Abstract
In Schizosaccharomyces pombe the RNAi machinery and proteins mediating heterochromatin formation regulate the transcription of non-coding centromeric repeats. These repeats share a high sequence similarity with telomere-linked helicase (tlh) genes, implying an ancestral relationship between the two types of elements and suggesting that transcription of the tlh genes might be regulated by the same factors as centromeric repeats. Indeed, we found that mutants lacking the histone methyltransferase Clr4, the Pcu4 cullin, Clr7 or Clr8, accumulate high levels of tlh forward and reverse transcripts. Mutations and conditions perturbing histone acetylation had similar effects further demonstrating that the tlh genes are normally repressed by heterochromatin. In contrast, mutations in the RNAi factors Dcr1, Ago1 or Rdp1 led only to a modest derepression of the tlh genes indicating an alternate pathway recruits heterochromatin components to telomeres. The telomere-binding protein Taz1 might be part of such a redundant pathway, tlh transcripts being present at low levels in Δtaz1 mutants and at higher levels in Δtaz1 Δdcr1 double mutants. Surprisingly, the chromodomain protein Chp1, a component of the Ago1-containing RITS complex, contributes more to tlh repression than Ago1, indicating the repressive effects of Chp1 are partially independent of RITS. The tlh genes are found in the subtelomeric regions of several other fungi raising the intriguing possibility of conserved regulation and function.
INTRODUCTION
The heterochromatic regions associated with the centromeres of many higher eukaryotes contain an abundance of repeats originating from transposable elements. The epigenetic association of these centromeric regions with particular sets of proteins is in some respects more critical to centromere function than specific DNA sequences as indicated e.g. by the properties of neocentromeres [reviewed in (1)].
In spite of their relatively small size of ∼40–100 kb, the three Schizosaccharomyces pombe centromeres resemble those of higher eukaryotes [reviewed in (2)]. Their central cores (cnt) are associated with the CENH3/CenpA protein, a hallmark of eukaryotic centromeres defining the region of kinetochore attachment. The central cores are flanked by repeated sequences packaged into heterochromatin, the inner most (imr) and outer repeats (otr). The otr can in turn be subdivided into two types of repeats whose relative positions differ between the three centromeres (3–8), designated here dh and dg repeats (Figure 1A). Both inner and outer repeats are required for efficient chromosome segregation (9,10) as are components of heterochromatin [reviewed in (2)]. The origin of centromeric repeats is not known.
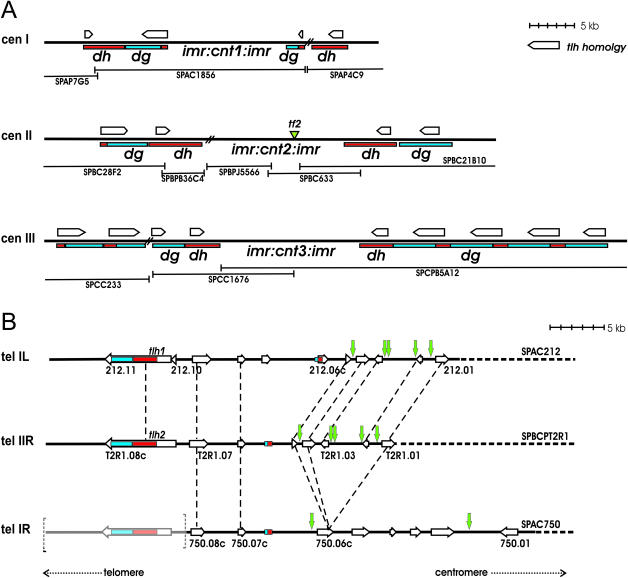
Figure 1.
Regions of sequence homology in S.pombe centromeres and subtelomeres. (A) Schematic representation of S.pombe centromeres. dh and dg repeats are indicated in red and blue, respectively. Regions of homology (>60% sequence identity) between tlh genes and centromeric repeats are indicated by open arrows. These regions map within 3921 bp in the 5662 bp tlh1 ORF, 1102 bp (seven segments; <2% gap) of tlh1 displaying an identity to centromeric sequences >75%. Centromeric repeats that have not been sequenced are indicated by double bars. dh and dg repeats were placed according to published data (8) and our own sequence comparisons. The green arrowhead indicates a previously unidentified fragment from a tf2-retrotransposable element. Available cosmid sequences (8) are represented by bars below each centromere. (B) Schematic representation of S.pombe subtelomeric regions. The subtelomeric cosmids SPAC212, SPBCPT2R1 and SPAC750 are represented. Dashed lines between SPAC212, SPBCPT2R1 and SPAC750 point to regions of synteny. The tlh homology to centromeric repeats is represented in red (dh) or blue (dg). Green arrows indicate the presence of LTRs. Open arrows denote ORFs. Bracketed grey lines in the left portion of tel1R represent a hypothetical tlh gene that is not part of the SPAC750 sequence. Its presence and position are suggested by hybridization experiments (35) and by the syntenic relationship between the three represented telomeres (see text).
A dh-dg repeat unlinked to centromeres is found in the silenced part of the mating-type region. This particular element has been designated cenH, for centromeric homology (11). cenH facilitates heterochromatin formation in the mating-type region by recruiting the histone methyltransferase (HMT) Clr4 and the chromodomain protein Swi6 (12–15). All or parts of cenH can also promote heterochromatin formation at an ectopic site (16) as can fragments derived from centromeric DNA (17).
The centromeric dh and dg repeats and cenH have been implicated in RNAi-dependent heterochromatin formation (15,18–20). In S.pombe the RNAi pathway involves a member of the Argonaute family (Ago1), an RNase III-like enzyme (Dcr1) and an RNA-dependent RNA polymerase (Rdp1). Ago1 is part of a ribonucleoprotein complex called RITS for RNA-induced initiation of transcriptional silencing complex. In addition to Ago1, RITS contains the Tas3 protein, the chromodomain protein Chp1 and 20–22 nt RNA molecules (siRNA) mostly originating from centromeric dh and dg repeats (21–23). Rdp1 participates in a different complex, the RNA-directed RNA polymerase complex or RDRC, capable of interacting with RITS and containing the RNA helicase Hrr1 and Cid12, a member of the poly(A) polymerase family (22). Rdp1, Dcr1, Ago1, Chp1, Tas3, Hrr1 and Cid12 are major players in the formation of centromeric heterochromatin (19,21,22,24–26). According to all available evidence, double-stranded RNAs produced by either the bi-directional transcription of centromeric repeats, or by RDRC acting on centromeric transcripts, are processed into siRNA by Dcr1 and one strand is incorporated into the RITS complex. Early models proposed that the primary function of RITS and its components was to direct heterochromatin formation to chromosomal regions with homology to the small single-stranded RNA molecules contained in the complex (15,21). It has become increasingly clear however that the association of RITS with chromatin depends on histone H3 lysine 9 methylation (H3K9Me) whereas H3K9Me can be formed in cells lacking RNAi components (27,28). The current view is therefore that H3K9Me serves as an anchor for Chp1, tethering RITS and RDRC to heterochromatin and allowing these complexes to propagate existing heterochromatin rather than, or in addition to, establishing its formation (17,22,29,30). Histones are deacetylated in the marked regions by the Clr3 and Clr6 deacetylases and histone H3 is methylated at K9 by the Clr4 methyltransferase, the molecular mechanisms linking these events to RITS or RDRC remaining unknown.
The RITS component Chp1 associates not only with centromeric heterochromatin but also with the mating-type region and telomeres (23,28,29). In contrast to its role at centromeres, however, Chp1 is not essential to heterochromatin formation or transcriptional silencing in the mating-type or telomeric regions (25,28,29). In the mating-type region, Chp1 facilitates silencing in a manner redundant with the DNA-binding protein Atf1 (31,32). Similarly, Chp1 might promote silencing in telomeric and subtelomeric regions in a manner redundant with some other factor, possibly the telomeric-repeat binding protein Taz1 (33).
We report here our characterization of subtelomeric sequences sharing a high similarity with the dh-dg centromeric repeats and cenH element. These sequences are part of the open reading frames (ORFs) of putative telomere-linked helicases (Tlh) of the RecQ family. Expression of the tlh genes is induced in cells undergoing telomere crisis caused by the loss of telomerase (34). Furthermore, the ectopic expression of part of a tlh ORF speeds up recovery from crisis (35) indicating a role in telomere rescue under this extreme condition. The level of tlh RNA is naturally low in wild-type cells that are not undergoing crisis and this low level has been proposed to be brought about by RNAi (34). We examined the transcriptional regulation of the tlh genes in greater detail, determining the relative contributions of histone-modifying enzymes, RNAi components, and of the telomere-binding protein Taz1 to their expression level. Furthermore, we determined that related ORFs are present in the subtelomeric regions of other fungi, suggesting an evolutionary-conserved function and mode of regulation for the tlh genes.
MATERIALS AND METHODS
S.pombe strains and media
The strains used in this study and their genotypes are listed in Table 1. Media were as described previously (13).
Table 1.
Strains and their genotypes
| J11 | h+Nleu1-32 ura4-D18 ade6-M216 taz1::ura4+ |
| PG2870 | h+Nleu1-32 ura4-D18 ade6-M210 chp1::LEU2 |
| PG3039 | h−Sura4-D18 dcr1::kanR |
| PG3267 | h−Sleu1-32 ade6-DN/N [ura4+-tlh2-ade6+] |
| PG3269 | h−Sleu1-32 ade6-DN/N [ura4+-2hlt-ade6+] |
| PG3273 | h−Sleu1-32 ade6-DN/N [ura4+-tlh1-ade6+] |
| PG3275 | h−Sleu1-32 ade6-DN/N [ura4+-1hlt-ade6+] |
| PG3389 | h+Notr1R(SphI)::ura4+ leu1-32 ura4-DS/E ade6-M210 clr8Δ::kanR |
| PG3435 | h90ura4-D18 pcu4::ura4+ |
| PTI3 | h90leu1-32 ura4-DS/E ade6-DN/N |
| PTI14 | h−Sleu1-32 ade6-DN/N [ura4+-ade6+] |
| SPA15 | h−Sotr1R(SphI)::ura4+leu1-32 ura4-DS/E ade6-M210 clr7Δ::kanR |
| SPK10 | h−S |
| SPK20 | h−Sura4-D18 clr4::ura4 |
| SPK27 | h−Sclr6-1 clr3::kanR |
| SPK50 | h+Nura4-D18 leu1-32 dcr1::kanR |
| SPK51 | h−Nura4-D18 ade6-M216 taz1:: ura4+ |
| SPK52 | h+Nura4-D18 leu1-32 dcr1::kanR |
| SPK53 | h−Nura4-D18 ade6-M216 taz1:: ura4+ |
| SPK54 | h−Sura4-D18 ade6-M216 |
| SPK55 | h+Nura4-D18 leu1-32 dcr1::kanR taz1:: ura4+ |
| SPK56 | h+Nura4-D18 ade6-M216 leu1-32 |
| SPK57 | h−Sura4-D18::kanR taz1:: ura4+ |
| SPK61 | h−Sura4-D18 ade6-M216 dcr1::kanR taz1:: ura4+ |
| TV292 | h−Sura4-DS/E ago1::kanR |
| TV293 | h−Sura4-DS/E dcr1::kanR |
| TV296 | h−Sura4-DS/E rdp1::kanR |
Cultures for RNA extractions
Unless indicated otherwise, cells were propagated in 25–50 ml YES medium at 30°C and collected while in their exponential growth phase by a 3 min centrifugation at 3000 r.p.m. in a Heraeus megafuge 2R. The cell pellets were frozen in liquid nitrogen immediately after centrifugation and stored at −80°C until further processing. For the nitrogen starvation experiments, 200 ml cultures growing exponentially in YES (∼5 × 106 cells/ml) were spun down by a 3 min centrifugation at 3000 r.p.m. The cells were washed once in 25 ml MSL-N pre-warmed to 30°C, resuspended at ∼5 × 106 cells/ml in MSL-N supplemented with 100 mg/l adenine, 100 mg/l uridine and 200 mg/l leucine, and placed back on a shaking platform at 30°C. At each time point ∼1 × 108 cells were collected by centrifugation for RNA extraction.
Treatments with Trichostatin A (TSA)
TSA treatments were performed by adding 35 µg/ml TSA (US Biologicals) to exponentially growing 5 ml YES cultures that had reached ∼5 × 106 cells/ml and incubating the cultures for 24 h on a shaking platform at 30°C. The cells were then spun down by a 3 min centrifugation at 3000 r.p.m., washed once in 10 ml YES pre-warmed to 30°C, and used to inoculate 25 ml YES recovery cultures. Cells in recovery cultures were examined daily under a microscope, counted and used to inoculate fresh cultures. The cultures were diluted ∼250-fold each day. Cells (∼1 × 108) were collected for RNA extractions at the indicated time points. In the case of PG3267 and PG3273 suspensions of cells in their repressed state were divided in two. One half was treated with TSA for 24 h as described above while the other half was mock treated. Following treatment, cells were plated on AA medium lacking adenine or containing 15 mg/l adenine. The plates were incubated at 33°C for 4 days and photographed. The experiment was conducted in triplicate using three independent cultures of each PG3267 and PG3273.
Plasmid and strain constructions
pPTI1, a plasmid containing a full-length ade6+ gene flanked by DNA originating from the ura4+ locus, was created by ligating into Bluescript SKII(−) (Stratagene) (i) a 645 bp SacI–EagI fragment created by PCR with GTO-212 (GGGAGCTCGAGGGTATTATACAAGGC), GTO-213 (GGACGGCCGTGTGGTAATGTTGTAGGAG) and a 1.8 kb HindIII template containing ura4+ and flanking sequences (36); (ii) the 2735 bp EaeI–SpeI fragment of pAS1 (37) containing the ade6+ gene, and (iii) a 201 bp SpeI–KpnI fragment created by PCR with GTO-214 (GGACTAGTATCTTTTCTCTGGATTGAC), GTO-215 (GGGGTACCTGACGAAACTTTTTGACATC) and the 1.8 kb ura4+ template. The predicted ORFs of tlh1 and tlh2 were amplified from genomic DNA of S.pombe 968 cells (h90) using OKR63 (TTAGGCCTCGGCCGATGGTCGTCGCTTCAGAAATTGC) and OKR64 (TTAGGCCTCGGCCGCGATATGAAACGTTGTCTTGCATCAG) or OKR65 (TTAGGCCTCGGCCGGCGTGTACTATGGCAACTGGTGGTGTTGGCTTG) and OKR64 and the Expand PCR system (Roche). The PCR products were cloned into the pCRII-TOPO vector (Invitrogen), excised by EagI digestion, and inserted in either orientation into the EagI site of pPTI1 to create pKR44 (OKR63-OKR64 fragment with OKR64 close to ura4+); pKR45 (OKR63-OKR64 fragment with OKR63 close to ura4+); pKR48 (OKR64-OKR65 fragment with OKR64 close to ura4+); and pKR49 (OKR64-OKR65 fragment with OKR65 close to ura4+). The four plasmids and pPTI1 were digested with SacI and KpnI to release their inserts and used to transform the S.pombe strain PTI1. Ade+ transformants were selected on AA-ade plates. To take into account the possibility that the tlh genes might repress transcription of ade6+, both fast and slow growing Ade+ transformants were examined by Southern blot. Several transformants containing correct chromosomal integrations at the ura4+ locus were identified in this way for each construct including PG3267 with pKR49 (ura4+-tlh2-ade6+), PG3269 with pKR48 (ura4+-2hlt-ade6+), PG3273 with pKR45 (ura4+-tlh1-ade6+), PG3275 with pK44 (ura4+-2hlt-ade6+), and PTI14 with pPTI1 (ura4+-ade6+).
Microscopy
DIC images were obtained using a Zeiss Axio Imager fluorescence microscope equipped with a Hamamatsu Orca-ER digital camera and Volocity 3.5.1 software.
RNA extraction, RT–PCR and real-time PCR
RNA was extracted with a hot-phenol procedure as described previously (38). Detection of centromeric transcripts by RT–PCR was performed as in Ref. (19) using GTO-223 (GAAAACACATCGTTGTCTTCAGAG) and GTO-226 (TCGTCTTGTAGCTGCATGTGA). tlh dh transcripts were amplified with OKR40 (TCGTCTTGTAGCAGCATGTGA) and OKR41 (GAGATGAACGTATCTCTATCGAC); tlh DEAH transcripts with OKR44 (CTGGAGGTGGAAAGTCTTTGTCG) and OKR45 (AACCGAAGACCACTGTTGGTTAATGC); actin transcripts with OKR70 (GGCATCACACTTTCTACAACG) and OKR71 (GAGTCCAAGACGATACCAGTG). OKR70 and OKR71 were also used for −RT controls. Strand specificity was achieved by using a single primer in the reverse transcription: GTO-226 for the centromeric forward transcript, GTO-223 for the centromeric reverse transcript; OKR41 for the tlh dh forward transcript; OKR40 for the tlh dh reverse transcript; OKR45 for the tlh DEAH forward transcript; and OKR44 for the tlh DEAH reverse transcript. For the amplification of centromeric or telomeric transcripts 27 PCR cycles were performed, and 25 cycles for the amplification of actin transcripts.
Real-time PCR was performed with a BioRad iCycler using the Quantitect SYBR Green RT–PCR mixture (Qiagen) supplemented with 10 nM Fluorescein (BioRad). Reverse transcriptions of ∼90 ng total RNA were allowed to take place at 50°C for 30 min in 25 µl reactions. The reverse transcription step was followed by 15 min at 95°C to inactivate the RT and activate the HotStarTaq polymerase. cDNAs were then amplified by 40 cycles of (30s 94°C; 45s 56°C; 60s 72°C). The OKR40 and OKR41 primers were used for tlh amplification, and OKR70 and OKR71 for actin amplification. All primers were used at a final concentration of 0.6 µM. The melting curve of each sample was determined after amplification. Reactions were set up in triplicate. Three-fold dilution series of genomic DNA were used to determine the efficiency of amplification for each primer pair by plotting standard curves. Mean normalized expression (MNE) values were calculated according to Equation 1 and standard errors on the mean (SEMNE) according to Equation 2 (39).
| 1 |
| 2 |
where Ereference is the efficiency of actin amplification, Etarget the efficiency of tlh amplification, CTreference, mean the mean cycle threshold value for actin, and CTtarget, mean the mean cycle threshold value for tlh.
DNA and protein sequence analyses
Sequence analyses were performed using online available BLAST (40) services from the Sanger Institute (www.sanger.ac.uk), Broad Institute (www.broad.mit.edu), Genolevures (cbi.labri.u-bordeaux.fr/Genolevures) and NCBI (www.ncbi.nlm.nih.gov). Multiple alignments were created using ClustalW (41) at EBI (www.ebi.ac.uk). The cladogram presented in Figure 2 was manually edited using NJPLOT (42).
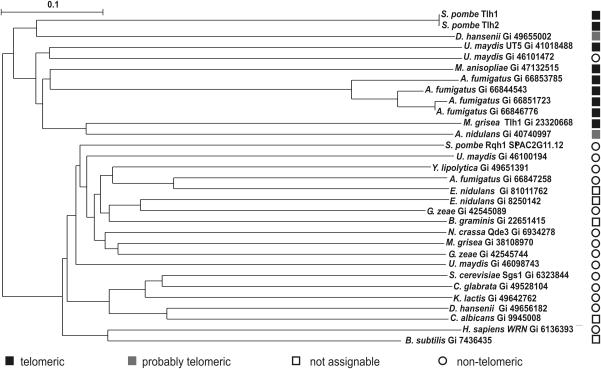
Figure 2.
Conservation of tlh genes in fungi. The displayed subtree was created from a full alignment including the product of 58 S.pombe helicase genes and 42 RecQ helicases from other species (see text). Black squares mark the products of telomeric ORFs (according to annotations); grey squares mark ORFs that are first or second in contigs >500 kb; open circles mark ORFs that are more than two ORFs away from the end of a DNA segment; and open squares mark single ORFs not assigned to any segment, or bacterial ORFs.
RESULTS AND DISCUSSION
The centromeric dh and dg repeats share sequence homology with subtelomeric helicase genes
The dh and dg repeats found at centromeres and in the mating-type region display extensive sequence homology with large telomeric ORFs encoding putative helicases of the recQ family (Figure 1) (8,34). The telomere-linked helicase (tlh) ORFs are the last ORFs in the subtelomeric regions of chromosome I (SPAC212.11 or tlh1) and chromosome II (SPBCPT2R1.08c or tlh2). In addition to the high sequence similarity shared by tlh1 and tlh2 a significant conservation is observed between the ORFs neighboring tlh1 and tlh2 indicating the two subtelomeric regions resulted from a duplication (Figure 1B). A region partly similar to the duplicated sequence is also found in the right subtelomere of chromosome I suggesting that ORFs homologous to tlh1/tlh2 might exist there as well in a region whose sequence has not been completed yet. This hypothesis is supported by hybridization experiments (34). Similar ORFs are not present in the sequenced chromosome III, the telomeric repeats in chromosome III being immediately adjacent to rDNA repeats (8).
The sequence similarity between tlh1 and tlh2 is greater than 99% in a 5757 nt long region. The high sequence conservation between tlh1 and tlh2 might be maintained by frequent gene conversions between subtelomeric regions that are known to cluster early in meiosis promoting the alignment of homologous chromosomes [reviewed in (43)]. Consistent with frequent physical exchanges during meiosis, we observed 3:1 segregations of restriction fragment length polymorphisms at the tlh genes in tetrad dissections (data not shown).
In addition to SPAC212.11 and SPBCPT2R1.08c, truncated tlh copies are present near the full-length genes in the two subtelomeric regions. These truncated ORFs are unlikely to encode functional proteins. However, they might serve as structural components similar to the otr repeats at centromeres.
Conservation of tlh genes in fungi
We used ClustalW to compare Tlh1 with a set of related proteins. This set comprised 40 protein sequences identified in a BLAST search performed with Tlh1 against the Fungi database at NCBI (Matrix: BLOSUM62; Gap Costs: Existence: 11, Extension:1; E-value ≤ 1 × 10−13), the protein products of all S.pombe helicase genes (58 sequences), RecQ from Bacillus subtilis, and the human RecQ homologue WRN. All RecQ helicases clustered in a subtree apart from the non-recQ helicases of S.pombe (data not shown). The RecQ subtree could be divided into two main branches (Figure 2), one containing among others human WRN and S.pombe Rqh1, and the other primarily containing proteins encoded by telomere-linked genes: S.pombe Tlh1 and Tlh2; Ustilago maydis UT5 (44); Metarhizium anasopliae TAH1 (45); Magnaporthe grisea Tlh1 (46); and four telomeric helicases from Aspergillus fumigatus (Gi 66853785, 66844543, 66851723 and 66846776). In addition, the latter branch contained two proteins from Aspergillus nidulans (Gi 40740997) and Debaryomyces hansenii (Gi 49655002), respectively, both encoded by the second last ORF in segments >500 kb. According to their position in the contigs and sequence similarity with tlh1 and tlh2 these two genes might be novel tlh genes encoding homologues of S.pombe tlh1 and tlh2 in A.nidulans and D.hansenii.
The subtelomeric Y′ elements of Saccharomyces cerevisiae also encode helicases (47,48). The Y′ ORFs are very distantly related to the S.pombe tlh genes; however, they might fulfill a similar function since their expression is increased in a class of telomere-crisis survivors (48), as are the S.pombe tlh genes (34). In both cases the helicases encoded by the subtelomeric genes possibly contribute to the resolution of defective telomere structures. Y′ elements help counteract telomere erosion in other ways by being substrates for amplification (49). Their amplification occurs by unequal recombination events (50) or following mobilization by Ty1 retrotransposons (51) and it compensates for telomere loss. The plasticity of the tlh genes suggests that similar mechanisms might operate in S.pombe.
The tlh genes are strictly repressed during various growth conditions
We investigated whether the tlh genes were expressed under stresses other than the stress caused by the loss of telomeric DNA. A large proportion of subtelomeric genes are upregulated during nitrogen starvation and repressed by heterochromatin in the presence of a good nitrogen source (52,53) suggesting that the tlh genes might be under a similar control. We observed however that expression of the tlh genes did not increase upon nitrogen starvation (Supplementary Figure 1). We also measured tlh expression in wild-type cells entering stationary phase in either minimal medium (EMM2) or rich medium (YES). No increase in tlh gene expression occurred under these conditions either (Supplementary Figure 1). This tight repression distinguishes the tlh genes from other subtelomeric ORFs.
The tlh genes are repressed by heterochromatin
tlh1 and tlh2 displaying a strong sequence homology to well-characterized silenced regions, we investigated whether their expression was controlled by known silencing factors. tlh transcripts were easily detected in mutants lacking the histone acetyltransferases (HDACs) Clr3 and Clr6 or the HMT Clr4, establishing that the tlh genes are repressed by heterochromatin in wild-type cells (Figure 3).
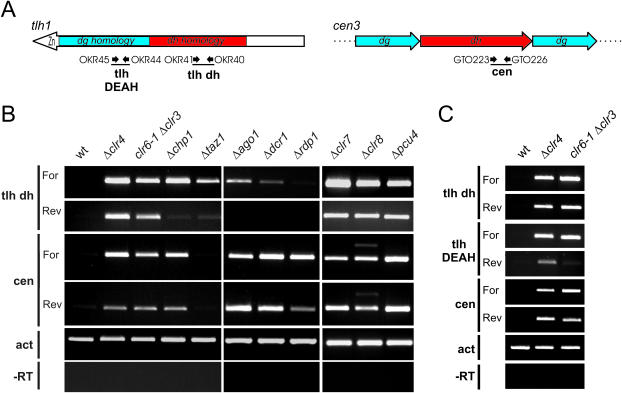
Figure 3.
Transcriptional regulation of the S.pombe tlh genes. (A) Schematic representation of the tlh1 gene and of a portion of centromere 3 indicating the location of primers used in (B and C) and fragments amplified with these primers. (B) Abundance of forward and reverse tlh transcripts in heterochromatin, RNAi and Taz1 mutants. Strand specific RT–PCR was performed to estimate the levels of tlh RNA by amplifying a region of tlh with high sequence similarity to the centromeric dh repeats (tlh dh). Reverse transcription was primed with OKR41 to detect the tlh forward strand (For), or with OKR40 to detect the tlh reverse strand (Rev). Centromeric forward (cen For) and reverse (cen Rev) RNAs were detected as described previously (19). The bands observed above the centromeric forward and reverse PCR products in the Δclr8 panels originate from the mating-type region in h90 strains. The actin transcript (act) was amplified to estimate the amount of total RNA in the samples. −RT was performed with actin primers. wt:SPK10; Δclr4:SPK20; clr6-1Δclr3:SPK27; Δchp1:PG2870; Δtaz1:J11; Δago1:TV292; Δdcr1:TV293; Δrdp1:TV296; Δclr7:SPA15; Δclr8:PG3389; Δpcu4:PG3435. (C) Variations in the abundance of reverse strand along the tlh ORF. RT–PCR was performed as in (A). In addition, a region of tlh RNA encoding the DEAH helicase domain (tlh DEAH) was amplified, the reverse transcription being primed with OKR45 to detect the forward strand (For) or with OKR44 to detect the reverse strand (Rev).
A protein complex containing the Pcu4 cullin, Clr7 and Clr8 was shown recently to mediate an early step in heterochromatin formation (54–57). This complex possesses ubiquitin-ligase activity (54) and it is required for H3K9Me at centromeres and in the mating-type region (54–57). We found that the transcriptional repression of the tlh genes was strongly dependent on Pcu4, Clr7 and Clr8 (Figure 3B). Deletion of any of these factors resulted in an increase in tlh forward and reverse transcript comparable with the increase in cells lacking Clr4. This derepression is consistent with Pcu4, Clr7 and Clr8 controlling histone methylation and transcription of the tlh genes similar to the control they exert in the mating-type and centromeric regions.
Transcripts originating from both strands could be detected in Δclr4, clr6-1Δclr3, Δpcu4, Δclr7 and Δclr8 mutants (Figure 3), reminiscent of the situation at centromeres (19). When using primers amplifying the region with highest dh homology (OKR40 and OKR41), we detected similar levels of forward and reverse strand transcripts in derepressed mutants (Figure 3C). However, when using primers further toward the 3′ end of the tlh genes where the sequence similarity to centromeric repeats is less pronounced (OKR44 and OKR45) we observed significantly lower levels of the reverse strand transcript than of the forward transcript, suggesting that reverse strand synthesis is initiated within the tlh ORF and primarily occurs near the strongest dh homology.
Derepresssion of the tlh genes by TSA
Treatment with the histone deacetylase inhibitor TSA caused an accumulation of tlh transcripts similar to the level observed in the clr6-1Δclr3 HDAC double mutant (Figure 4). This derepression further supports the notion that histone deacetylation is a critical determinant of tlh gene regulation. TSA treatment had a similar effect upon centromeric silencing, leading to an accumulation of non-coding transcripts from centromeric repeats (Figure 4).
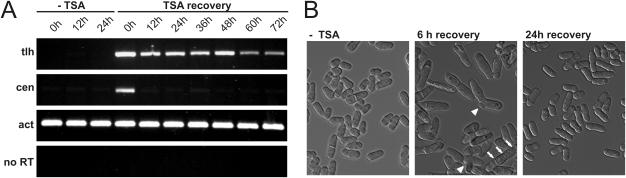
Figure 4.
Recovery from TSA treatment. (A) Centromeric and subtelomeric RNAs in cells recovering from TSA treatment. RT–PCR was performed on wild-type cells (SPK10) not treated with TSA (−TSA), or treated with TSA and maintained in culture for the indicated times following TSA removal (+TSA). (B) Normal cell morphology (−TSA) is perturbed by TSA treatment (6 h recovery) and re-established within a few generations following TSA removal (24 h recovery). White arrowheads point to cases of pseudohyphal growth and white arrows to multiple septa in a single cell.
Following TSA removal, the kinetics of repression differed markedly at centromeres and telomeres. Centromeric silencing was re-established within 12 h, whereas the tlh genes were still expressed as long as 72 h after TSA removal. Dramatic changes in cell morphology accompanied TSA treatment. Six hours after TSA removal nearly all cells were elongated with multiple septa and a substantial fraction were undergoing pseudo-hyphal growth. These morphological changes likely reflect global changes in gene expression resulting from the loss of HDAC activity (53). Cell morphology had returned to normal 24 h after TSA removal indicating the global effects of TSA treatment had faded at that time, consistent with the restored centromeric silencing. Hence, the long-term derepression of the tlh genes by TSA treatment appears very specific for these genes. This effect points to histone deacetylation as a key epigenetic mark controlling the expression of the tlh genes, and to an inefficient mechanism of heterochromatin establishment in subtelomeric regions.
Heterochromatin formation at the tlh loci
The levels of tlh transcripts were reported recently to be controlled by RNAi (34). We found that in contrast to the deletion of histone-modifying enzymes, deletion of the RNAi components Ago1 or Dcr1 caused a relatively minor accumulation of the forward tlh transcript, the reverse strand transcript remaining undetectable (Figure 3B). Strains lacking Rdp1 did not display a noticeable increase in tlh forward or reverse transcripts. This is in sharp contrast with the effects of rdp1 deletion at centromeres where non-coding transcripts accumulate (Figure 3B) (19).
Ago1 associates with the chromodomain protein Chp1 in the RITS complex (21). We assayed therefore the effect of Chp1 on tlh RNA levels in our attempts to assess the role of RNAi in the control of tlh expression. Previous reports showed that Chp1 is present at the tlh loci in wild-type cells, together with the H3K9Me mark and Swi6 (23,28). H3K9Me and Swi6 remain associated with tlh, and telomeric reporter genes remain silent in Δchp1 cells, supporting models where a repressive chromatin structure is formed in the absence of Chp1 (23,25,28). Surprisingly considering these previous observations, we found that forward tlh transcripts were relatively abundant in Δchp1 cells (Figure 3A). The abundance of tlh transcripts in Δchp1 cells in spite of the presence of Swi6 at the tlh genes indicates that forward transcription of the tlh genes is partially refractory to transcriptional silencing by Swi6. Surprisingly as well, the derepression caused by deleting Chp1 was more pronounced that the derepression caused by deleting Ago1 suggesting that Chp1 does not act solely in the context of RITS. One common effect of deleting Chp1 or RNAi components was that reverse transcripts were not detected in these backgrounds.
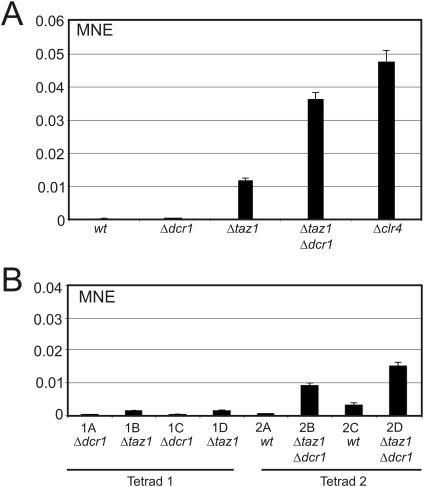
Since RNAi did not appear to be the sole, or even predominant, mechanism recruiting heterochromatin to subtelomeric regions we tested other candidates. The DNA-binding protein Taz1 associates with telomeric repeats and represses reporter genes artificially introduced in regions immediately adjacent to telomeric repeats (33,58). Thus, it appeared likely that Taz1 might affect tlh expression as well, possibly by recruiting silencing factors to the region. Indeed, we observed an upregulation of tlh transcripts in Δtaz1 cells, primarily of the forward transcript (Figure 3B). The transcriptional repression mediated by Taz1 is therefore not restricted to the immediate vicinity of telomeres but also affects genes located ∼10 kb away from the telomeric repeats. This observation supports a model where a repressive chromatin structure nucleated by Taz1 bound to telomeric repeats spreads from the telomeres into adjacent subtelomeric regions. The derepression of the tlh genes in Δtaz1 cells was not as strong as the derepression in clr6-1Δclr3 or Δclr4 cells (Figure 3B) indicating a Taz1-independent pathway mediates tlh repression in addition to Taz1. Indeed, we found that deleting both taz1 and dcr1 had a cumulative derepressive effect on the tlh genes (Figure 5A). We further examined the expression of the tlh genes in the progeny of a cross between a Δtaz1 and a Δdcr1 strain (Figure 5B). A slight derepression of the tlh genes was observed in some of the wild-type progeny (i.e. spore 2C in Figure 5B). This slight derepression reveals that silencing is slowly re-established after having been compromised by a trans-acting mutation, consistent with the slow re-establishment observed in cells recovering from TSA treatment (Figure 4). The tlh transcript levels were consistently higher in Δtaz1Δdcr1 double mutants than in single mutants reinforcing the conclusion that RNAi and Taz1 participate in redundant silencing pathways in telomeric regions. Noticeably though, the derepression observed in Δtaz1Δdcr1 double mutants was yet not as pronounced as the derepression in a Δclr4 mutant (Figure 5A). Other factors, such as Arp6 (59), or the products of uncharacterized genes (58) might contribute to tlh repression. This multiple redundancy is similar to the redundancy of silencing mechanisms in the mating-type region, where the transcription factor Atf1 and a third pathway work in parallel with RNAi to insure a full transcriptional repression of the region (31,32,60).
Figure 5.
tlh RNA levels in Δtaz1, Δdcr single and double mutants. MNE values were obtained for tlh transcripts by real-time RT–PCR analysis, using actin transcripts for the normalization as described in Materials and Methods. (A) wt:SPK10; Δclr4:SPK20; Δtaz1Δdcr1:SPK61; Δtaz1:J11; Δdcr1:TV293. (B) Tetrad analysis of a cross between J11 and PG3039: Tetrad 1 (parental ditype) [Δdcr1:SPK50; Δtaz1:SPK51; Δdcr1:SPK52; Δtaz1:SPK53], Tetrad 2 (non-parental ditype) [wt:SPK54; Δtaz1Δdcr1:SPK55; wt:SPK56; Δtaz1Δdcr1:SPK57].
The tlh genes repress ade6 at an ectopic site
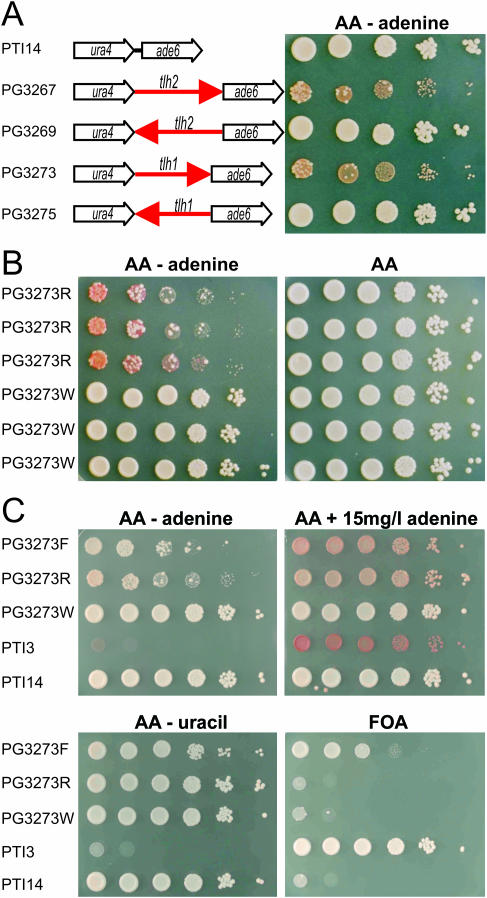
We investigated whether the tlh ORFs were sufficient to establish silencing and influence the expression of neighbor genes by cloning tlh1 and tlh2, respectively, upstream of an ade6+ marker gene introduced near the native ura4+ gene (Figure 6A and B). Both tlh genes repressed ade6+, however only when the tlh ORF was placed in the same orientation as ade6+. No detectable repression was observed when the ade6+ and tlh ORFs diverged from each other (Figure 6A). The ura4+ gene was still expressed following nearby insertion of the tlh genes. Clones with a repressed ura4+ gene could be obtained by FOA selection (Figure 6C); however, the frequency of FOA-resistant colonies was low suggesting spreading of transcriptional silencing to ura4+ is inefficient or alternatively raising the possibility that these clones contain additional mutations affecting silencing. The epigenetic states imparted on the ade6+ gene by tlh1 or tlh2 were relatively stable. The two epigenetic states conferred easily distinguishable phenotypes on media containing a low concentration of adenine, or lacking adenine (Figure 6B). Cells expressing ade6+ form white colonies on plates containing limiting adenine concentrations while cells in which ade6+ is repressed form red colonies. This allowed to estimate the rates of interconversion between the two states by counting half-sectored colonies. We estimated the rate of change from repressed to derepressed state at ∼0.05 event per cell division (13/273 for PG3267 and 11/217 for PG3273) and the rate of change from derepressed to repressed state at ∼0.001 event per cell division (1/829 for PG3267 and 1/852 for PG3273).
Figure 6.
The tlh genes affect the expression of neighboring genes. (A) Schematic representation of constructs integrated at the ura4 locus and phenotypes of corresponding strains. Ten-fold serial dilutions of cell suspensions were spotted on the indicated media, allowed to grow for 4 days at 33°C, and photographed. (B) Three individual PG3273 colonies in the red (PG3273R) or white (PG3273W) epigenetic state were spotted on the indicated media. (C) A FOA-resistant isolate of PG3273 (PG3273F) was plated along with a red and white isolate of the same strain and two tester strains, PTI3 (Ade− Ura−) and PTI14 (Ade+ Ura+).
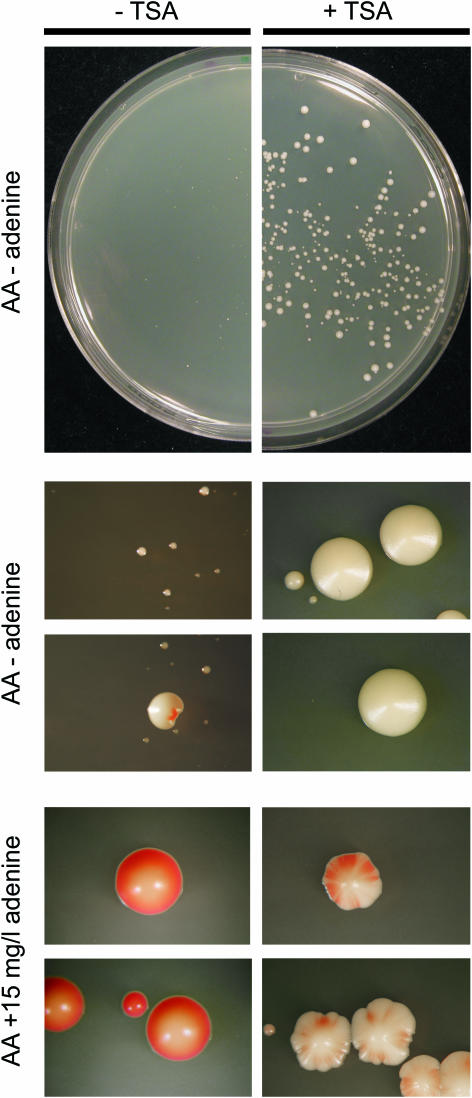
The existence of two interconverting epigenetic states indicated to us that the repression of ade6+ by the tlh genes was mediated by heterochromatin rather than due to transcriptional interference/promoter occlusion. To confirm that this was the case, we treated cells containing an ectopic ade6+ gene with TSA. Both PG3267 and PG3273 cells were treated and plated on medium lacking adenine and medium with a low adenine concentration (Figure 7 and data not shown). TSA treatment derepressed tlh-ade6+ in both strains, allowing cells to form full-sized colonies on plates lacking adenine and largely white colonies on limiting adenine. This derepression is consistent with tlh-ade6+ being regulated by histone deacetylation like the telomeric tlh genes.
Figure 7.
Sensitivity of an ectopic tlh2-ade6+ reporter to histone acetylation. PG3263 cells were treated with TSA (+TSA) or mock treated (−TSA), plated on the indicated selective or indicator media, incubated for 4 days at 33°C, and plates were scored and photographed.
Common ancestry of S.pombe tlh genes and centromeric repeats
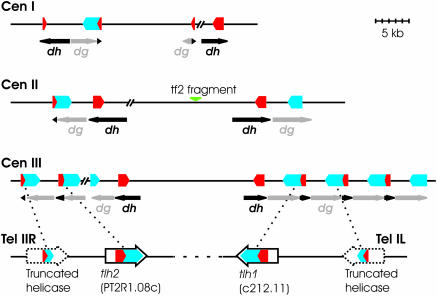
The dh and dg repeats in centromeric and mating-type region and the telomeric tlh genes clearly originate from a common ancestor. Several models can be contemplated regarding the duplicative events that created these repeats. A possibility suggested by the presence of transposable elements in the pericentromeric heterochromatin of higher eukaryotes is that the tlh genes might be parts of transposons that preferentially transpose to heterochromatic regions. One class of putative helicase-encoding transposons called helitrons has been proposed to replicate by a rolling-circle mechanism (61). Rolling-circle replication could create tandem repeats similar those observed at S.pombe centromeres. Alternatively, tlh genes might have been mobilized with the help of retrotransposons similar to the propagation of Y′ elements in S.cerevisiae cells undergoing telomerase crisis (51), and the tlh cDNAs might have subsequently recombined with centromeric regions. A second type of event capable of creating arrangements similar to those found at centromeres would be the integration at internal chromosomal loci of elements similar to the sod2 amplicons (62,63). The sod2 amplicons are produced from telomeric regions and display a symmetry similar to the symmetry of centromeric regions. Yet another possibility suggested by the arrangement of the repetitive elements at centromeres and telomeres would be that S.pombe centromeres arose from telomere fusions (Figure 8). Numerous examples of telomere fusions have taken place in evolution, e.g. during hominoid evolution the generation of human chromosome 2 involved a telomere to telomere fusion [reviewed in (64)]. Telomeric repeats have been observed within or adjacent to pericentric regions in several avian species (65) or Arabidopsis thaliana (66). Similar observations were made in the genus of Taterillus (67). S.pombe cells can survive in the absence of telomerase activity by forming circularized chromosomes (68). In all characterized classes of such survivors the fused ends contain the tlh genes and are able to maintain heterochromatic properties in the absence of Taz1-binding sequences (69), demonstrating that such end fusions confer some of the properties required for centromere function in S.pombe. The ability of tlh1 or tlh2 to induce heterochromatin formation in a context different from telomeres is also demonstrated by their effects at an ectopic site. A similar origin can be imagined for the cenH element in the mating-type region. In hemiascomycetes with triplicated mating-type loci, the two silent mating-type cassettes are located in subtelomeric regions (70). The mat2-P and mat3-M loci might also once have been subtelomeric in an ancestor of S.pombe and been internalized together with a tlh copy creating the mating-type region.
Figure 8.
Model for the generation of a centromere by telomere fusion. Partial tlh ORFs are found in subtelomeric regions in addition to the full-length genes, suggesting that tlh genes were once present in several copies at these sites (dotted arrows). A dashed line shows how a telomeric end to end fusion of the PT2R1 (Tel IIR) and c212 (Tel IL) regions would create a symmetrical structure like those at centromeres.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Rob Martienssen for bringing literature reports of helitrons to our attention and Janne Verhein-Hansen for technical help with some of the experiments. Our research was supported by the Danish Research Council and by a PhD fellowship from the University of Copenhagen to K.R.H. Funding to pay the Open Access publication charges for this article was provided by the Danish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Henikoff S., Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Allshire R. Centromere and kinetochore structure and function. In: Egel R., editor. The Molecular Biology of Schizosaccharomyce pombe. Berlin: Springer-Verlag; 2004. pp. 149–169. [Google Scholar]
- 3.Clarke L., Amstutz H., Fishel B., Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakaseko Y., Kinoshita N., Yanagida M. A novel sequence common to centromere regions of Schizosaccharomyces pombe chromosomes. Nucleic Acids Res. 1987;12:4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishel B., Amstutz H., Baum M., Carbon J., Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M. Composite motifs and repeat symmetry in S.pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 7.Murakami S., Matsumoto T., Niwa O., Yanagida M. Structure of the fission yeast centromere cen3: direct analysis of the reiterated inverted region. Chromosoma. 1991;101:214–221. doi: 10.1007/BF00365153. [DOI] [PubMed] [Google Scholar]
- 8.Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 9.Clarke L., Baum M.P. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol. Cell. Biol. 1990;10:1863–7182. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal S.I., Klar A.J. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–1238. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grewal S.I., Klar A.J. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 13.Thon G., Friis T. Epigenetic inheritance of transcriptional silencing and switching competence in fission yeast. Genetics. 1997;145:685–696. doi: 10.1093/genetics/145.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama J., Klar A.J., Grewal S.I. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 15.Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 16.Ayoub N., Goldshmidt I., Lyakhovetsky R., Cohen A. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics. 2000;156:983–994. doi: 10.1093/genetics/156.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge J.F., Scott K.S., Bannister A.J., Kouzarides T., Allshire R.C. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- 18.Reinhart B.J., Bartel D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 19.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 20.Martienssen R.A., Zaratiegui M., Goto D.B. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 23.Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 24.Partridge J.F., Borgstrøm B., Allshire R.C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 25.Thon G., Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provost P., Silverstein R.A., Dishart D., Walfridsson J., Djupedal I., Kniola B., Wright A., Samuelsson B., Radmark O., Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl Acad. Sci. USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nature Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 28.Sadaie M., Iida T., Urano T., Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrie V.J., Wuitschick J.D., Givens C.D., Kosinski A.M., Partridge J.F. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol. Cell. Biol. 2005;25:2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S.I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia S., Noma K., Grewal S.I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 32.Kim H.S., Choi E.S., Shin J.A., Jang Y.K., Park S.D. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J. Biol. Chem. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- 33.Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 34.Mandell J.G., Bähler J., Volpe T.A., Martienssen R.A., Cech T.R. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 2005;6:R1. doi: 10.1186/gb-2004-6-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandell J.G., Goodrich K.J., Bähler J., Cech T.R. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J. Biol. Chem. 2005;280:5249–5257. doi: 10.1074/jbc.M412756200. [DOI] [PubMed] [Google Scholar]
- 36.Bach M.L. Cloning and expression of the OMP decarboxylase gene URA4 from Schizosaccharomyces pombe. Curr. Genet. 1987;12:527–534. doi: 10.1007/BF00419562. [DOI] [PubMed] [Google Scholar]
- 37.Szankasi P., Heyer W.D., Schuchert P., Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J. Mol. Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 38.Lyne R., Burns G., Mata J., Penkett C.J., Rustici G., Chen D., Langford C., Vetrie D., Bähler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon P. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrière G., Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 43.Harper L., Golubovskaya I., Cande W.Z. A bouquet of chromosomes. J. Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez-Alonso P., Guzman P. Organization of chromosome ends in Ustilago maydis. RecQ-like helicase motifs at telomeric regions. Genetics. 1998;148:1043–1054. doi: 10.1093/genetics/148.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inglis P.W., Rigden D.J., Mello L.V., Louis E.J., Valadares-Inglis M.C. Monomorphic subtelomeric DNA in the filamentous fungus Metarhizium anisopliae contains a RecQ helicase-like gene. Mol. Genet. Genomics. 2005;4:79–90. doi: 10.1007/s00438-005-1154-5. [DOI] [PubMed] [Google Scholar]
- 46.Gao W., Khang C.H., Park S.Y., Lee Y.H., Kang S. Evolution and organization of a highly dynamic, subtelomeric helicase gene family in the rice blast fungus Magnaporthe grisea. Genetics. 2002;162:103–112. doi: 10.1093/genetics/162.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louis E.J., Haber J.E. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada M., Hayatsu N., Matsuura A., Ishikawa F. Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J. Biol. Chem. 1998;273:33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- 49.Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 50.Teng S.C., Zakian V.A. Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maxwell P.H., Coombes C., Kenny A.E., Lawler J.F., Boeke J.D., Curcio M.J. Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 2004;24:9887–9898. doi: 10.1128/MCB.24.22.9887-9898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mata J., Lyne R., Burns G., Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nature Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 53.Hansen K.R., Burns G., Mata J., Volpe T.A., Martienssen R.A., Bähler J., Thon G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horn P.J., Bastie J.N., Peterson C.L. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia S., Kobayashi R., Grewal S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nature Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 56.Li F., Goto D.B., Zaratiegui M., Tang X., Martienssen R., Cande W.Z. Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 57.Thon G., Hansen K.R., Altes S.P., Sidhu D., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J.S. Genetics. Vol. 171. 2005. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe; pp. 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nimmo E.R., Pidoux A.L., Perry P.E., Allshire R.C. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 59.Ueno M., Murase T., Kibe T., Ohashi N., Tomita K., Murakami Y., Uritani M., Ushimaru T., Harata M. Fission yeast Arp6 is required for telomere silencing, but functions independently of Swi6. Nucleic Acids Res. 2004;32:736–741. doi: 10.1093/nar/gkh234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thon G., Bjerling K.P., Nielsen I.S. Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics. 1999;151:945–963. doi: 10.1093/genetics/151.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapitonov V.V., Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patterson T.E., Albrecht E.B., Nurse P., Sazer S., Stark G.R. Effects of genome position and the DNA damage checkpoint on the structure and frequency of sod2 gene amplification in fission yeast. Mol. Biol. Cell. 1999;10:2199–2208. doi: 10.1091/mbc.10.7.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albrecht E.B., Hunyady A.B., Stark G.R., Patterson T.E. Mechanisms of sod2 gene amplification in Schizosaccharomyces pombe. Mol. Biol. Cell. 2000;11:873–886. doi: 10.1091/mbc.11.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mewborn S.K., Lese Martin C., Ledbetter D.H. The dynamic nature and evolutionary history of subtelomeric and pericentromeric regions. Cytogenet. Genome Res. 2005;108:22–25. doi: 10.1159/000080798. [DOI] [PubMed] [Google Scholar]
- 65.Nanda I., Schrama D., Feichtinger W., Haaf T., Schartl M., Schmid M. Distribution of telomeric (TTAGGG)(n) sequences in avian chromosomes. Chromosoma. 2002;111:215–227. doi: 10.1007/s00412-002-0206-4. [DOI] [PubMed] [Google Scholar]
- 66.Uchida W., Matsunaga S., Sugiyama R., Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002;77:63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- 67.Dobigny G., Ozouf-Costaz C., Bonillo C., Volobouev V. Evolution of rRNA gene clusters and telomeric repeats during explosive genome repatterning in TATERILLUS X (Rodentia, Gerbillinae) Cytogenet. Genome Res. 2003;103:94–103. doi: 10.1159/000076296. [DOI] [PubMed] [Google Scholar]
- 68.Nakamura T.M., Cooper J.P., Cech T.R. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 69.Sadaie M., Naito T., Ishikawa F. Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 2003;17:2271–2282. doi: 10.1101/gad.1112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.