was calculated for a region extending 10 bp 5′ to 10 bp 3′ to the TATA boxes, and is shown in Figure 4. Table 2 compares the information content and the consensus sequences of the selected sequences to those of natural promoters found in the EPD.
Figure 4.
Information content of TATA boxes and their flanking sequences found in the EPD (35). (a) MLP-like TATA boxes from all eukaryotic promoters. (b) MLP-like TATA boxes from human promoters. (c) E4-like TATA boxes from eukaryotic promoters. Rseq is the reduction in uncertainty at each position of the binding site. See Figure 3 for details.
Kinetic analysis of selected sequences
We measured the dissociation kinetics of individual sequences selected by kinetic criteria in vitro (Figure 2). The selected sequence motifs were embedded in hairpin constructs, similar to those used in our previous study [(11) see Materials and Methods for details]. Here, we analyze dissociation kinetics data using a bi-exponential equation (Table 1), because it has been previously shown that such a fit represents better the observed dissociation data than a single exponential equation (39–41). The analysis by a bi-exponential equation of the studied sequences (Table 1) showed a well-determined slow dominant phase (70–90% of the molecules were caught at this stage upon adding competitor DNA), and a not well-determined (having a large standard error) fast phase. The half-life values presented in Table 1 and discussed here are only those of the well-determined slow phase.
The results for individual sequences from the selected (T-A)4 pool shows high variability of binding stabilities (Table 1), ranging from 147 ± 5 min (E4k36) to 333 ± 14 min (E4k28). The half-life of complexes of TBP to two E4-related sequences (E4k28 and E4k60) is significantly larger than the half-life of TBP complexes with sequences from the MLP pool, a known strong basal promoter of high binding stability. The sequences from the E4 pool are all bona fide (T-A)4-like sequences and the flanking sequences do not contain any DNA element that can convert the molecules to an MLP-like TATA box, or any other TATA box of known high complex stability. The results for individual sequences selected from the MLP pool show less pronounced variability in their dissociation kinetics from TBP, spanning the range from 206 ± 17 min (MLPk88) to 271 ± 20 min (fsMLP).
To assess the range of half-life values that can be observed as a function of the sequences flanking the (T-A)4 core TATA box, we measured the dissociation of TBP from the (T-A)4 box embedded in its natural flanking sequences as found in adenovirus (wtE4, Table 1). The results (Figure 2 and Table 1) show that in this sequence context the E4 TATA box is a kinetically weak binding site, which is consistent with its physiological role during the adenovirus life cycle (42,43). This shows that for the E4 TATA box there is a large variability of binding stabilities as a function of the sequences flanking the (T-A)4 box. This span of half-life values is as large as that shown by different consensus-like core TATA-box variants (Table 1) [(11); Hana Faiger, Marina Ivanchenko, Ilana Cohen and Tali E. Haran, manuscript in preparation).
Analysis of the binding affinity of selected sequences
We measured the equilibrium dissociation constants of individual sequences selected for high binding affinity (Figure 5). Here too the selected sequences were embedded in hairpin constructs, as described above. The binding affinity of sequences selected by the thermodynamic criteria show that here too there is high variability between the binding affinities of individual selected sequences, and between the selected sequences and the natural E4 TATA box from adenovirus. The highest binding affinity (2.4 ± 0.3 nM, E4t10, Table 1) is about 3-fold higher than that of E4t6 (7.5 ± 0.4 nM, Table 1), and about 8-fold higher than that of wtE4 (19 ± 2 nM, Table 1). Thus, the results show that in its natural adenovirus sequence context E4 TATA box is a relatively weak binding site thermodynamically, and that by changing only the flanking sequences it can become a much higher-affinity binding site.
Kinetic stability of mononucleotide-based consensus sequences
We constructed a mononucleotide-based consensus sequence for the E4 pool selected for high binding stability by taking the most frequent base at each position (E4kcon-mono, Table 1). The half-life of the complex of TBP with E4kcon-mono is 145 ± 5 min (Table 1), which shows that this complex is significantly less stable than complexes with all but one of the studied individual sequences selected from the (T-A)4 pool. No mononucleotide-based consensus sequences have been designed for the MLP pool selected for high binding stability, or the E4 pool selected for high affinity, since the degeneracy at this level is too high.
Frequency of occurrence of higher-order sequence motifs in the selected flanking region
DNA structure is now known to be influenced by long-range interactions (15,30,44–46). Hence, our assumption in the analysis carried out so far in this paper, of the independent occurrence of mononucleotides, may not be accurate. Information content analysis assumes independent occurrence of mononucleotides. This could be one reason why no recognizable pattern is observed in the selected sequences in the flanking regions of the various constructs (Figure 3). In principal, information content analysis can be extended to include cooperative occurrence of larger sequence tracts, such as dinucleotides, trinucleotides and tetranucleotides. However, more sequence data than is available from this study are needed for a reliable estimate of such correlations. Instead, the statistical significance of these motifs was established using several significance tests. First, we determined the most frequent motif at each position and calculated the deviation of the observed motif from that expected based on their mononucleotide components (36). Since the flanking regions were originally random, we assumed equal occurrence of all bases at each position. This gives us the Z-statistic for this motif, which states how many standard deviations this motif falls away from that expected based on additive mononucleotides. We then calculated the standard normal probability that these motifs will occur. All calculations were compared with similar calculations made on a set of random sequences, with the same number of sequences and the same number of random positions. In the random set dinucleotides appeared six or five times, in sets of size 47 sequences or smaller, respectively. Trinucleotides appeared three times at most, in all set sizes examined, and tetranucleotides motifs appeared at most twice. The probability that a specific tetranucleotide motif will occur three times in a dataset containing 40–47 sequences is 5 × 10−12 to 4 × 10−10, respectively, and higher for its occurring four times. On the trinucleotide level, a motif will occur four times with a probability of 3 × 10−4 to 5 × 10−5 (for sets of 47 or 40 sequences, respectively), and the probability of its occurring six times is 2 × 10−9 to 3 × 10−11 (for sets of 47 and 40 sequences, respectively).
In Table 3, we present the observed occurrences of statistically significant dinucleotides, trinucleotides and tetranucleotides sequence motifs at each position in the two 10 bp segments around the two (T-A)4 boxes and for the MLP pool selected for high binding stability. As one can appreciate from the data in Table 3, most positions in the two E4 pools, and especially in the more conserved downstream region, contain statistically significant dinucleotides, trinucleotides and tetranucleotides. The region on the upstream side of the MLP box selected for high binding stability shows a conserved higher-order pattern as well, with many statistical significant motifs. Based on these statistically significant motifs we constructed a new mononucleotide-based consensus sequence, as follows. In each higher-order level we looked at the most frequent nucleotide at each position and then summed them in all three levels. The advantage of this consensus sequence (relative to that based on mononucleotide-based information content) is that it strengthens the mononucleotide signal if and when it appears in several linked and statistically significant cooperative higher-order motifs. Indeed, these higher-order consensus sequences (Table 2) have more stringent sequence requirements and are actually a subset in most positions of the regular mononucleotide-based consensus sequence. This consensus sequence shows more clearly the sequence conservation in the 5′ side of the MLP pool selected for binding stability, and the asymmetry of sequence conservation in the two E4 pools.
Table 3.
Statistically significant higher-order motifs in the selected DNA pools
| Positiona | E4 kinetic selection (40 sequences) | E4 thermodynamic selection (47 sequences) | MLP kinetic selection (42 sequences) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conb | Statistically significant dinucleotidec | Zd | Statistically significant trinucleotidec | Zd | Statistically significant tetranucleotidec | Zd | Conb | Statistically significant dinucleotidec | Zd | Statistically significant trinucleotidec | Zd | Most frequent tetranucleotidec | Zd | Conb | Statistically significant dinucleotidec | Zd | Statistically significant trinucleotidec | Zd | Statistically significant tetranucleotidec | Zd | |
| 1 | n | n | n | ||||||||||||||||||
| 2 | n | n | n | ||||||||||||||||||
| 3 | n | n | n | ||||||||||||||||||
| CA 6 | 2.2 | ||||||||||||||||||||
| GT 7 | 2.8 | ||||||||||||||||||||
| 4 | n | n | n | CAT 5 | 5.4 | ||||||||||||||||
| GTT 5 | 5.4 | ||||||||||||||||||||
| AT 9 | 4.1 | CATT 3 | 7.0 | ||||||||||||||||||
| GTTT 3 | 7.0 | ||||||||||||||||||||
| 5 | n | AGG 4 | 4.3 | n | k | ATT 6 | 6.6 | ||||||||||||||
| GG 7 | 2.9 | TT 12 | 6.0 | TTTG 3 | 7.0 | ||||||||||||||||
| 6 | g | n | t | TTG 7 | 7.9 | ||||||||||||||||
| TC 9 | 3.7 | GTCC 3 | 6.6 | TG 11 | 5.3 | TTGG 4 | 9.5 | ||||||||||||||
| TTCG 3 | 7.0 | ||||||||||||||||||||
| 7 | g | s | n | TGG 7 | 7.9 | ||||||||||||||||
| CCCG 3 | 7.2 | CC 8 | 3.1 | TCCG 3 | 6.6 | GG 9 | 4.1 | TGGG 4 | 9.5 | ||||||||||||
| TGGC 3 | 7.0 | ||||||||||||||||||||
| 8 | n | CCG 4 | 4.3 | n | CCG 7 | 7.4 | n | GGG 4 | 4.2 | ||||||||||||
| GCG 4 | 4.3 | GGC 4 | 4.2 | ||||||||||||||||||
| CG 11 | 5.6 | GGTG 3 | 7.2 | CG 10 | 4.3 | CCGC 3 | 6.6 | ||||||||||||||
| CCGT 3 | 6.6 | ||||||||||||||||||||
| 9 | G | CGC 5 | 5.6 | G | CGT 5 | 5.0 | s | ||||||||||||||
| CGC 4 | 3.8 | ||||||||||||||||||||
| GC 13 | 6.9 | CGCT 5 | 5.6 | GT 9 | 3.7 | CGCT 4 | 3.8 | GG 7 | 2.8 | ||||||||||||
| CGTT 5 | 5.0 | ||||||||||||||||||||
| 10 | C | GCT 13 | 6.9 | n | GTT 9 | 3.7 | n | GGT 7 | 2.8 | ||||||||||||
| CT 20 | 3.7 | GCTA 13 | 6.9 | TT 16 | 1.4 | GT 15 | 1.6 | ||||||||||||||
| 11 | T | T | T | ||||||||||||||||||
| 18 | A | A | G | ||||||||||||||||||
| AC 17 | 2.6 | AC 19 | 2.4 | GT 17 | 2.3 | ||||||||||||||||
| 19 | c | ACA 8 | 3.6 | c | AGG 7 | 2.4 | k | GTT 7 | 2.8 | ||||||||||||
| ACG 7 | 2.9 | GGT 6 | 2.2 | ||||||||||||||||||
| CA 8 | 3.6 | ACGC 7 | 8.1 | GG 7 | 2.4 | TT 7 | 2.8 | ||||||||||||||
| CG 7 | 3.0 | ACAC 4 | 4.3 | ||||||||||||||||||
| AGTG 4 | 4.3 | ||||||||||||||||||||
| 20 | g | CGC 7 | 8.1 | g | n | ||||||||||||||||
| CAC 4 | 4.3 | ||||||||||||||||||||
| GTG 4 | 4.3 | ||||||||||||||||||||
| GC 10 | 4.9 | CGCG 3 | 7.2 7.2 | GT 9 | 3.7 | CCCT 3 | 6.6 | ||||||||||||||
| TG 8 | 3.8 | CGCC 3 | |||||||||||||||||||
| 21 | s | GCG 5 | 5.6 | n | GTT 6 | 6.2 | n | ||||||||||||||
| GCT 4 | 3.8 | ||||||||||||||||||||
| CCT 4 | 3.8 | ||||||||||||||||||||
| CG 8 | 3.6 | GCGG 3 | 7.2 | CT 11 | 4.9 | GTTG 4 | 8.9 | ||||||||||||||
| GG 7 | 3.0 | TT 9 | 3.7 | ||||||||||||||||||
| GC 7 | 3.0 | GG 7 | 2.4 | ||||||||||||||||||
| 22 | g | GCG 4 | 4.3 | T | TTG 6 | 6.2 | r | TAG 5 | 5.4 | ||||||||||||
| GGG 4 | 4.3 | GGC 5 | 5.0 | ||||||||||||||||||
| CGG 4 | 4.3 | CTG 5 | 5.0 | ||||||||||||||||||
| GG 10 | 4.9 | CGGC 3 | 7.2 | TG 15 | 7.3 | TTGG 4 | 8.9 | GG 7 | 2.8 | ||||||||||||
| GGGG 3 | 7.2 | GGCA 3 | 6.6 | ||||||||||||||||||
| 23 | G | GGG 6 | 6.9 | s | TGG 6 | 6.2 | s | ||||||||||||||
| GCG 4 | 4.3 | TGA 5 | 5.0 | ||||||||||||||||||
| GGC 4 | 4.3 | GGA 4 | 3.8 | ||||||||||||||||||
| GG 13 | 6.9 | GG 8 | 3.1 | TCCC 4 | 8.9 | ||||||||||||||||
| GC 7 | 3.0 | GA 8 | 3.1 | TACC 3 | 6.6 | ||||||||||||||||
| TGGC 3 | 6.6 | ||||||||||||||||||||
| 24 | G | GGC 6 | 6.9 | n | CCC 4 | 4.3 | k | CTG 4 | 4.2 | ||||||||||||
| GGC 4 | 4.3 | ||||||||||||||||||||
| GC 8 | 3.6 | CC 9 | 3.6 | CGGC 3 | 6.6 | CTGA 3 | 7.0 | ||||||||||||||
| AC 7 | 2.4 | ||||||||||||||||||||
| 25 | n | GGG 4 | 4.3 | c | n | TGA 4 | 4.2 | ||||||||||||||
| TGG 4 | 4.3 | GCT 4 | 4.2 | ||||||||||||||||||
| GG 9 | 4.2 | TGGG 3 | 7.2 | CG 7 | 2.4 | GGCG 3 | 6.6 | GG 7 | 2.8 | GCTC 3 | 7.0 | ||||||||||
| 26 | g | GGG 6 | 6.9 | n | CCC 5 | 5.0 | n | ||||||||||||||
| CGG 4 | 4.3 | CAC 5 | 5.0 | ||||||||||||||||||
| CGC 4 | 3.8 | ||||||||||||||||||||
| GG 12 | 6.2 | GGGG 4 | 9.9 | CC 8 | 3.1 | CACG 3 | 6.6 | GT 7 | 2.8 | ||||||||||||
| 27 | g | GGG 7 | 8.3 | b | ACC 4 | 3.8 | k | ||||||||||||||
| GG 7 | 3.0 | CG 8 | 3.1 | GC 7 | 2.8 | ||||||||||||||||
| 28 | n | n | n | ||||||||||||||||||
aPosition in the sequence.
bMononucleotide-based consensus sequence. For details on the lettering see Table 2.
cDNA tracts (2, 3 or 4 bp long), which are statistically significant at each position. Dinucleotides are positioned between the two bases that constitute it, trinucleotides on the central base, and tetranucleotides between the second and third base. Here lettering is not an indication on the frequency of occurrence. The subscript numbers are the number of occurrences of each motif.
dZ-statistics or the deviation of the observed frequency of DNA tracts from that expected based on its mononucleotide composition. It is calculated by subtracting from the observed number of occurrences of the most frequent motif, the expected number of occurrences based on the mononucleotide frequency of the respective base pairs, and then dividing this value by the expected standard deviation (25). Statistically significant motifs are those that appear with frequency higher than that observed in a completely random sequence set, of similar size, in which there is an equal representation of each nucleotide in each position.
Kinetic stability and binding affinity of TBP complexes to consensus sequences based on higher-order DNA motifs
The data in Table 3 show that in many positions there is a linkage between the occurrence of two statistically significant mononucleotides and the occurrence of a statistically significant dinucleotide. Similarly, in many positions two statically significant dinucleotides are contained in the statistically significant trinucleotide and two trinucleotides in a statistically significant tetranucleotide. Another type of linkage is that between partially overlapping adjacent dinucleotides, or trinucleotides or tetranucleotides, all of them being the most significant in their respective positions. This is noticeable in particular at the more conserved downstream side of the two E4 pools, and in the upstream part of the MLP pool selected for binding stability.
The two types of linkages between sequence motifs discussed above also point to higher-order DNA structures as responsible for the modulating effects that the flanking sequences have on TBP/TATA-box interaction. We therefore constructed a new consensus sequence for the E4 pool selected for high binding stability, based on linked higher-order motifs (‘E4kcon-high’ in Table 1) and measured the dissociation kinetics of TBP from this target. E4kcon-high was constructed by looking for the most significant linked dinucleotide, trinucleotide and tetranucleotide arrangement at each side. Thus, at the 5′ side the sequence CCCGC(T) is formed by three linked dinucleotides, three linked trinucleotides and two linked tetranucleotide motifs, which are all statistically significant and are all the most frequent at each position. At the 3′ side the sequence (A)CGCGGGG is formed by six linked dinucleotides, five linked trinucleotides and four linked tetranucleotides, all of which are statistically significant and all but one are the most frequent at their position. The half-life of the complex of this target with TBP is 182 ± 8 min, which is significantly higher than that of E4kcon-mono, but significantly lower than the half-life of E4k28, E4k30, Ek56 or E4k60.
A consensus sequence based on higher-order motifs was constructed also for the E4 pool selected for high binding affinity, based on the same principle (E4tcon, Table 1). The binding affinity of TBP to this sequence (3.3 ± 0.3 nM, Table 1) is similar to the binding affinity of most selected sequences. A similar consensus sequence was constructed for the MLP pool selected for binding stability. On the 3′ side of the MLP TATA box there are no statistically significant conserved tetranucleotides, and not many conserved di- and trinucleotide motifs. Nevertheless, the half-life of the complex of TBP to this sequence (232 ± 12 min, Table 1) is intermediate to the values obtained with the other MLP-related TATA boxes studied here.
The effect of the flanking sequences is dependent on the identity of the base pair at position 8
So far, we have studied two different core TATA boxes, in which the modulation of binding stability by the flanking sequences is significant, (T-A)4 and T5T7 (Table 1). Do same flanking sequences influence these two TATA boxes similarly? To answer this question we synthesized a DNA sequence with the (T-A)4 core and the original flanking sequences [fs(T-A)4, Table 1] used in our previous study (11). The half-life of the complex of TBP with the two targets is similar [163 ± 6 versus 157 ± 5 min, for (T-A)4 and fs(T-A)4, respectively, Table 1]. Thus, the effect of the flanking sequences on ‘malleable’ TATA boxes is different as a function of the core TATA box sequence. This is further discussed below.
DISCUSSION
We carried out selection experiments probing the effects of sequences flanking two well-studied TATA boxes, adenovirus MLP and E4. We asked several related questions. First, can we find any flanking sequence that influences the dissociation kinetics of TBP from the MLP TATA box to the extent observed with TATATATN boxes? Second, do different flanking sequences have different mechanistic effects on TBP-TATA-box interactions? I.e. will unique flanking sequences be chosen if we select for kinetically stable complexes, and will they be different from those selected for high binding affinity? To address these questions we have carried out two kinds of selections. In the first one, the oligonucleotides were selected based on high binding affinity to TBP, and in the second on high kinetic stability.
Flanking sequence effects are sequence specific, selection specific and TATA-box dependent
The differential effect of the flanking sequences on TBP/TATA-box interaction, first observed in our previous study (11), is shown here to be a general phenomenon. On the whole, the interaction of TBP with the E4 target is significantly more affected by the identity of the adjoining sequences than the interaction with the MLP target. Thus, we have validated our previous hypothesis that the change in TBP/TATA-box interactions, as a function of the flanking sequences, will be more significant for ‘malleable’ sequences, i.e. those that have a structure that is easily changed by the sequence context (such as those of the form TATATATN).
Sequence motifs found in the flanking regions of the studied E4 and MLP TATA boxes are also ‘selection specific’. For example, A- or T-tracts (defined as at least A4, T4, A3T, or A2T2,) (46) appear significantly in the MLP pool selected for high binding affinity (8/42 sequences, mostly in the 3′ side), but not in the MLP pool selected for high binding stability (3/42 sequences). G-tracts (defined as CnGm or GmCn, n + m > 4) (30) appear significantly in the E4 pool selected for high complex stability (12/40 sequences), but not in the pool selected for high binding affinity (3/47 sequences).
Sequence specificity of the effect of the flanking sequences on TBP/TATA-box interaction is 2-fold. First, the whole phenomenon is significant only for certain TATA boxes. Second, sequence motifs in flanking regions that influence TBP/TATA-box interaction are usually unique to a certain TATA-box sequence and are different for different core TATA-box sequences. The two studied TATATATN boxes, (T-A)4 here and T5T7 before (11), are differentially affected by the sequences flanking them on either side. Sequence changes that reduce the kinetic stability of the TBP/T5T7-box complex [i.e. alternating (G-C)n tracts to homopolymeric G-tracts] do not affect the interaction of TBP with the (T-A)4 box and are observed in the E4 population selected for high binding stability.
Wong and Bateman (47) selected TBP-binding sequences from a pool of random duplex DNA. The majority of the selected DNA sequences contained the TATATAAG box. When we analyzed the information content of the data presented by Wong and Bateman (47), for the pattern surrounding the TATATAAG box, we got the consensus sequence aGaGTATATAAGG (The core TATA box is underlined, for notations see the legend to Table 2). This shows that flanking sequences affect the binding of TBP to TATA boxes of the form TATATANN as well, and moreover that the sequence pattern in the flanking sequences is different from that observed for (T-A)4 pool in general, and that selected for high binding affinity, in particular. We observe here that the specifics of the effect that the flanking sequences exert on pliable TATA boxes depend on the nature of the base pairs at positions 7 and 8 of TATA boxes, the terminal base pair step. This step (together with the T1-A2 step) is where two phenylalanines residues from each ‘stirrup’ of the saddle-shaped TBP structure intercalate and kink the DNA double helix (20,24–28). We have previously shown that the identity of this step is linked with the stability of TBP/TATA-box complexes (11).
Flanking sequence effects are directional
An unexpected finding of the present study is that one side of the E4-like target, selected by either criterion, is much more conserved, and hence has much higher information content, than the other side. Our basic idea is that the sequences flanking the (T-A)4 target modulate its structure, such that TBP actually bind to different DNA structures as a function of the identity of the flanking sequence, and that there is a selective advantage for certain motifs in the flanking sequences because the TBP/TATA-box complex formed at these targets is more kinetically stable, or that the TATA box thus formed has higher affinity to TBP. However, the remaining question is how an asymmetric pattern arises from a totally symmetric TATA box, and for a protein that is known to bind in both orientations (38)? If both orientations of TBP on the symmetric (T-A)4 box are equally populated, or nearly so, then the preferred sequences of the N10 on the 5′ end of the top strand should be equal to those of N10 on the 5′ side of the bottom strand.
Parkhurst et al. (39,41) suggested that TBP binds to TATA boxes in several steps, and that the mechanism of binding includes two intermediate species. In the first intermediate complex, neither pairs of phenylalanine has intercalated into the DNA (at positions 1–2 and 7–8 of the 8 bp TATA box). In the second intermediate complex one side has intercalated phenylalanines, whereas the other side has not. In the final complex both sides have intercalated phenylalanines, as observed in the crystal structure (20,24–28). If we assume that the first intercalation event is facile and reversible, but that the second intercalation step occurs readily only for certain base-pair combinations, then we would see a conservation of TATA box sequence/conformation as observed here. Similarly, if TBP dissociate in a mechanism involving several steps, with intermediates involving partly intercalated species, it could be that the first de-intercalation step is facile and especially is reversible, whereas the second de-intercalation step is not reversible (because TBP may rapidly fall off the DNA), and facile only for certain base-pair combinations. In this case, we will trap and select only those base pair combinations for which this second dissociation event is not facile. Hence, the larger sequence stringency observed on one adjacent side only, even though the TATA-box sequence itself is fully symmetric. There is no way, however, to assign the two binding steps to the two sides of the symmetric (T-A)4 sequence based on the results from this study.
Looking at Table 3, it seems that the flanking sequences of the MLP box selected for binding stability also shows an asymmetric pattern of DNA sequence motifs, with the 5′ side (the TATA side) having a more conserved pattern. However, in this case both sides have similar information content because, even though an occurrence of a tetranucleotide motif twice in 42 sequences set is not significant (and hence not shown in Table 3), its occurrence five times, for different tetranucelotide motifs, is significant. There are two such positions in the 3′ side of the MLP pool selected for binding stability (positions 20–23 and 21–24, Table 3). This renders the 3′ side to be as informative as the 5′ side of this TATA box.
Flanking sequence effects are determined by long-range cooperative DNA conformation
It is now well established that sequence-dependent DNA structure is influenced by long-range interactions rather than by nearest-neighbor ones (15,30,44–46). We propose here that it is the structure of the whole DNA region in the flanking sequences that modulates the structure of the DNA in the TATA box, and thus it is a novel form of indirect readout. The cooperative nature of DNA structure can explain why there are no recognizable patterns in the mononucleotide-derived information content of the flanking sequences (Figure 3) and in the sequence logos derived from them (data not shown). Alternatively, flanking sequences could be favored based on their intrinsic structural characteristics, and especially on their bendability, without any concomitant influence on the adjacent TATA box. Since TBP bends the TATA box significantly, sequences that are pre-bent, or more easily bendable in a particular orientation could be more energetically favored. This probably contributes to the observed pattern in all sequences. However, this on its own cannot fully explain the asymmetry observed in the selected sequences, as discussed above, nor the difference in the extent of influence that the flanking sequences have in the MLP versus the E4 sequence pool. When we designed a TBP binding site based on the most frequent base at each position of the E4 pool selected for high binding stability (E4kcon-mono, Table 1), this consensus sequence formed a complex with TBP that was significantly less stable than complexes with most of the individually studied sequences from the selected pool. A consensus sequence based on linked higher-order motifs (E4kcon-high, Table 1) forms a more stable complex with TBP than E4kcon-mono. It does not form as stable a complex with TBP, as do several selected sequences; however, it does approach the average half-life of the E4 pool selected for high binding stability when we include in the average all studied E4-related sequences. The binding characteristics of consensus sequences based on linked higher-order motifs for the MLP pool selected for high binding stability and the E4 pool selected for high binding affinity represent intermediate values to those obtained with individual members of these pools. We stopped our analysis at tetranucleotides because of the paucity of data for higher-order analysis, but also because tetranucleotides can already form independent cooperatively built DNA units (48), such as those observed in the structure of A-tracts (46,49) and G-tracts (30). There is a difference between the E4 pool selected for high binding affinity versus the E4 pool selected for high binding stability, with respect to the correlation between the occurrences of frequently observed higher-order motifs and binding characteristics. There is no correlation between the occurrence of frequently observed dinucleotides, trinucleotides or tetranucleotides and stability of binding to TBP. The flanking sequences in the highest kinetically stable complexes are formed mostly from sequence motifs that are unique to these sequences and are not the most frequent ones at any position. However, some frequent motifs do appear in complexes of intermediate stability (Table 3). On the other hand, in the E4 pool selected for high binding affinity there is a strong correlation between the occurrence of frequently-observed high-order motifs in general, and tetranucelotide in particular, and the affinity of binding.
This leads us to suggest that a particular combination of at least 4 bp, but perhaps the whole 10 bp DNA segment, at both sides of the TATA box, is responsible for the observed effect of flanking sequences on TBP/TATA-box interactions, and especially for binding stability. It is the exact sequence that matters, not an average of an ensemble. Particular base pair combinations confer to the whole region a particular structure that influences the conformation in the adjacent TATA box. Similar long-range effects have been previously observed in sequences flanking A-tracts (30). Furthermore, at many positions there is more than one choice of significant long-range structural motifs (mono to tetranucleotides). This means that optimal flanking sequences are not unique. There can be several base pair combinations that will result in an optimal binding site, as can be observed in the flanking sequences of the individual examples studied from each DNA pool (Table 1).
We looked for correlations between observed stabilities of TBP/TATA-box complexes and various published parameter sets for bending and/or bendability at the tri- or tetranucleotide level (50,51) and could not find any. This may be another indication that the structural properties of the sequences flanking TATA boxes are derived from the characteristics of the entire region, or at least a region larger than a tetranucleotide. We are currently examining the global structure and mechanical properties of TATA boxes and their flanking sequences using cyclization kinetics of minicircles (52).
Comparison to natural promoters containing the MLP and E4 TATA boxes
The information content of natural promoters containing the MLP-like TATA box is slightly higher than that of the two selected pools containing the MLP TATA box (Table 2). This may be due to additional information that is located in the sequences flanking the core 8 bp TATA box in natural promoters (the core TATA box is invariant and hence the information content is a function of the pool size only). We show here that binding of TBP to MLP-like TATA boxes is not very sensitive to the nature of the flanking sequences. Thus, the additional information at natural promoters may not be related to TBP binding per se, but to binding of other factors involved in transcription initiation, such as TFIIB. Indeed, upstream of the MLP TATA box in eukaryotic promoters as well as in human promoters there is a guanine nucleotide at position −3 relative to the TATA box (in 44 and 60% of the eukaryotic and human promoters, respectively), which is highly conserved in BRE sites (53). On the other hand, we note that the sequence pattern surrounding the E4 TATA box selected for binding stability is very similar to the sequence pattern surrounding natural MLP promoters (eukaryotic and human). Thus, it could be that the selected sequence pattern converged to the pattern observed in the strong MLP promoter. Indeed, Wolner and Gralla (12) showed that exchanging the flanking sequences surrounding the E4 promoter by those surrounding the MLP promoter decreased the dissociation of TBP from this hybrid promoter.
E4-like targets, selected for high binding affinity or high kinetic stability, have higher information content than natural eukaryotic promoters containing the E4 box (Table 2). This can mean that the ability to modulate the level of TBP binding as a function of the flanking sequences is not always used in natural promoters to the extent observed under in vitro conditions. However, it could also mean that the low sensitivity of MLP-like promoters to the identity of the flanking sequences with respect to TBP binding enables the information on the binding of other factors to come forth. High information content usually indicates a binding site for a unique protein factor. In the flanking sequences of E4-like TATA boxes there may be sequence information for the binding of various protein activators (18), and also for the modulation of TBP binding. This overlap of information may preclude its manifestation through the information content parameter.
Implications for gene regulation
The ability to modulate the binding of TBP to TATA boxes, as a function of the flanking sequences, can be used to change the TATA-box identity. Since TBP contact TATA boxes using 8 bp only, which are mostly A,T-rich, the number of different sequence combinations is not that large and is much lower than the number of genes in the human genome (or even genes that contain a TATA box). Using the flanking sequences is one approach to extent the specificity of TBP/TATA-box recognition, without the need to directly contact more than 8 bp.
We could not surpass the binding stability of the wild-type AdMLP target (wtMLP, Table 1). Thus, even though the effect of the flanking sequences on MLP TATA-boxes is about 30% at most, all the sequences studied individually show the same or reduced binding stability relative to wtMLP. Hence, it seems that Nature has optimized the intrinsic binding stability of AdMLP. The opposite situation is observed at the AdE4 site, which was selected to be a weak binding site in adenoviruses. However, the AdE4 promoter is a weak basal promoter not because of the (T-A)4 sequence, but because of the choice of base pairs in the flanking sequences. This allows the added level of regulation through the binding of protein activators to these same regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Z.S. Juo (Yale) for purified yTBPc. We thank Martin Ackerman and Yael Mandel-Gutfreund for help with the statistical calculations. We thank Mike Fried for critical reading and helpful discussions. This work was supported by the Israel Science Foundation (T.E.H.). Funding to pay the Open Access publication charges for this article was provided by funds for the promotion of research in the Technion.
Conflict of interest statement. None declared.
REFERENCES
- 1.Seeman N.C., Rosenberg J.M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D.K., Horikoshi M., Roeder R.G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- 3.Starr D.B., Hawley D.K. TFIID binds in the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- 4.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. Duality of TBP, the universal transcription factor. Nature. 1994;263:1103–1104. doi: 10.1126/science.8108728. [DOI] [PubMed] [Google Scholar]
- 7.Burley S.K. The TATA box binding protein. Curr. Opin. Struct. Biol. 1996;6:69–75. doi: 10.1016/s0959-440x(96)80097-2. [DOI] [PubMed] [Google Scholar]
- 8.Burley S.K., Roeder R.G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Stargell L.A., Struhl K. Mechanism of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 10.Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Ann. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 11.Bareket-Samish A., Cohen I., Haran T.E. Signals for TBP/TATA box recognition. J. Mol. Biol. 2000;299:965–977. doi: 10.1006/jmbi.2000.3797. [DOI] [PubMed] [Google Scholar]
- 12.Wolner B.S., Gralla J.D. TATA-flanking sequences influence the rate and stability of TBP and TFIIB. J. Biol. Chem. 2001;275:6260–6266. doi: 10.1074/jbc.M008273200. [DOI] [PubMed] [Google Scholar]
- 13.Crothers D.M., Haran T.E., Nadeau J.G. Intrinsically bent DNA. J. Biol. Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 14.Haran T.E., Kahn J.D., Crothers D.M. Sequence elements responsible for DNA curvature. J. Mol. Biol. 1994;244:135–143. doi: 10.1006/jmbi.1994.1713. [DOI] [PubMed] [Google Scholar]
- 15.Yuan H., Quintana J., Dickerson R.E. Alternative structures for alternating poly(dA–dT) tracts: the structure of the B-DNA decamer C-G-A-T-A-T-A-T-C-G. Biochemistry. 1992;31:8009–8021. [PubMed] [Google Scholar]
- 16.Dickerson R.E., Goodsell D.S., Neidle S. ‘…the tyranny of the lattice…’. Proc. Natl Acad. Sci. USA. 1994;91:3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Librizzi M.D., Brenowitz M., Willis I.M. The TATA element and its context affect the cooperative interaction of TATA-binding protein with the TFIIB-related factor, TFIIIB70. J. Biol. Chem. 1998;273:4563–4568. doi: 10.1074/jbc.273.8.4563. [DOI] [PubMed] [Google Scholar]
- 18.Wolner B.S., Gralla J.D. Roles for non-TATA core promoter sequences in transcription and factor binding. Mol. Cell. Biol. 2000;20:3608–3615. doi: 10.1128/mcb.20.10.3608-3615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Kim J.L., Nikolov D.B., Burley S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 21.Nikolov D.B., Hu S.H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N.H., Roeder R.G., Burley S.K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 22.Nikolov D.B., Burley S.K. 2.1 A resolution refined structure of a TATA box-binding protein (TBP) Nature Struct. Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- 23.Chasman D.I., Flaherty K.M., Sharp P.A., Kornberg R.D. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl Acad. Sci. USA. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y., Geiger J.H., Hahn S., Sigler P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.L., Burley S.K. 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nature Struct. Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- 26.Patikoglou G.A., Kim J.L., Sun L., Yang S.H., Kodadek T., Burley S.K. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolov D.B., Chen H., Halay E.D., Hoffman A., Roeder R.G., Burley S.K. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl Acad. Sci. USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juo Z.S., Chiu T.K., Leiberman P.M., Baikalov I., Berk A.J., Dickerson R.E. How proteins recognize the TATA box. J. Mol. Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 29.Starr D.B., Hoopes B.C., Hawley D.K. DNA bending is an important component of site-specific recognition by the TATA binding protein. J. Mol. Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 30.Merling A., Sagaydakova N., Haran T.E. A-tract polarity dominate the curvature in flanking sequences. Biochemistry. 2003;42:4978–4984. doi: 10.1021/bi020662w. [DOI] [PubMed] [Google Scholar]
- 31.Bareket-Samish A., Cohen I., Haran T.E. Repressor assembly at trp binding sites is dependent on the identity of the intervening dinucleotide between the binding half sites. J. Mol. Biol. 1997;267:103–117. doi: 10.1006/jmbi.1996.0826. [DOI] [PubMed] [Google Scholar]
- 32.Brenowitz M., Jamison E., Majumdar A., Adhya S. Interaction of the Escherichia coli Gal repressor protein with its DNA operators in vitro. Biochemistry. 1990;29:3374–3383. doi: 10.1021/bi00465a033. [DOI] [PubMed] [Google Scholar]
- 33.Bailey T.L., Elkan C., editors. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, CA: AAAI Press; 1994. [PubMed] [Google Scholar]
- 34.Haran T.E. Statistical and structural analysis of trp binding sites: comparison of natural and in vitro selectd sequences. J. Biomol. Struct. Dyn. 1998;15:689–701. doi: 10.1080/07391102.1998.10508985. [DOI] [PubMed] [Google Scholar]
- 35.Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg O.G., von Hippel P.H. Selection of DNA binding sites by regulatory proteins. Statistical–mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 37.Schneider T.D., Stormo G.D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 38.Cox J.M., Hayward M.M., Sanchez J.F., Gegnas L.D., van der Zee S., Dennis J.H., Sigler P.B., Schepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc. Natl Acad. Sci. USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkhurst K.M., Richards R.M., Brenowitz M., Parkhurst L.J. Intermediate species possessing bent DNA are present along the pathway to formation of a final TBP-TATA complex. J. Mol. Biol. 1999;289:1327–1341. doi: 10.1006/jmbi.1999.2835. [DOI] [PubMed] [Google Scholar]
- 40.Powell R.M., Parkhurst K.M., Brenowitz M., Parkhurst L.J. Marked stepwise differences within a common kinetic mechanism characterize TATA-binding protein interactions with two consensus promoters. J. Biol. Chem. 2001;276:29782–29791. doi: 10.1074/jbc.M104099200. [DOI] [PubMed] [Google Scholar]
- 41.Powell R.M., Parkhurst K.M., Parkhurst L.J. Comparison of TATA-binding protein recognition of a variant and consensus DNA promoters. J. Biol. Chem. 2002;277:7776–7784. doi: 10.1074/jbc.M110147200. [DOI] [PubMed] [Google Scholar]
- 42.Fessler S.P., Young C.S. Control of adenovirus early gene expression during the late phase of infection. J. Virol. 1998;72:4049–4056. doi: 10.1128/jvi.72.5.4049-4056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginsberg H.S., editor. Vol. 1. New York: Plenum; 1984. The adenoviruses. [Google Scholar]
- 44.Yanagi K., Prive G.G., Dickerson R.E. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J. Mol. Biol. 1991;217:201–214. doi: 10.1016/0022-2836(91)90620-l. [DOI] [PubMed] [Google Scholar]
- 45.Quintana J.R., Grzeskowiak K., Yanagi K., Dickerson R.E. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. Mol. Biol. 1992;225:379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- 46.Haran T.E., Crothers D.M. Cooperativity in A-tract structure and bending properties of composite TnAn blocks. Biochemistry. 1989;28:2763–2767. doi: 10.1021/bi00433a003. [DOI] [PubMed] [Google Scholar]
- 47.Wong J.M., Bateman E. TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saenger W. Principles of Nucleic Acid Structure. In: Cantor C.R., editor. Forces stabilizing associations between bases: hydrogen bonding and base stacking, Springer Advanced Texts in Chemistry. NY: Springer-Verlag; 1984. [Google Scholar]
- 49.Nadeau J.G., Crothers D.M. Structural basis for DNA bending. Proc. Natl Acad. Sci. USA. 1989;86:2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brukner I., Sanchez R., Suck D., Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Packer M.J., Dauncey M.P., Hunter C.A. Sequence-dependent DNA structure: tetranucelotide conformational maps. J. Mol. Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Crothers D.M. High-throughput approach for detection of DNA bending and flexibility based on cyclization. Proc. Natl Acad. Sci. USA. 2003;100:3161–3166. doi: 10.1073/pnas.0530189100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.