Abstract
Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein, which binds both single-stranded (ss) and double-stranded (ds) DNA and facilitates the formation of higher order protein–DNA complexes in vitro. LrpC binds at different sites within the same DNA molecule promoting intramolecular ligation. When bound to separate molecules, it promotes intermolecular ligation, and joint molecule formation between a circular ssDNA and a homologous ssDNA-tailed linear dsDNA. LrpC binding showed a higher affinity for 4-way (Holliday) junctions in their open conformation, when compared with curved dsDNA. Consistent with these biochemical activities, an lrpC null mutant strain rendered cells sensitive to DNA damaging agents such as methyl methanesulfonate and 4-nitroquinoline-1-oxide, and showed a segregation defect. These findings collectively suggest that LrpC may be involved in DNA transactions during DNA repair and recombination.
INTRODUCTION
The Lrp family of transcriptional regulators appears to be widely distributed in bacteria and archaea [reviewed in (1)], but the molecular basis of Lrp action is poorly understood. A direct interaction of the Lrp protein with components of the transcription initiation machinery has not been reported. The consensus DNA-binding motif of the family was found to be a palindromic sequence with a central TTT; however, this consensus motif is not conserved among all the family [reviewed in (1)]. The members of this family are small DNA-binding proteins with predicted molecular masses of ∼15 kDa and whose multimeric state includes dimers, tetramers, octamers and hexadecamers [reviewed in (1)]. The Escherichia coli Lrp (EcoLrp), which is the best characterized member of the family, is a global regulatory protein that either activates or represses the expression of a high number of genes [reviewed in (2,3)]. Recent DNA microarray analysis revealed that, depending on growth conditions, between ∼100 and 400 E.coli genes are directly affected by EcoLrp, ∼10% of all genes are indirectly affected, and most of them upon entrance into stationary phase (4,5). EcoLrp often acts in concerted action with other global regulators, such as EcoCRP, EcoIHF, EcoH-NS, EcoFNR and EcoHU (6–8). Self-association seems to govern the way how the protein binds to DNA. The EcoLrp protein self-associates to a hexadecamer or an octamer, the hexadecamer being the predominant species at micromolar concentrations. At low concentrations (nM range), it dissociates to a dimer (9). This is consistent with the crystal structure of Pyrococcus furiosus LrpA, which forms a homodimer mainly through interactions between the antiparallel β-sheets of the C-terminal, and further interactions lead to octamer formation (10). The recently published structure of another family member, the archaeal protein, FL11 (11), also reports the existence of dimers and octamers for this protein.
EcoLrp seems to coordinate cellular metabolism with the general nutritional state by sensing the L-leucine concentration in the cell, resulting in a difference in the number of Lrp molecules/cell, depending on growth state and conditions. There are reportedly about 300–500 octamers per cell when grown in rich medium, and these values are 3- to 4-fold higher when the cells are growing in minimal medium. In rich medium, stationary-phase cells contain ∼2-fold higher levels of EcoLrp than log-phase cells (12,13). This variation in the amount of EcoLrp under different growth conditions and rate, together with the high discrimination between specific and non-specific binding to DNA, favor the view that EcoLrp is a global regulatory protein (6,9,14). However, other Lrp-homologs, such as EcoAsnC or Haemophilus influenzae LrfB appear to exert a more specific control (1).
Global regulators represent one of the most diverse classes of prokaryotic transcription factors. The EcoCRP, EcoIHF, EcoH-NS, EcoFNR, EcoArcA, EcoNarL and EcoLrp global regulators are sufficient for directly modulating the expression of 51% of E.coli genes (6–8), but many of these regulators do not have their counterpart in Bacillus subtilis. The nucleotide sequence analysis of the B.subtilis genome revealed the presence of only one gene coding for a sequence-independent chromatin-associated Hbsu protein (counterpart of EcoHU protein). However, genes coding for sequence-specific architectural proteins and global regulators, such as EcoIHF, EcoFis, EcoH-NS, EcoStpA, EcoCRP, EcoHha and EcoHfq could not be predicted (15). Furthermore, analysis of the B.subtilis DNA sequence revealed an unexpected high number of Lrp-homologs, the non-essential LrpA, LrpB, LrpC, AzlB, YezC, YwrC and YugG proteins, whose degree of identity to the EcoLrp protein ranges from 25 to 34% (1,16–18). Very little is known about the role of these proteins, but considering their abundance, a more specific role, as the one described for EcoLrp, has to be considered, at least for some of them. Between these Lrp-like proteins, the best characterized is the LrpC protein. All of the following information refers to B.subtilis genes and products, unless otherwise indicated.
LrpC exists in glucose minimal medium at intracellular levels of ∼12 octamers per cell both in exponential and stationary-phase conditions. The addition of leucine did not modify these levels. In rich medium, the LrpC expression level in exponentially growing cells is ∼6 octamers per cell, and increases ∼6-fold when the cells are in stationary-phase (19). LrpC is a sequence-independent DNA-binding and DNA-bending protein, which preferentially binds curved double-stranded (ds) DNA (20,21). DNase I footprinting studies revealed that LrpC binds curved dsDNA and further bends it, as judged for the number of hypersensitive sites covering an AT rich DNA stretch. Nevertheless, area protected against cleavage, as expected for a sequence-specific DNA-binding protein, were not observed (20). Similar results were observed in the presence or the absence of L-leucine. The binding of LrpC to two or more sites in the same DNA molecule (DNA looping) (20), and to long DNA substrates forming nucleosome-like structures (21) was also observed.
DNA looping is a widespread mechanism to deliver transcriptional repressors or activators to target sites (22,23). The discovery that members of the Lrp family have the capacity to bend DNA upon binding (24), wrap the DNA (21) and contribute to chromosomal packaging (25) has led some authors to propose not only a role as transcriptional regulators for the Lrp proteins, but also to act as chromatin-associated proteins that may work in concerted action with other chromatin-associated proteins (e.g. IHF and H-NS) (1,25). Furthermore, the LrpC protein was shown to be an architectural protein that modulates the DNA-binding activity of the chromatin-associated Hbsu protein (20), which has been shown to bind bent and kinked DNA preferentially (26,27).
In this paper, we investigate the role of LrpC in DNA transactions during DNA recombination. We show that LrpC also binds single-stranded (ss) DNA, supercoiled dsDNA, and with high affinity 4-way (Holliday) and Y-junctions in their open conformations. LrpC facilitates intermolecular or intramolecular ligation depending on protein and DNA concentration by favoring the formation of a higher order protein–DNA complex. By promoting co-aggregation, LrpC enhances spontaneous DNA annealing. Consistent with these results, an lrpC null mutant strain renders cells moderately sensitive to DNA damaging agents such as methyl methanesulfonate (MMS) or 4-nitroquinoline-1-oxide (4NQO), and a segregation defect is observed. These findings suggest a role for LrpC in DNA transactions, and specifically during DNA repair and recombination.
MATERIALS AND METHODS
Bacterial strains and plasmids
E.coli strain JM103 was used for the amplification of plasmid DNA (28). B.subtilis YB886 strain (SPβ-free and non-inducible for PBSX), and its isogenic derivatives ΔrecU (BG427), hbs4755 (BG405), ΔrecA (BG190), recF15 (BG127), addA5 addB72 (BG189), ΔrecS (BG425) and ΔrecQ (BG705) have been described previously (29–34). Plasmid pCB528 contains the lrpC gene interrupted at the 5′ region by a 410 bp DNA segment coding for a phleomycin resistance gene. Linear plasmid pCB528 DNA was used to transform YB886 (wild type) or BG427 (ΔrecU) competent cells, by a double crossing-over event, with selection for phleomycin resistance, as described previously (33), generating thereby, the ΔlrpC (BG657) and the ΔrecU ΔlrpC (BG663) strains, respectively. In this construction, both lrpC and phleomycin promoters, which are poorly utilized (16,35), are in the same direction of transcription, and the topB gene which is downstream of lrpC, should be transcribed from these promoters. If not stated otherwise, the phagemids pGEM-3Zf(+) and pGEM-3Zf(−) (Promega) were used as a source of ssDNA and dsDNA.
Chemical treatment
Treatment of exponentially growing mutant strains with 10 mM MMS (Merck) or 100 µM 4NQO (Sigma) was performed as described previously (32).
Fluorescence microscopy of B.subtilis cells
Exponentially growing cells were obtained by inoculating overnight cultures in fresh Luria–Bertani media and grown to an OD560 = 0.4 at 37°C. Mid log-phase cells were then fixed with 2% formaldehyde, 4′,6′-diamino-2-phenylindole (DAPI) (1 µg/ml) was added for nucleoid visualization, and cells were analyzed by fluorescence microscopy as described previously (36).
Enzymes
T4 DNA ligase was from NEB. LrpC was purified as described previously (20). Previously tetramers in solution were detected (20), but recently the crystal structure of LrpC revealed the existence of octamers (P. Thaw and J. B. Rafferty, unpublished data), similarly to other members of the protein family. Therefore, its concentration is expressed as octamers. Bacteriophage SPP1 G35P recombinase, and G34.1P 5′–3′ exonuclease were purified as described previously (37,38), and their concentration are expressed as dimers. B.subtilis RecA and RecU were a gift from Begoña Carrasco (CNB, CSIC), and their concentrations are expressed as monomers and dimers, respectively.
Protein-crosslinking experiments were performed incubating 0.8 µM LrpC with 2 µM RecU in buffer A (50 mM Tris–HCl, pH 7.5, 75 mM NaCl, glycerol 10% and 0.5 mM suberic acid) for 10 min at 37°C. After stopping the reaction by heating the samples in cracking buffer, the reactions were analyzed by 10% SDS–PAGE.
Measurements of LrpC–DNA complexes
The amount of DNA is expressed as moles of DNA molecules. Binding of LrpC to ssDNA and plasmid DNA was analyzed by 0.8% agarose gel electrophoresis in TAE buffer followed by ethidium bromide staining. Briefly, 10 nM pGEM-3Zf(+) DNA was incubated with increasing concentrations of LrpC in buffer B (50 mM Tris–HCl, pH 7.5, 25 mM NaCl and glycerol 5%) containing 5 mM EDTA or 5 mM MgCl2 for 10 min at 37°C. The reaction was stopped by the addition of loading buffer (50 mM Tris–HCl, pH 7.8, glycerol 30%, bromophenol blue 0.25% and xylene cyanol 0.25%) following electrophoresis at room temperature.
For the construction of the 4-way, Y-junction and the forked structure, a combination of oligos #1 to #6 was used (39). Annealing of oligos #1 to #4 render the 4-way junction, annealing of #1, #4 and #6 the Y-junction, and #1 and #5, 50 bp duplex-DNA. In all constructs, oligo #1 was γ-32P-radiolabeled, and after annealing, the products were gel purified. Binding to these substrates was analyzed by incubating 0.5 nM of γ-32P-labeled DNA substrate with increasing concentrations of LrpC in buffer C (25 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.05 mg/ml BSA, 5% glycerol and 50 mM NaCl) containing 5 mM EDTA or 5 mM MgCl2 for 10 min at 37°C. The reaction was stopped by the addition of loading buffer following fractionation by non-denaturing 8% PAGE, run in TBE and autoradiography.
Aggregation of DNA by LrpC was performed in buffer B containing 5 mM EDTA or 5 mM MgCl2 as described previously (20).
Joint molecules formation assay
Linear dsDNA molecules having 3′ ssDNA termini (3′-tailed dsDNA) were prepared by incubation of HincII-linearized (3197 bp) pGEM-3Zf(+) dsDNA for 45 s with 7 nM G34.1P (a 5′–3′ exonuclease). Then, the enzyme was heat inactivated as described previously (38). The assay measures the formation of a stable complex between circular pGEM ssDNA (5 nM) and linear 3′-tailed pGEM dsDNA or blunt-ended dsDNA (5 nM). Both substrates were incubated with increasing concentrations of LrpC (45 nM to 1.5 µM) or G35P recombinase (700 nM) in buffer C containing 2.5 mM MgCl2 during 15 min at 30°C. As a control, blunt-ended dsDNA was incubated with circular ssDNA and B.subtilis RecA (2 and 4 µM) in buffer C containing 10 mM magnesium acetate and 2 mM dATP for 60 min at 30°C. The reactions were stopped by addition of 20 mM EDTA, 0.5% SDS and 0.5 mg/ml proteinase K. Then, the samples were loaded in a 0.8% agarose gel and run overnight at 2 V/cm.
ssDNA annealing by LrpC was also analyzed by incubating 5 nM of EcoRI-linearized pGEM-3Zf(+) ssDNA with 5 nM circular pGEM-3Zf(−) ssDNA and increasing concentrations of LrpC (22–180 nM) in buffer B containing 0.1 mM MgCl2 for 15 min at 30°C. The reactions were stopped by addition of EDTA, SDS and proteinase K, loaded in a 0.8% agarose gel and run overnight at 2 V/cm.
Ligation assay
EcoRI-linearized pUC18 or pGEM-3Zf(+) dsDNA was incubated in ligase buffer (66 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 10 mM DTT and 1 mM ATP) with 3 U of T4 DNA ligase and increasing concentrations of LrpC at room temperature for 3 h. The reactions were stopped with EDTA, SDS and proteinase K, concentrated, dialyzed, and fractionated by 0.8% agarose gel electrophoresis in TBE, and then stained with ethidium bromide. To favor intramolecular ligations, a DNA concentration of 0.3 nM was used, whereas in intermolecular ligation assays, a DNA concentration of 1.8 nM was used. To define the nature of the ligation products, supercoiled pUC18 DNA (where Form I, and in minor percentage Form II, are present), and EcoRI-linearized SPP1 molecular marker (GIBCO-BRL) were also loaded in one of the agarose gels.
RESULTS AND DISCUSSION
LrpC binds ssDNA and supercoiled DNA
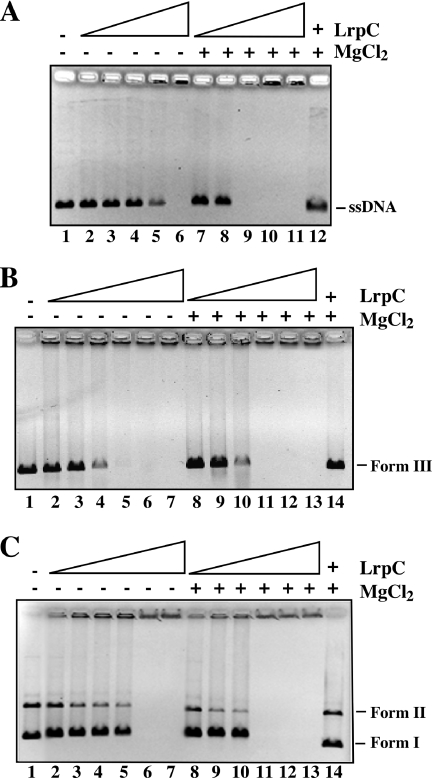
Previously, it has been shown that LrpC binds, in a L-leucine-independent manner, preferentially to a ∼500 bp curved dsDNA with an apparent binding constant (Kapp, protein concentration required to reach 50% of the DNA bound) of ∼35 nM when compared to non-curved dsDNA (Kapp ∼350 nM) (20). Several faint bands and high molecular weight (HMW) complexes that did not enter the agarose gel were observed under conditions that give relative unstable protein–DNA complexes (e.g. high NaCl concentrations, 75 mM), (20). These complexes could represent binding and looping on the same DNA molecule, or binding to two or more linear dsDNA molecules (sandwich molecules or intermolecular complexes), respectively. To learn whether LrpC also forms looped and/or sandwich structures with other DNA substrates, such as circular ssDNA and supercoiled dsDNA, DNA-binding experiments were performed under low ionic strength conditions (25 mM NaCl), and compared with the binding to linear dsDNA under these conditions. Since Mg2+ affects the folding of ssDNA and other DNA susbtrates (see below), the binding in the absence or the presence of 5 mM MgCl2, was assayed (Figure 1).
Figure 1.
Effect of Mg2+ in the binding of LrpC to DNA. The binding of increasing concentrations of LrpC to 10 nM pGEM-3Zf(+) circular ssDNA (A), pGEM-3Zf(+) EcoRI-linearized dsDNA (B) and pGEM-3Zf(+) supercoiled dsDNA (C) was assayed in the presence of 5 mM EDTA or 5 mM MgC12 by agarose gel electrophoresis followed by ethidium bromide staining. LrpC concentrations are 50, 100, 150, 200 and 400 nM in (A); 100, 200, 400, 500, 600 and 700 nM in (B); 200, 400, 500, 600, 700 and 800 nM in (C). In the last lane of all the parts, the maximal LrpC concentration was incubated with DNA followed by addition of 1% SDS.
LrpC bound ssDNA with slightly higher affinity in the presence of Mg2+ than in its absence. The LrpC concentration for ssDNA half-saturation (Kapp) was ∼187 nM and ∼125 nM in the absence and presence of 5 mM MgCl2. At protein concentrations higher than 200 nM (−Mg2+) or 100 nM (+Mg2+) HMW complexes that did not enter in the agarose gel were observed (Figure 1A). The HMW material could be the product of large looped LrpC–ssDNA complexes and/or intermolecular complexes, where two, or more, ssDNA molecules are bridged by LrpC. To learn whether the protein introduces any modification on the DNA, the protein complex of lane 11 (10 nM ssDNA plus 400 nM LrpC) was treated with 1% SDS and the reaction mixture separated by agarose electrophoresis (Figure 1A, lane 12). The sample ran in the same position as the non-treated DNA.
The binding of LrpC to supercoiled and linear ‘non-curved’ dsDNA in the presence or absence of 5 mM MgCl2 was also measured (Figure 1B and C, respectively). As described previously LrpC forms upon binding to linear dsDNA higher order complexes that do not migrate into the gel and such protein–DNA interaction is cooperative (20,21). In the absence of Mg2+, the protein concentration midpoint (half-saturation) of LrpC for linear dsDNA was ∼300 nM, whereas for negatively supercoiled DNA was ∼650 nM. In the presence of 5 mM MgCl2, the binding to linear dsDNA (∼300 nM) was not altered and slightly enhanced for negatively supercoiled dsDNA (∼550 nM). In quantitative terms the stoichiometry of LrpC for ssDNA, linear dsDNA and negatively supercoiled dsDNA was estimated to be ∼15, ∼30 and ∼60 octamers per DNA molecule, respectively, in the presence of 5 mM MgCl2. It is likely that the linear dsDNA used is a genuine non-curved DNA, because it showed a 8-fold lower affinity than to curved DNA (20). Since the same type of complexes were observed with all the substrates tested, and it has been shown previously that LrpC forms loops and sandwich molecules on curved dsDNA (20,21), we propose that LrpC also forms DNA loops and intermolecular complexes with circular ssDNA and negatively supercoiled DNA.
LrpC promotes looped and intermolecular complexes
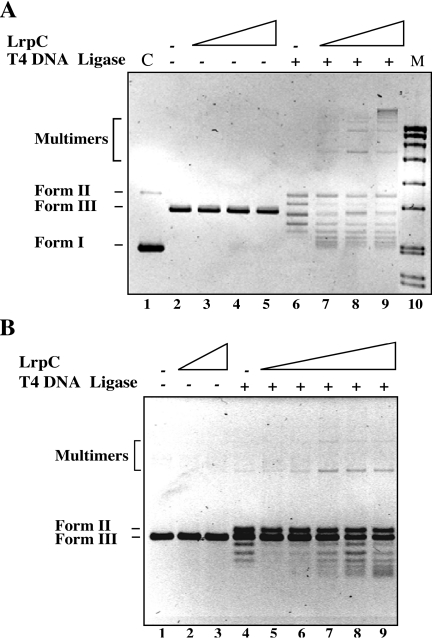
We further investigated the preference of LrpC for forming loops or sandwich molecules with dsDNA. Since DNA looping is conceptually similar to ring closure of restriction fragments (40) and in trans or intermolecular complexes to the ligation of two separate DNA fragments, a ligation assay, under conditions that may favor the in trans reaction (intermolecular ligation, 1.8 nM DNA, Figure 2A) or the in cis (intramolecular or self-ligation, 0.3 nM DNA, Figure 2B) reaction, was performed. First, EcoRI-linearized dsDNA (1.8 nM) was incubated in ligase buffer with increasing concentrations of LrpC (20–80 nM) and with T4 DNA ligase during 3 h at room temperature. Then, the samples were deproteinized, the DNA was precipitated and separated by agarose gel electrophoresis. In the presence of LrpC, but in the absence of T4 DNA ligase, the migration of deproteinized linear dsDNA remained unaltered (Figure 2A, lanes 3–5). T4 DNA ligase converted linear dsDNA (Form III) into a relaxed circle (Form II), and some circular forms, which move faster than Form III DNA (Figure 2A, lane 6). At low LrpC ratios (∼10 LrpC octamers per dsDNA molecule, corresponding to 1 LrpC octamer every 280 bp) Form II, a high amount of faster moving forms, and few oligomeric forms were observed (Figure 2A, lane 7), whereas at LrpC ratios of ∼20:1 and ∼40:1, less amount of faster moving forms and high-order oligomers accumulated (Figure 2A, lanes 8 and 9). It is likely, therefore, that T4 DNA ligase converts the looped complexes formed by LrpC with linear dsDNA into monomeric covalently closed circles with a different degree of superhelicity (see below) and the sandwich complexes into multimers. Similar results were observed when linearized blunt-ended dsDNA was used (data not shown). It is likely, therefore, that the micro-homology (4 bp) present at the EcoRI site plays no role in the intermolecular ligation.
Figure 2.
LrpC promotes looped and transcomplexes on dsDNA. EcoRI-linearized pUC18 dsDNA [1.8 nM (A) or 0.3 nM (B)] was incubated with increasing concentrations of LrpC in the presence of 3 U of T4 DNA ligase for 3 h a room temperature. The deproteinized DNA was analyzed by agarose gel electrophoresis followed by ethidium bromide staining. LrpC concentrations are 20–80 nM in (A) (lanes 3–5 and 7–9); 80 and 160 nM in (B) (lanes 2 and 3) and 10–160 nM (lanes 5–9). Positions of Form II, Form III and multimers are indicated. In (A), the controls are shown (Form I and II in lane 1, and molecular marker in lane 10).
Conditions that may favor self-ligation were then used. EcoRI-linearized non-curved dsDNA (0.3 nM) was incubated with increasing concentrations of LrpC (10–160 nM) and with T4 DNA ligase during 3 h at room temperature. After sample deproteination we found that T4 DNA ligase converted a fraction of the linearized dsDNA into Form II, and some faster moving forms (Figure 2B, lane 4). At LrpC ratios of 30:1 and 60:1 (which correspond to one LrpC octamer per 95 and 48 bp, respectively) Form II and faster moving forms were observed (Figure 2B, lanes 5 and 6), whereas at LrpC ratios of 120:1 and 240:1 a high amount of faster moving products accumulated (Figure 2B, lanes 7 and 8). Under these conditions, only few dimers were observed, and higher order multimers were barely visible. It is likely, therefore, that the DNA concentration dictates the type of complexes formed by LrpC, probably by dictating the probability of intermolecular collision of different DNA molecules, since when the DNA concentration was low, even at LrpC:DNA ratios as high as 240:1, multimers were almost not detected.
The appearance of faster moving forms after LrpC treatment and ligation might correspond to molecules with a different degree of superhelicity, since it has been shown that LrpC, by wrapping around the DNA, constrains positive supercoils [(21), and G. López-Torrejón, unpublished data]. In some cases, the long-range interactions are not constrained to a particular topology and accommodate a variety of DNA configurations in the looped structure [e.g. LacI repressor and the SfiI endonuclease, (40,41)]. In the case of LrpC, the looping interaction might be preferentially arranged in a particular topology.
LrpC promotes ssDNA compaction
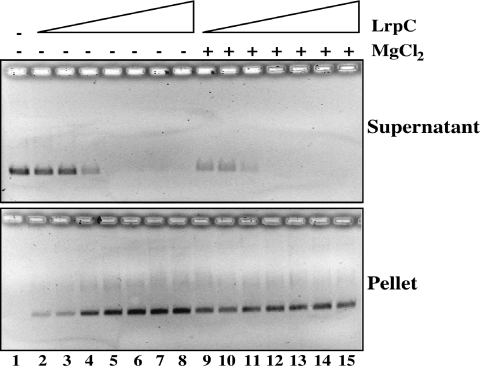
LrpC forms high-order complexes with circular ssDNA, and non-curved linear dsDNA (Figure 1A and C) and previously it was shown that LrpC promotes compaction or bridging of linear dsDNA (20). To address whether LrpC also promotes compaction of ssDNA, circular ssDNA (5 nM) was incubated with increasing concentrations of LrpC (12.5–500 nM, or LrpC ratios of 2.5:1 to 100:1) in the absence or the presence of 5 mM MgCl2 for 10 min on ice. Then the reaction mixture was centrifuged at 12 000 g for 20 min. Both supernatant and pellet (Figure 3) were deproteinized and analyzed by 0.8% agarose gel electrophoresis. In the absence of Mg2+, ∼25% of the DNA can be pelleted by low-speed centrifugation in the presence of 2.5–5 LrpC octamers per ssDNA molecule (Figure 3, lanes 2 and 3), and ∼90% of the ssDNA substrate was precipitated at ratios 20:1 with the concomitant disappearance from the supernatant (Figure 3, lane 5). Similar results were observed in the presence of 5 mM MgCl2. Here, however a lower LrpC concentration was required to reach similar results. This is consistent with the observed higher affinity of LrpC for ssDNA in the presence of 5 mM MgCl2 (Figure 1A). Moreover, it suggests that LrpC also promotes bridging of ssDNA molecules.
Figure 3.
LrpC promotes ssDNA-bridging. pGEM-3Zf(+) and introduce a space between pGEM and ssDNA (5 nM) was incubated with increasing concentrations of LrpC (12.5–500 nM) in the presence of 5 mM EDTA or 5 mM MgC12 for 10 min on ice, and then centrifuged at 12 000 g for 20 min. After addition of 0.5% SDS, the supernatant (upper panel) and the pellet (lower panel) were analyzed in agarose gel electrophoresis followed by ethidium bromide staining.
LrpC enhances strand annealing
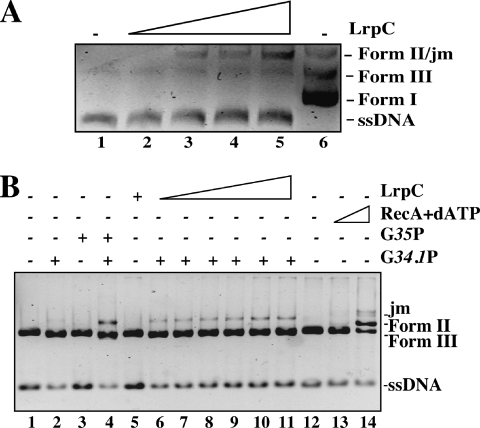
To address whether LrpC-promoted intermolecular complexes between two ssDNA molecules might promote the annealing of complementary strands, linear ssDNA (5 nM) and complementary circular ssDNA (5 nM) were incubated at 0.1 mM MgCl2 with increasing concentrations of LrpC (22–180 nM, Figure 4A) for 15 min at 30°C. The reaction was deproteinized and the DNA molecules separated by 0.8% agarose gel electrophoresis. Annealed DNA was observed from LrpC:ssDNA ratios of 1 LrpC octamer per 360 nt (Figure 4A, lane 3) and increased by increasing protein concentration (maximal ratio tested 1:90 nt, Figure 4A, lane 5). All DNA annealed species entered the agarose gel (data not shown). Under the conditions used, a spontaneous annealing in the absence of LrpC was not observed (Figure 4A, lane 1).
Figure 4.
DNA annealing by LrpC. (A) linearized pGEM-3Zf(+) ssDNA (5 nM) and circular pGEM-3Zf(−) ssDNA (5 nM) were incubated with increasing concentrations of LrpC (22–180 nM) in the presence of buffer B containing 0.1 mM MgCl2 for 15 min at 30°C. The deproteinized DNA was analyzed by agarose gel electrophoresis followed ethidium bromide staining. (B) HincII-linearized pGEM-3Zf(+) was treated, when indicated, with 7 nM of the SPP1-encoded 5′–3′ exonuclease G34.1P for 45 s to generate 3′-tailed dsDNA. Then the enzyme was inactivated, and the DNA was incubated with circular pGEM-3Zf(+) ssDNA (5 nM) and increasing concentrations of LrpC (lane 5, 1.5 µM; lanes 6–11, 45–1.5 µM) for 15 min at 30°C in buffer C containing 2.5 mM MgCl2. As controls, the SPP1-encoded G35P ATP-independent recombinase, that anneals only 3′-tailed DNA (700 nM, lanes 3 and 4) and the B.subtilis RecA protein, that performs strand exchange on blunt-ended DNA (2 µM, lane 13, and 4 µM lane 14) were used. The deproteinized DNA was analyzed by agarose gel electrophoresis followed ethidium bromide staining. Running positions of the joint molecules (jm), Form I, II and III DNA, and ssDNA (circular and lineal) are indicated. Plus and minus denote the presence and absence of the indicated protein.
We then examined whether LrpC might also promote DNA annealing and strand exchange at higher MgCl2 concentrations. The formation of joint molecules between circular ssDNA and a homologous linear dsDNA was analyzed in the presence of 2.5 mM MgCl2 using as control two bona fide strand exchange proteins [namely the B.subtilis dATP-dependent RecA protein, that catalyzes full strand exchange with the formation of open circular DNA (42), and the SPP1-encoded ATP-independent G35P recombinase, that catalyzes the formation of alpha and sigma structures, but fails to render the open circular product (37)]. When the highest LrpC concentration (1.5 µM) was incubated with circular ssDNA and homologous linear blunt-ended dsDNA, the protein failed to catalyze the formation of joint molecules (Figure 4B, lane 5), whereas RecA protein was able to render the final product of the reaction (Figure 4B, lane 14). A homologous 3′-tailed dsDNA was generated as described in Materials and Methods, and used to measure the LrpC-promoted joint molecule formation. In the presence of LrpC, the accumulation of a discrete new band that migrated slower than relaxed and linear dsDNA was observed. The addition of ATP did not affect joint molecules formation (data not shown). The mobility of the joint molecules formed in the presence of LrpC was indistinguishable from those formed in the presence of G35P and in both cases a 3′-tailed dsDNA was strictly required (Figure 4B, lanes 3–11) (37).
Formation of joint molecules was almost not visible when LrpC was omitted (Figure 4B, lane 2). Furthermore, joint molecules formation was not observed when a heterologous 3′-tailed dsDNA was used (data not shown). It is likely that LrpC catalyzes the aggregation (annealing) of a circular ssDNA with a homologous linear 3′-tailed dsDNA, but the final products of the strand exchange reaction (relaxed dsDNA, and linear ssDNA), as detected by the presence of RecA protein, did not accumulate (Figure 4B).
LrpC binds to 4-way and Y-junctions
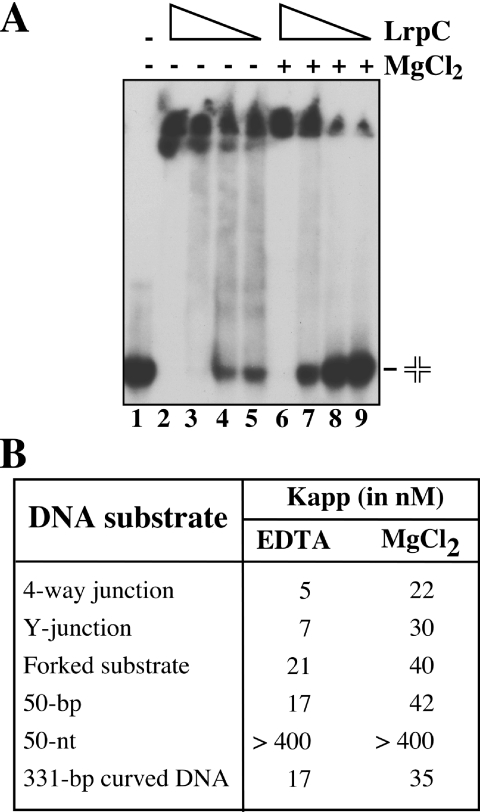
Many architectural proteins have been shown to be involved in DNA recombination, and in some cases, such as in HU, their preference for binding recombination intermediates has been shown (27). The 4-way DNA junction, also termed Holliday junction, is the key intermediate in nearly all homologous recombination processes. A stacked arrangement of the arms of the Holliday junction has been observed for free junctions in the presence of divalent cations, whereas in the presence of EDTA, the junction adopts an open square planar conformation (43). The 3-way or 3-armed junction (also termed Y-junction), which consists of three double-helical arms connected at the junction point, resembles a replicated fork, whereas the forked substrate (consisting of a duplex region with a double ssDNA arm) resembles a non-replicated fork. In DNA, Y-junctions arise during recombination involving phages (44) or if the DNA contains triplet repeats (45). They also adopt, apart from the open conformation, stacked conformations in the presence of multivalent cations, at least in some cases (46). Therefore, the binding of LrpC to different recombination intermediates was tested in the presence of 5 mM EDTA or 5 mM MgCl2, and compared with the binding to 50 bp DNA, 50 nt ssDNA and to curved DNA (Figure 5).
Figure 5.
DNA-binding specificity of LrpC. (A) Holliday-junction DNA (0.5 nM) was incubated with increasing concentrations of LrpC (6–50 nM) protein in presence of 5 mM of MgCl2 or 5 mM EDTA. Protein–DNA complexes were visualized by 6% native PAGE followed autoradiography. (B) The LrpC concentration to reach half-saturation (Kapp) with the different DNA substrates used at 5 mM MgCl2 or 5 mM EDTA is indicated. The values are the average of at least three independent experiments.
LrpC (6–50 nM) was incubated with 0.5 nM DNA in the presence of 5 mM EDTA or 5 mM MgCl2 for 10 min at 37°C, and the complexes separated by non-denaturing PAGE. HMW species that did not enter the gel were observed with all DNA probes used (Figure 5A and data not shown). Similarly to the previous results, the binding to the forked substrate or to the 50 bp DNA was almost not affected by the presence or absence of divalent cations, but the absence of divalent cations that favors the open conformation stimulated 4- to 5-fold the binding of LrpC to the 4-way and to the Y-junction (Figure 5B). The Kapp values were around 5 nM, which are similar to the ones observed for the chromatin-associated protein HU/Hbsu (27). Open square planar conformation of the Holliday junction may occur in vivo due to the binding of recombination enzymes that, either specifically bind to it, or promote branch migration, or catalyze its resolution. For example, EcoRuvA, which specifically binds to the Holliday junction, has been shown to organize it into a 4-fold symmetric form (47), and this has been also observed for the B.subtilis RecU Holliday-junction resolvase, which catalyzes its resolution (48,49).
LrpC bound the 50 bp susbtrate with an affinity similar to the 331 bp curved DNA (Figure 5B), but the binding of LrpC to the 50 nt ssDNA was very poor (Figure 5B), both in the presence and the absence of Mg2+. This was confirmed with other short ssDNA substrates (data not shown), whereas binding with an affinity similar to DNA was observed when the pGEM-3Zf(+) ssDNA was used (Figure 1A). At present, it is unknown whether a minimal ssDNA length is required for LrpC binding, and whether the presence of folded regions on the ssDNA molecule (i.e. dsDNA, owing to the presence of Mg2+ ions), might contribute to the observed activity.
ΔlrpC renders cells sensitive to DNA damaging agents
Many of the activities associated with LrpC such as binding to dsDNA and ssDNA enhancing DNA ligation and promoting DNA annealing and joint molecule formation, as well as binding to recombination intermediates, such as Holliday junctions and Y-junctions [(19,20) and this study] have been associated with proteins that catalyze strand exchange (RecA), or are RecA modulators, such as the RecU Holliday-junction resolvase (42,49), and the EcoRecBCD (counterpart of B.subtilis AddAB) and EcoRecFOR complexes (50,51). Furthermore, cells impaired in the essential chromatin-associated Hbsu protein (hbs4755 mutation) (31) or lacking the EcoHU protein (52), which also specifically bind recombination intermediates, are deficient in DNA repair. To address whether LrpC could be involved in recombinational DNA repair, we constructed an lrpC null mutant (ΔlrpC) strain and exposed it to the lethal effect of MMS or 4NQO. DNA lesions generated by the alkylating agent MMS, which reacts with single reactive groups in adenine (N3-alkyladenine) and guanine (N7-alkylguanine), are different to the ones generated by 4NQO, which is a potent mutagen that induces two main guanine adducts, at positions C8 and N2 (53). Once the DNA damage is removed, recombinational repair is able to re-establish the collapsed or stalled replication fork (54)
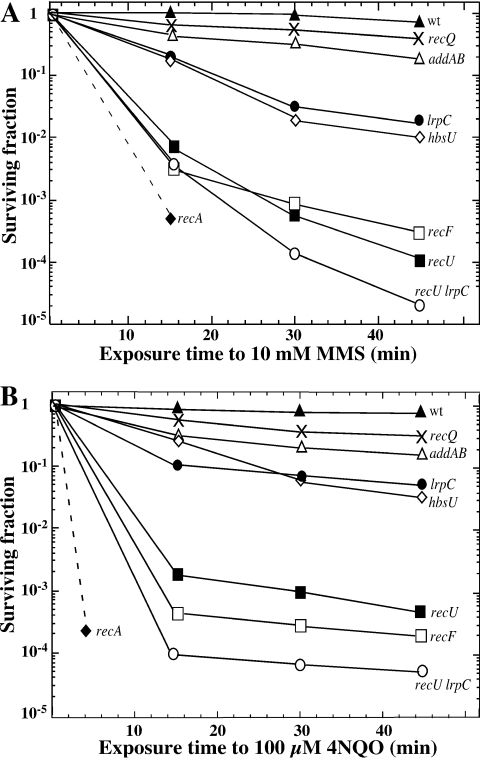
Mutant strains impaired in the modulation of RecA (ΔrecU, recF15, addA5 addB72), in RecA (ΔrecA) or in Hbsu (hbs4755) were selected as controls. In B.subtilis lrpC forms an operon with topB (coding for the topoisomerase III enzyme) (15). In E.coli, topoisomerase III has been shown to work in concert with the RecQ helicase (55). In B.subtilis, two RecQ homologs exist [the RecS and RecQ proteins, (15)]. In our ΔlrpC strain, the topB gene should be transcribed from its own promoter, or from the phleomycin promoter. In order to rule out any defect in the transcription of the downstream topB gene, the survival to MMS and 4NQO of the ΔlrpC strain was also compared with the one of the ΔrecQ and the ΔrecS strains. The ΔlrpC strain was more sensitive to the killing action of 10 mM MMS (Figure 6A) or 100 µM 4NQO (Figure 6B) than the addA5 addB72 strain or the ΔrecQ or ΔrecS strain. The ΔrecQ and ΔrecS strains show a similar degree of sensitivity to MMS or to 4NQO, hence only the former strain was shown in Figure 6. With both DNA damaging agents, the sensitivity was similar to the sensitivity of hbs4755 cells, and less sensitive than the recF15, ΔrecU and ΔrecA cells. It is likely, therefore, that the LrpC protein displays a specialized function in B.subtilis cells, because the presence of the other Lrp-like proteins [namely LrpA, LrpB, AzlB, YezC, YwrC and YugG, (15)], in the ΔlrpC background, could not compensate for the absence of LrpC in the MMS or 4NQO survival experiments.
Figure 6.
LrpC is involved in DNA repair. Survival of B.subtilis strains following exposure to 10 mM MMS (A) and to 100 µM 4NQO (B). The survival of wild-type YB886 (closed triangles), recQ (crosses), addA5addB72 (open triangles), ΔlrpC (closed circles), hbs4755 (open diamonds), recF15 (open squares), ΔrecU (closed squares), ΔrecU ΔlrpC (open circles) and ΔrecA (closed diamond) is shown.
Recently, it has been shown that RecU binds ssDNA and dsDNA catalyzing strand annealing. RecU binds with high affinity and cleaves Holliday junctions (49). Except Holliday-junction cleavage, many of these activities were also present in the LrpC protein. Therefore the ΔlrpC ΔrecU double-mutant strain was constructed and challenged to the killing action of MMS or 4NQO. As expected for non-epistatic functions, the double-mutant strain was more sensitive to 10 mM MMS or 100 µM 4NQO than the most sensitive parental strain (Figure 6). Despite the biochemical similarities, RecU and LrpC are unrelated in function, and this was confirmed by protein–protein crosslinking with suberic acid. No hetero-complex was detected, but the presence of dimers for RecU, and dimers and higher order complexes for LrpC could be observed (data not shown).
Nucleoid and cell morphology phenotypes of ΔlrpC cells
The absence of architectural proteins, such as EcoHU and EcoFis, causes cell filamentation, anucleated cells and aberrant nucleoid segregation (56,57), and these phenotypes have been also observed for the bacterial structural maintenance of chromosomes homologs, which also cause DNA condensation (58). Furthermore, mutations in genes acting on Holliday-junction intermediates, as ruvA or recU also show a defect in segregation (36). To address whether LrpC is involved in chromosomal segregation, nucleoids were stained with DAPI and examined using fluorescence microscopy. In wild-type cells, compact and condensed one or two regular nucleoid bodies were seen in fixed cells, and absence of DAPI stained material was rare (∼0.05% of 3600 cells counted, Figure 7 and data not shown). However, when ΔlrpC cells were analyzed, the typical phenotypes associated with defects in chromosome segregation as (i) anucleated cells, (ii) extension and anomalous positioning of the nucleoid in elongated cells and (iii) splitting of the nucleoid by a septum in dividing cells (guillotine effect) were observed (Figure 7). In the ΔlrpC strain, the percentage of cells lacking DAPI-stained material was ∼1.9% (total cell counted 2698) which accounts for an increase in the defect in segregation of >35-fold when compared with the wild type control.
Figure 7.
Nucleoid morphologies of ΔlrpC cells. Exponentially growing cells were fixed, stained with DAPI and analyzed by fluorescence microscopy to visualize the nucleoids. The white arrows point to misplaced and linked nucleoids, anucleate cells, and cells with a guillotine phenotype. In the lower panel, wild-type cells (wt) are shown as a control.
Nucleoid and cell morphology have been also analyzed in EcotopB mutants. No defect is observed in topB single mutants (59), what is consistent with our data of B.subtilis RecQ homologs: in ΔrecS, ΔrecQ and ΔrecS ΔrecQ cells, absence of DAPI-stained material was rare (<0.1%), and nucleoids were similar to the ones observed in wild-type cells [(36) and data not shown]. Abnormal nucleoids and elongated cells have been only observed by combination of the topB mutation with another topoisomerase mutation, as with a mutation in Topoisomerase I (59), or with a mutation in topoisomerase IV (60).
CONCLUSIONS
Genetic evidence suggests that LrpC plays a significant, but unknown role, in the repair of DNA damages and in chromosomal segregation in B.subtilis cells (this study). We previously hypothesized that LrpC is an architectural protein that facilitates the formation of nucleoprotein structures (20). The biochemical experiments reported here and previously suggest a direct role of LrpC in diverse activities that might be achieved through its participation in high-order nucleoprotein complexes, such as DNA loops and trans complexes with both ssDNA and dsDNA [(20) and this study]. LrpC constrains positive supercoils by wrapping the DNA in a right-handed superhelix, forming nucleosome-like structures [(20,21,61) and this study]. The overwinding activity of LrpC might compensate the activity of other chromosomal associated proteins [e.g. Hbsu/EcoHU or EcoH-NS, that constrain negative supercoils, (62)]. This is consistent with the observations that LrpC cooperatively increased DNA binding of the Hbsu protein (20).
A parallel can be built between the octameric LrpC, the dimeric sequence-independent chromatin-associated Hbsu protein and the monomeric eukaryotic HMG box protein. The three proteins have been shown to bind specifically to Holliday junctions, and in the case of LrpC and HMG box proteins, to the open conformations of the Holliday junctions [(63) and this study]. Furthermore, the three proteins play a significant, but unknown role, in the repair of DNA damages in B.subtilis cells [(31,64) and this study]. However, considering the result that LrpC is not an abundant protein in the cells under normal growth conditions (19), a role as a global chromosome organizer has to be taken with caution.
Recently a molecular mechanism that introduces a strong bias in the direction of Holliday-junction processing towards non-cross-over products (monomeric circular chromosomes), during the repair of double-strands breaks, has been uncovered both in E.coli (65) and in B.subtilis cells (36). LrpC has many of the activities associated with RecU (namely it binds ssDNA, dsDNA and with high affinity Holliday junctions and promotes joint molecule formation), but is not involved in the cleavage of Holliday junctions. Furthermore, similarly to ΔrecU cells and ΔruvAB cells, the presence of linked nucloeids was observed in ΔlrpC cells. We suggest that LrpC is required to promote, either non-cross-over products, or to avoid resolution of Holliday junctions towards cross-over products (dimeric chromosomes), by binding to the Holliday junctions. Experiments are in progress to address this hypothesis.
Acknowledgments
We thank Juan C. Alonso, Paul Thaw and John B. Rafferty for critical reading of the manuscript. This work was supported by grant BMC-2003-01969 from DGICYT to S.A. G.L. was a holder of a fellowship of DGICYT (PB 96-0817). Funding to pay the Open Access publication charges for this article was provided by grant BMC-2003-01969 by the Spanish Ministry of Science and Education.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brinkman A.B., Ettema T.J., De Vos W.M., Van Der Oost J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 2.Calvo J.M., Matthews R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman E.B., Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in Escherichia coli. Annu. Rev. Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 4.Hung S.P., Baldi P., Hatfield G.W. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 2002;277:40309–40323. doi: 10.1074/jbc.M204044200. [DOI] [PubMed] [Google Scholar]
- 5.Tani T.H., Khodursky A., Blumenthal R.M., Brown P.O., Matthews R.G. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl Acad. Sci. USA. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Antonio A., Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Madan Babu M., Teichmann S.A. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmon K., Hung S.P., Mekjian K., Baldi P., Hatfield G.W., Gunsalus R.P. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Hao Z., Bieniek E., Calvo J.M. Modulation of Lrp action in Escherichia coli by leucine: effects on non-specific binding of Lrp to DNA. J. Mol. Biol. 2001;314:1067–1075. doi: 10.1006/jmbi.2000.5209. [DOI] [PubMed] [Google Scholar]
- 10.Leonard P.M., Smits S.H., Sedelnikova S.E., Brinkman A.B., de Vos W.M., van der Oost J., Rice D.W., Rafferty J.B. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koike H., Ishijima S.A., Clowney L., Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc. Natl Acad. Sci. USA. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf J.R., Wu J., Calvo J.M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Rosner M.H., Calvo J.M. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 2001;312:625–635. doi: 10.1006/jmbi.2001.4955. [DOI] [PubMed] [Google Scholar]
- 15.Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Beloin C., Ayora S., Exley R., Hirschbein L., Ogasawara N., Kasahara Y., Alonso J.C., Hegarat F.L. Characterization of an lrp-like (lrpC) gene from Bacillus subtilis. Mol. Gen. Genet. 1997;256:63–71. doi: 10.1007/s004380050546. [DOI] [PubMed] [Google Scholar]
- 17.Dartois V., Liu J., Hoch J.A. Alterations in the flow of one-carbon units affect KinB-dependent sporulation in Bacillus subtilis. Mol. Microbiol. 1997;25:39–51. doi: 10.1046/j.1365-2958.1997.4491805.x. [DOI] [PubMed] [Google Scholar]
- 18.Belitsky B.R., Gustafsson M.C., Sonenshein A.L., Von Wachenfeldt C. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 1997;179:5448–5457. doi: 10.1128/jb.179.17.5448-5457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beloin C., Exley R., Mahe A.L., Zouine M., Cubasch S., Le Hegarat F. Characterization of LrpC DNA-binding properties and regulation of Bacillus subtilis lrpC gene expression. J. Bacteriol. 2000;182:4414–4424. doi: 10.1128/jb.182.16.4414-4424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapias A., Lopez G., Ayora S. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 2000;28:552–559. doi: 10.1093/nar/28.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beloin C., Jeusset J., Revet B., Mirambeau G., Le Hegarat F., Le Cam E. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 2003;278:5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- 22.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 23.Petrascheck M., Escher D., Mahmoudi T., Verrijzer C.P., Schaffner W., Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Calvo J.M. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993;12:2495–2501. doi: 10.1002/j.1460-2075.1993.tb05904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D' Ari R., Lin R.T., Newman E.B. The leucine-responsive regulatory protein: more than a regulator? Trends Biochem. Sci. 1993;18:260–263. doi: 10.1016/0968-0004(93)90177-o. [DOI] [PubMed] [Google Scholar]
- 26.Pontiggia A., Negri A., Beltrame M., Bianchi M.E. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 27.Kamashev D., Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sanchez H., Kidane D., Reed P., Curtis F.A., Cozar M.C., Graumann P.L., Sharples G.J., Alonso J.C. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171:873–883. doi: 10.1534/genetics.105.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez S., Sorokin A., Alonso J.C. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez S., Rojo F., Alonso J.C. The Bacillus subtilis chromatin-associated protein Hbsu is involved in DNA repair and recombination. Mol. Microbiol. 1997;23:1169–1179. doi: 10.1046/j.1365-2958.1997.3061670.x. [DOI] [PubMed] [Google Scholar]
- 32.Alonso J.C., Tailor R.H., Luder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 1988;170:3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso J.C., Stiege A.C., Luder G. Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 1993;239:129–136. doi: 10.1007/BF00281611. [DOI] [PubMed] [Google Scholar]
- 34.Ceglowski P., Luder G., Alonso J.C. Genetic analysis of recE activities in Bacillus subtilis. Mol. Gen. Genet. 1990;222:441–445. doi: 10.1007/BF00633853. [DOI] [PubMed] [Google Scholar]
- 35.Bravo A., Alonso J.C., Trautner T.A. Functional analysis of the Bacillus subtilis bacteriophage SPP1 pac site. Nucleic Acids Res. 1990;18:2881–2886. doi: 10.1093/nar/18.10.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrasco B., Cozar M.C., Lurz R., Alonso J.C., Ayora S. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction-processing functions in chromosome segregation. J. Bacteriol. 2004;186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayora S., Missich R., Mesa P., Lurz R., Yang S., Egelman E.H., Alonso J.C. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 2002;277:35969–35979. doi: 10.1074/jbc.M204467200. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Jimenez M.I., Alonso J.C., Ayora S. Bacillus subtilis bacteriophage SPP1-encoded gene 34.1 product is a recombination-dependent DNA replication protein. J. Mol. Biol. 2005;351:1007–1019. doi: 10.1016/j.jmb.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 39.Ayora S., Rojo F., Ogasawara N., Nakai S., Alonso J.C. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 1996;256:301–318. doi: 10.1006/jmbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- 40.Mehta R.A., Kahn J.D. Designed hyperstable Lac repressor DNA loop topologies suggest alternative loop geometries. J. Mol. Biol. 1999;294:67–77. doi: 10.1006/jmbi.1999.3244. [DOI] [PubMed] [Google Scholar]
- 41.Watson M.A., Gowers D.M., Halford S.E. Alternative geometries of DNA looping: an analysis using the SfiI endonuclease. J. Mol. Biol. 2000;298:461–475. doi: 10.1006/jmbi.2000.3676. [DOI] [PubMed] [Google Scholar]
- 42.Carrasco B., Ayora S., Lurz R., Alonso J.C. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho P.S., Eichman B.F. The crystal structures of DNA Holliday junctions. Curr. Opin. Struct. Biol. 2001;11:302–308. doi: 10.1016/s0959-440x(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 44.Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986;5:181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson C.E., Wang Y.H., Griffith J.D., Sinden R.R. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu B., Girard F., van Buuren B., Schleucher J., Tessari M., Wijmenga S. Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold. Nucleic Acids Res. 2004;32:3228–3239. doi: 10.1093/nar/gkh645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada K., Ariyoshi M., Morikawa K. Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr. Opin. Struct. Biol. 2004;14:130–137. doi: 10.1016/j.sbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 48.McGregor N., Ayora S., Sedelnikova S., Carrasco B., Alonso J.C., Thaw P., Rafferty J. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure (Camb) 2005;13:1341–1351. doi: 10.1016/j.str.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 49.Ayora S., Carrasco B., Doncel E., Lurz R., Alonso J.C. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl Acad. Sci. USA. 2004;101:452–457. doi: 10.1073/pnas.2533829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chedin F., Ehrlich S.D., Kowalczykowski S.C. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J. Mol. Biol. 2000;298:7–20. doi: 10.1006/jmbi.2000.3556. [DOI] [PubMed] [Google Scholar]
- 51.Morimatsu K., Kowalczykowski S.C. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 52.Boubrik F., Rouviere-Yaniv J. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl Acad. Sci. USA. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedberg E.C., Walker G.C., Siede W. DNA Repair and Mutagenesis. Washington DC: ASM Press; 1995. [Google Scholar]
- 54.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 55.Harmon F.G., DiGate R.J., Kowalczykowski S.C. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 56.Dri A.M., Rouviere-Yaniv J., Moreau P.L. Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J. Bacteriol. 1991;173:2852–2863. doi: 10.1128/jb.173.9.2852-2863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filutowicz M., Ross W., Wild J., Gourse R.L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J. Bacteriol. 1992;174:398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soppa J., Kobayashi K., Noirot-Gros M.F., Oesterhelt D., Ehrlich S.D., Dervyn E., Ogasawara N., Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Q., Pongpech P., DiGate R.J. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez C.R., Yang S., Deibler R.W., Ray S.A., Pennington J.M., Digate R.J., Hastings P.J., Rosenberg S.M., Zechiedrich E.L. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol. Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Torrejon G. Madrid, Spain: Universidad Autónoma de Madrid; 2002. Estudio de proteinas que intervienen en procesos de recombinación genética en Bacillus subtilis: LrpC y Hbsu. PhD Thesis. [Google Scholar]
- 62.Oberto J., Drlica K., Rouviere-Yaniv J. Histones, HMG, HU, IHF: Meme combat. Biochimie. 1994;76:901–908. doi: 10.1016/0300-9084(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 63.Pohler J.R., Norman D.G., Bramham J., Bianchi M.E., Lilley D.M. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritt C., Grimm R., Fernandez S., Alonso J.C., Grasser K.D. Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry. 1998;37:2673–2681. doi: 10.1021/bi972620r. [DOI] [PubMed] [Google Scholar]
- 65.Cromie G.A., Leach D.R. Control of crossing over. Mol. Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]