Abstract
Fluorescence techniques are commonly and powerfully applied to monitor biomolecular folding. In a limited fashion, the fluorescence emission intensity of covalently attached pyrene has been used as a reporter of RNA conformational changes. Here, we pursue two goals: we examine the relationship between tether identity and fluorescence response, and we determine the general utility of pyrene fluorescence to monitor RNA folding. The P4–P6 domain of the Tetrahymena group I intron RNA was systematically modified at multiple nucleotide positions with pyrene derivatives that provide a range of tether lengths and compositions between the RNA and chromophore. Certain tethers typically lead to a superior fluorescence signal upon RNA folding, as demonstrated by equilibrium titrations with Mg2+. In addition, useful fluorescence responses were obtained with pyrene placed at several nucleotide positions dispersed throughout P4–P6. This suggests that monitoring of tertiary folding by fluorescence of covalently attached pyrene will be generally applicable to structured RNA molecules.
INTRODUCTION
Applications of fluorescent probes in vitro and in vivo are an integral part of modern biochemistry (1,2). The use of fluorescence to study protein folding is well established (3,4), and the naturally occurring tryptophan residue often provides a useful chromophore (5,6). When tryptophan is absent, it may be engineered into a protein by site-directed mutagenesis [e.g. (7–10)]. For fluorescence studies of RNA folding, none of the four standard nucleobases is appreciably fluorescent, thereby demanding the introduction of an unnatural chromophore. The nonstandard nucleobase 2-aminopurine has been used (11,12), but the magnitude of its fluorescence change upon RNA folding is often small, and it reports optimally only when substantial changes occur in its base stacking (13). Other modified nucleobases have also been applied (14,15). We (16–19) and others (20,21) have used covalently attached pyrene in a limited fashion to monitor RNA conformational changes. For unknown structural reasons, the pyrene emission intensity often increases considerably—usually by more than a factor of two—upon folding of the RNA to which it is connected. Because this fluorescence response is orders of magnitude larger than the experimental detection limit, pyrene-labeled RNAs are an attractive target for RNA folding studies. The use of a single chromophore like pyrene has advantages over dual-chromophore (e.g. FRET) methods in that fewer chemical modifications to the RNA are required, and large distance changes between labeling sites are not necessary to generate a useful signal.
We previously used a covalently attached pyrene chromophore to monitor folding of the 160-nucleotide P4–P6 domain of the Tetrahymena group I intron RNA, a representative and well-studied RNA domain that adopts a characteristic tertiary structure (22,23). The pyrene was incorporated at a specific internal 2′-amino-2′-deoxynucleoside position (U107) via an amide-linked tether, denoted A5 in Figure 1 (16–19). When pyrene-labeled P4–P6 was titrated with Mg2+, which is generally required for RNA folding (24), the fluorescence intensity increased in two distinct phases (16). Based on several corroborating experiments, the first of these phases—with its [Mg2+]1/2 value (Mg2+ midpoint) on the order of 1 mM—was assigned to the global tertiary folding process. In contrast, the second phase (>10 mM Mg2+) was assigned to local folding changes that are independent of the overall tertiary folding process. This second undesired phase was more pronounced when an even shorter tether was used (A3 in Figure 1), suggesting that the fluorescence response could be optimized by more comprehensive examination of the tether length and composition.
Figure 1.
Pyrene derivatives of RNA examined in this study. (A) Uridine nucleotide with the A5 tether (16). (B) Pyrene derivatives A2–A7 and T5–T10. The A4 and T7 tethers were additionally examined in their unsaturated forms A4U and T7U. Pyrene-modified adenosines and cytidines were also examined in this study. (C) Preparative reactions of RNA amino groups with a pyrene STP ester. See Supplementary Data for all STP ester structures.
Considering the practical importance of fluorescence for studying biomolecular folding of all kinds, surprisingly few experiments have focused on the detailed relationship between the fluorescence signal and tether identity for either proteins or RNA. In our earlier report on P4–P6 folding using U107 pyrene fluorescence (16), the tether influence was not investigated fully, and other nucleotide positions were not examined within P4–P6. Subsequently, stopped-flow folding experiments using the key first phase of the U107 pyrene fluorescence change have aided our understanding of the P4–P6 folding landscape (17–19). From these experiments, we concluded that further development of the pyrene fluorescence approach to investigate RNA folding is warranted. In the present study, we examined how the fluorescence response changes when the tether between RNA and pyrene is altered systematically. We also determined how the fluorescence response depends on the placement of the pyrene probe within the RNA. The results reveal that the tether length and composition can affect the fluorescence response upon RNA folding and that generally applicable tethers can be identified. In addition, the results demonstrate that covalently attached pyrene provides a useful tertiary folding signal in many RNA contexts.
MATERIALS AND METHODS
Chemical structures and organic synthesis procedures of all pyrene labeling reagents (STP esters) are provided in Supplementary Data. See Figure 1 for pyr3-STP structure.
General considerations for nucleic acids manipulations
RNA and DNA oligonucleotides made by solid-phase synthesis were prepared at the UIUC W. M. Keck Center, Integrated DNA Technologies (Coralville, IA) or Dharmacon, Inc. (Lafayette, CO). Transcription of P4–P6 RNA sequences using T7 RNA polymerase and a linearized plasmid DNA template was performed as described previously (16). The template for P4–P6 nucleotides 117–261 (Δ15-P4–P6) was available from a previous study (16). The template for P4–P6 nucleotides 102–237 terminating with a 2′,3′-cyclic phosphate (P4–P6–24) was available from a previous study (25). The template for P4–P6 nucleotides 102–237 with the additional AA nucleotides inserted into the GAAA tetraloop (GAAAAA) was prepared from the parent P4–P6 plasmid (19) by linearization with BglII, which cleaves the plasmid DNA such that transcription ends after nucleotide C237. All DNA and RNA oligonucleotides were purified by PAGE as described previously (26,27). Splint ligation reactions to join pyrene-labeled RNA oligonucleotides and RNA transcripts were performed as described previously (16,17,28).
Pyrene labeling of 15-mer and 24-mer RNA oligonucleotides with an amino group (Figure 3A)
A common procedure was used for all derivatizations with pyrene, on the basis of our previous reports (16,17,19). The specific incubation conditions (pH, organic co-solvent, temperature and time) were optimized for each combination of amino-nucleoside and pyrene reagent, as described in Supplementary Data. A representative procedure for the preparation of a T8 derivative using pyr3-STP is as follows. A 250 μl solution containing 10 nmol 15-mer RNA oligonucleotide with a single 2′-(2-aminoethoxy) group in 100 mM sodium phosphate buffer (pH 8.0), 0.2 mM EDTA, 50% DMF and 25 mM pyr3-STP (added last from a 50 mM stock solution in DMF) was incubated at 37°C for 20 min. The solution was divided into two 125 μl portions in each of two 1.7 ml eppendorf tubes. To each sample was added 175 μl water, 30 μl of 3 M NaCl and 900 μl ethanol. The samples were mixed thoroughly, frozen on dry ice for 30 min and microcentrifuged at 13200 r.p.m. (16100 g) for 30 min. The supernatants were removed by pipette, and the pellets were dried under vacuum for 15 min. The pellets were redissolved in a combined volume of 100 μl water, and the pyrene-labeled RNA was isolated by 20% denaturing PAGE. Isolated yields ranged from 35 to 90% on the analytical scale without purification (see Supplementary Data) and from 30 to 50% on the preparative scale with PAGE purification. For ligation to form P4–P6 with pyrene at either U107 or A114 with any of the tether combinations (Figure 3B), the previously described procedure was followed (16,28). For the annealing procedure to assemble P4–P6 without ligation (Figure 3C), see the titration description below.
Matrix-assisted laser desorption ionization (MALDI) mass spectrometry data
MALDI-MS data were obtained for representative pyrene-derivatized oligonucleotides, as tabulated in Supplementary Data. All observed masses were within reasonable error (<0.2%) of the expected masses. Several full-length (160 nt) pyrene-labeled P4–P6 RNAs that were prepared according to Figure 3B were also analyzed by MALDI-MS. As tabulated in Supplementary Data, the observed masses were as expected.
Equilibrium fluorescence titrations with Mg2+ (Figures 4–7)
The titrations were performed generally as described previously (16) using a ThermoSpectronic AB2 fluorescence spectrometer for all measurements. The excitation wavelength λexc was 340 nm, and the fluorescence intensities plotted on each titration curve were determined at the emission λmax (380 ± 2 nm) for each particular sample. The emission λmax changed by <2 nm in all cases during any titration. The excitation and emission bandpass settings were 4 and 8 nm, and the instrument was set at 1 nm resolution. RNA samples were prepared in 1× TB buffer [89 mM each Tris and boric acid (pH 8.3)] and maintained at 35°C with a recirculating water bath. For samples prepared by path 1 of Figure 3 (derivatization-ligation), the RNA sample (650 μl) was prepared with 300–500 nM P4–P6 in 1× TB without annealing, which we have previously shown to be unnecessary (16,19). For samples prepared by path 2 of Figure 3 (derivatization-annealing), the RNA sample (650 μl) was prepared with 300 nM of pyrene-labeled 24-mer (nt 238–261) plus 900 nM of P4–P6–24 (nt 102–237) in 1× TB; this sample was annealed by heating at 60°C for 10 min and cooling at room temperature for 15 min. All samples were equilibrated at 35°C for at least 5 min before initiating the titration. During the titration, aliquots of either 100 or 900 mM MgCl2 in 1× TB as appropriate were added to the sample, which was mixed manually in a stoppered 1.5 ml UV cuvette and re-equilibrated to 35°C before measurement. Titration curves were fit by the two-component titration curve equation described previously (16). See Supplementary Data for tabulation of [Mg2+]1/2 and Hill coefficient (n) values. In Figure 4A, each spectrum was corrected for background fluorescence and normalized to 1.0 fluorescence units at 0 mM Mg2+, except the U107-T5 data were normalized to 1.0 at 65 μM Mg2+ to keep all spectra on the same y-axis scale. Similarly, in Figure 4B the U107-T5 titration data were normalized to 1.0 at 65 μM Mg2+; the non-normalized value of relative fluorescence intensity at 65 μM Mg2+ was 2.1.
Nondenaturing gel electrophoresis (Figure 8)
The native gel experiments were performed as described previously (19,28) except that samples were annealed at 60°C for 10 min before loading. Each gel experiment was performed using a buffer containing 1× TB [89 mM each Tris and boric acid (pH 8.3)] along with the desired concentration of Mg2+. Control lanes labeled ‘wt’ and ‘unf’ in Figure 8 refer to wild-type and unfolded control versions of P4–P6 (P4–P6-wt and P4–P6-bp), as described previously (19,28). See Supplementary Data for a tabulation of [Mg2+]1/2 and ΔΔG°′ values as determined from titration plots such as those in Figure 8B (28). For the samples prepared by annealing a 5′-32P-radiolabeled 136-nt RNA with several-fold excess of a 24-nt RNA (Figure 8C and D), an undesired band due to unannealed 136 nt RNA was generally observed (marked with an arrow in Figure 8C). The variable intensity of this band was greatest at low Mg2+ concentrations (<1.5 mM). The band persisted even when the concentration of unradiolabeled 24-mer RNA was increased several-fold (data not shown) and when the 24-mer lacked pyrene altogether. For all native gel experiments conducted at [Mg2+] ≥ 0.5 mM, a band due to fully assembled P4–P6 (136 + 24 = 160 nt) was clearly observed.
RESULTS
Design of tethers to connect RNA and pyrene
The pyrene-labeled uridine nucleotide shown in Figure 1A corresponds to the nucleotide we described previously for monitoring P4–P6 tertiary folding by fluorescence (16). Here, we used the 14 pyrene derivatives shown in Figure 1B. In each case, the pyrene moiety is connected to the RNA through an amide bond, either via a 2′-amino group (‘A’ series; A2–A7) or via a 2′-(2-aminoethoxy) group (tethered amine ‘T’ series; T5–T10). These derivatives collectively have between 2 and 10 non-hydrogen atoms between the ribose ring and the pyrene; the number of intervening atoms is placed after the A or T designation. Tether lengths of 5, 6 and 7 atoms are represented in both the A and T series, which allows us to test specifically for the influence of tether composition rather than length on the fluorescence response. To determine the roles of tether flexibility and also chromophore identity, the A4 and T7 tethers were additionally synthesized in their α,β-unsaturated forms denoted A4U and T7U, in which the pyrene π-system is conjugated with the carbonyl group through the tether.
For all of the pyrene derivatives, the covalent connection between RNA and pyrene was achieved by reaction of amino-modified RNA with a suitably activated pyrene-substituted carboxylic acid ester. Previously (16), we used the N-hydroxysuccinimidyl (NHS) esters of 1-pyrenebutanoic acid or 1-pyreneacetic acid to obtain the A5 or A3 tether, respectively. We have subsequently found that 4-sulfo-2,3,5,6-tetrafluorophenyl (STP) esters are superior to NHS esters for this purpose, due in part to their increased water solubility (29). For this reason, only the STP esters were used in this study, in the manner of Figure 1C. Syntheses of the required STP esters are described in Supplementary Data.
P4–P6 nucleotide positions for pyrene labeling
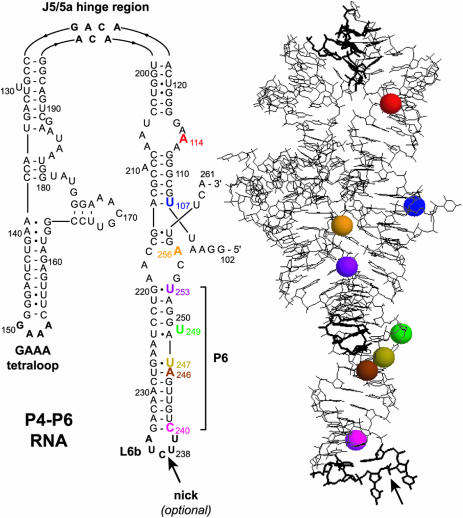
In the presence of at least millimolar Mg2+, the overall tertiary structure of P4–P6 is dominated by a bend of ∼180° at the J5/5a hinge region (22,30,31), which leads to a compact folded structure (Figure 2). Previously, covalent modification of U107 near the P4–P6 5′-terminus (G102) established that the 2′-position of U107 is a useful pyrene attachment site [P4–P6 encompasses nucleotides 102–261 because P4–P6 is a structured fragment of the larger group I intron RNA (32)]. In particular, 2′-modification at U107 does not substantially disrupt the P4–P6 tertiary structure (16), and pyrene attached at U107 is useful for monitoring the RNA conformation in both equilibrium and stopped-flow measurements (17–19). Here, we examined pyrene at several other nucleotides within P4–P6 to determine the generality of the fluorescence response. Based on the X-ray crystal structure (22), we chose seven P4–P6 nucleotides in addition to U107 (Figure 2). All of these new nucleotides were selected in part because the corresponding 2′-hydroxyl group in the X-ray crystal structure is pointing outward, suggesting that covalent modification will not be disruptive structurally. The same strategy was employed to choose U107 in our original study (16). Because U107 and the seven other nucleotides are widely dispersed throughout P4–P6 (Figure 2), the results were anticipated to reflect more broadly on the applicability of pyrene fluorescence for monitoring RNA tertiary folding.
Figure 2.
Secondary and tertiary structure of P4–P6 (22). Colored spheres highlight 2′-hydroxyls of the eight pyrene-labeled nucleotides that are marked on the secondary structure. The backbone may be nicked within the L6b loop without perturbing the tertiary folding (25), which permits assembly of full-length P4–P6 without covalent ligation of the 136-nt and 24-nt fragments (Figure 3).
Procedures for P4-P6 RNA assembly
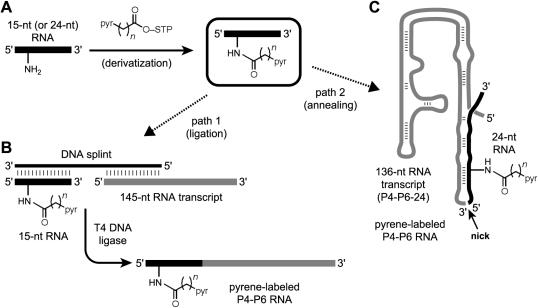
The eight nucleotide positions were also chosen based on the practical ability to assemble pyrene-derivatized P4–P6 RNAs with modifications at these sites. For both U107- and A114-modified P4–P6, the 160-nt pyrene-labeled RNAs were prepared in a two-step derivatization-ligation procedure (Figure 3, path 1). First, an amino-substituted oligoribonucleotide corresponding to the first 15 nt of P4–P6 was labeled with pyrene (Figure 3A). Elsewhere we have described preparation of the amino-modified RNA nucleoside phosphoramidites that were used during solid-phase oligoribonucleotide synthesis (33) (C.V. Miduturu and S.K. Silverman, manuscript in preparation). Second, pyrene-derivatized P4–P6 was obtained by ligation of this pyrene-labeled 15-mer to a 5′-phosphorylated 145-nt transcript using a complementary DNA splint and T4 DNA ligase (19,28,34–37) (Figure 3B). Ligation reactions for U107-modified RNA proceeded reasonably well, with 25–30% isolated yields after PAGE purification; these yields are comparable with those observed previously (16,28). Ligation reactions for A114-modified RNAs were poorer, with only 3–5% isolated yields. The lower yields in the latter case are attributed to the proximity of the pyrene modification at A114 to the ligation site (between G116 and G117), which interferes with T4 DNA ligase activity. It is important to note that this practical difficulty reflects a shortcoming of protein-mediated splint ligation rather than the fluorescence approach; here, we accepted obtaining enough material simply to test the viability of the fluorescence response. We emphasize that the low ligation yield with pyrene at A114 is a special case and clearly not representative of other ligation yields for assembling P4–P6 in this manner.
Figure 3.
Synthesizing pyrene-labeled P4–P6 by derivatization-ligation (path 1) or by derivatization-annealing (path 2). (A) A 15-mer or 24-mer oligoribonucleotide incorporating a single 2′-amino or 2′-(2-aminoethoxy) nucleotide is derivatized with pyrene as in Figure 1C and purified by PAGE. (B) To complete path 1, the 15-nt product from the derivatization step is ligated to the remaining 145 nt of P4–P6 using a DNA splint and T4 DNA ligase; the product is purified by PAGE. (C) To complete path 2, the 24-nt product from the derivatization step is annealed to the remaining 136 nt of P4–P6 without covalent ligation.
For the A3–A7 pyrene derivatives, the optimized labeling reactions of 2′-amino RNA with pyr1-STP through pyr5-STP provided 40–70% yield of pyrene-modified 15-mer as quantified by analytical PAGE. On the 10 nmol scale, the preparative yield were 10–30% after PAGE purification, which reflects typical losses during PAGE relative to the analytical-scale assays. These labeling reactions required 8–24 h incubations for optimal yield, and the reaction time and temperature were chosen to suppress secondary reactions of the pyrene-labeled product (see Supplementary Data). For reactions with pyr0-STP and pyr2U-STP to form A2 and A4U, in which the STP ester moiety is conjugated with the remainder of the molecule, the pyrene labeling reactions were slower than for the other derivatives, as anticipated based on chemical considerations (i.e. a conjugated ester is a less reactive electrophile than its nonconjugated analogue). Nevertheless, good yields of pyrene-labeled RNA were obtained within 24 h in all cases. For T6–T10, the yields of pyrene-derivatized RNA were generally higher than with the A series. Specifically, 65–90% yields were obtained on the analytical scale for reactions of 2′-(2-aminoethoxy)RNA with pyr1-STP through pyr5-STP; preparative yields were 20–75% after PAGE purification. For the modification reactions with pyr0-STP and pyr2U-STP to form T5 and T7U, the reactions were slower (as observed for A2 and A4U), but good yields of labeled RNA were obtained.
For P4–P6 derivatized with pyrene at the other six nucleotide positions of Figure 2, we took advantage of the fact that the 3′-terminal 24 nt of P4–P6 (U238–A261) can be annealed to the remaining 136 nt [denoted P4–P6–24 (25)] while leaving an unligated nick between the two fragments (Figure 2). Despite this discontinuity in the phosphodiester backbone, the bimolecular RNA:RNA complex adopts tertiary structure in a Mg2+-dependent manner like conventional P4–P6 and is even useful for crystallography (25). We therefore synthesized 24-mer oligoribonucleotides with individual amino group substitutions and separately derivatized each with pyrene as in Figure 3A. Yields were comparable with those obtained with the 15-mer oligonucleotide that corresponds to the 5′-terminus of P4–P6. Subsequently, full-length P4–P6 samples were assembled according to path 2 of Figure 3 merely by annealing each pyrene-derivatized 24-mer with a modest excess of the underivatized 136-nt P4–P6–24 (Figure 3C). No RNA ligation reactions are required in this latter approach, which therefore allows a rapid assessment of the fluorescence response using pyrene labeling at each nucleotide position.
Equilibrium fluorescence titrations of P4–P6 labeled with pyrene at U107: effect of tether length and composition
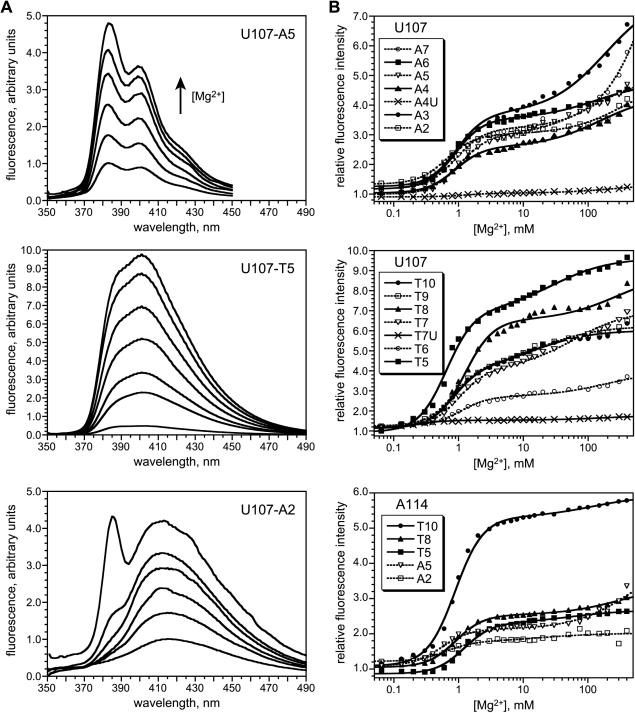
Because our earlier report specifically studied the A5 and A3 pyrene modifications at U107 (16), we first examined the complete series of pyrene derivatives from Figure 1 at this nucleotide position. Equilibrium fluorescence titrations were obtained by recording the fluorescence emission spectra of pyrene-labeled P4–P6 in the absence of Mg2+ and then titrating Mg2+ in small increments from 65 μM to 400 mM (Figure 4A). After correction for dilution, the relative fluorescence intensity as a function of [Mg2+] was fit to a standard multi-component titration curve (Figure 4B). Each of the U107 pyrene A3–A7 and T6–T10 derivatives showed significant fluorescence intensity increases, including a substantial increase at [Mg2+] ≈ 1 mM that corresponds to tertiary folding (16). As previously observed for A5 and A3 (16), each of the A3–A7 and T6–T10 derivatives has an additional high-Mg2+ titration component that does not correspond to tertiary folding. For each of these pyrene-labeled RNAs, the shape of the emission spectrum was essentially identical to that of pyrene free in solution (as determined using pyrenebutyric acid, which is the hydrolyzed version of pyr3-STP; data not shown).
Figure 4.
Equilibrium fluorescence titrations of P4–P6 derivatives labeled with pyrene at U107 and A114. (A) Fluorescence emission spectra (λexc = 340 nm) showing representative data from 0 to 400 Mg2+ (1× TB buffer, 35°C). The U107 A3–A7 and T6–T10 spectra were similar in shape to A5 (data not shown). The low-wavelength spectral feature for U107-A2 was observed with varying intensity at all other tested nucleotide positions (see Supplementary Data). For each of the five A114 tethers, the fluorescence spectra were qualitatively similar to those for U107 with the same tether (see Supplementary Data). (B) Fitted titration curves for the A and T series of tethers at U107 and A114. The relative fluorescence intensity was determined at λmax in all cases. See Materials and Methods for details of curve fitting. See Supplementary Data for [Mg2+]1/2 values.
The remaining tether combinations alter the pyrene chromophore itself relative to that found within A3–A7 and T6–T10. Specifically, A2 and T5 have a pyrenecarboxamide instead of a pyrenecarbonyl chromophore, whereas A4U and T7U have a vinylogous (conjugated) pyrenecarboxamide group (see structures in Figure 1B). The importance, if any, of changing the chromophore in these ways was unknown in advance. For the A2 and T5 derivatives, the shapes of the fluorescence emission spectra themselves are readily distinguishable from those of A3–A7 and T6–T10 (Figure 4A). In addition, A2 has a reproducible spectral feature of unknown origin near 385 nm that appears only at high Mg2+. When the A2 fluorescence emission intensity at λmax is plotted versus Mg2+ concentration (Figure 4B), the resulting titration curve is very similar to that of A5, and T5 also shows a large increase in fluorescence intensity for the tertiary folding component. In sharp contrast, A4U and T7U show relatively little change in emission intensity upon Mg2+ titration (Figure 4B). The shapes of the fluorescence emission spectra of the A4U- and T7U-labeled RNA (broad fluorescence spectra with λmax ≈ 480 nm and 465 nm; data not shown) are noticeably different from those of the other labeled RNAs, apparently due to the extended conjugation between the pyrene and tether. On the basis of their very small fluorescence responses with added Mg2+, the A4U and T7U chromophores were not examined further in this study.
Pyrene as an RNA structure probe at other P4–P6 nucleotides
The results for the series of pyrene substitutions at U107 suggest that the tether length and composition can strongly affect the fluorescence response upon Mg2+ titration. To determine the influence of chromophore location within the RNA, we examined a representative subset of the pyrene derivatives (A2, A5, T5, T8 and T10) at the seven other nucleotide positions of Figure 2. These five pyrene derivatives cover all tether lengths from 2 to 10 non-hydrogen atoms and also span the entire range of useful tether and chromophore combinations, including the qualitatively distinct A2 and T5 chromophores. The A114 pyrene derivatives were prepared using the same derivatization-ligation approach (Figure 3, path 1) that was used for the U107 derivatives. The A114 fluorescence responses depend on the tether identity, with the T10 derivative providing a particularly large response (Figure 4B, bottom panel).
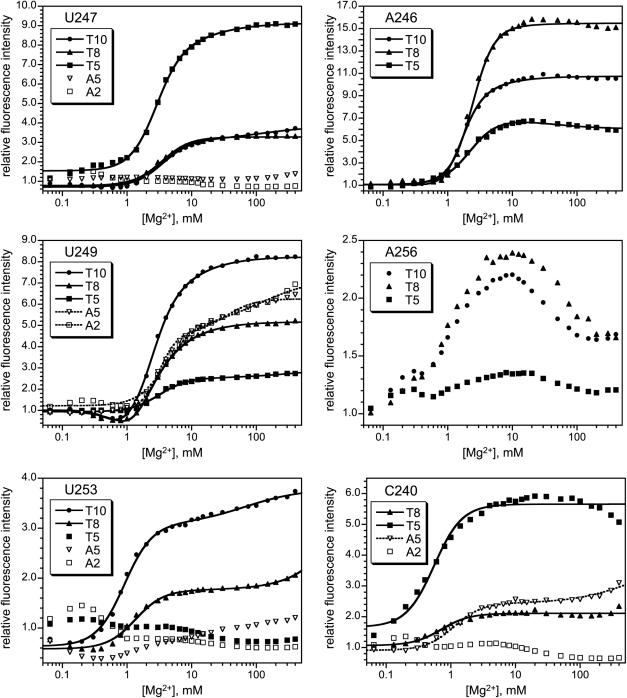
For positions U247, U249 and U253, the five representative pyrene derivatives were each tested using RNAs prepared by the derivatization-annealing approach (Figure 3, path 2). Each of the U249-labeled RNAs shows a substantial increase in pyrene fluorescence emission for all five of these tethers, whereas the U247 and U253 derivatives show intensity increases only for the longer T8 and T10 tethers (Figure 5). Additionally, U247 shows a significant response with T5 but not A5 (which are the same length), indicating that tether composition as well as length can be important. For all three positions U247, U249 and U253, the high-Mg2+ fluorescence component that does not correspond to tertiary folding was very small or undetected.
Figure 5.
Equilibrium fluorescence titrations of P4–P6 derivatives labeled with pyrene at U247, U249, U253, A246, A256 or C240. For each of the five tethers, the fluorescence spectra were qualitatively similar to those for U107 that are depicted in Figure 4 (data not shown). See Supplementary Data for [Mg2+]1/2 values.
Pyrene derivatization at nucleotide A246 was studied for the longer T5, T8 and T10 tethers (A2 and A5 were not examined). All three derivatives showed significant fluorescence responses with very little of the unwanted high-Mg2+ component (Figure 5). The A246-T8 tether provided a particularly strong signal upon Mg2+-dependent tertiary folding; its 15-fold fluorescence increase is several-fold larger than for U107-A5 and also much larger than for U107-T8. Thus, A246 appears to be a more responsive modification site than U107 for reporting on P4–P6 folding by pyrene fluorescence. Indeed, A246 is the most responsive of all positions tested to date within P4–P6.
The T5, T8 and T10 tethers were also placed at A256 (Figure 5). The A256-T5 derivative led to a poor fluorescence response that is not useful for monitoring RNA folding. For the T8 and T10 derivatives, a 2-fold increase in fluorescence was observed, with an unusual decrease in intensity for the second component. We consider these ambiguous responses. That is, the A256-T8 and A256-T10 derivatives might be useful for monitoring RNA folding, but other incorporation sites are better practical choices.
Finally, when pyrene was placed at C240 on any of the five representative tethers, the responses were variable (Figure 5). At C240, the A2 tether gave little signal change, whereas A5, T5 and T8 had more conventional-appearing responses. However, the data described in the next section show that these responses are probably not due to RNA tertiary folding. The C240-T10 combination was unusual in that pyrene excimer formation was observed; even the 24-mer C240-T10 oligonucleotide alone (i.e. not as part of P4–P6) showed excimer formation, as depicted in Supplementary Data. On the basis of distinct RNA concentration dependence, we ascribe these signals to aggregation, and thus the C240-T10 responses are not relevant to unimolecular RNA tertiary folding.
Demonstrating that Mg2+-dependent fluorescence changes are due to RNA tertiary folding
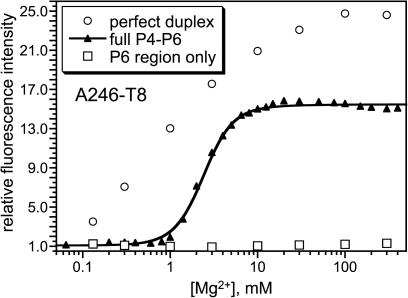
In previous work, we unambiguously established that the major component of fluorescence increase upon Mg2+ titration is due to RNA tertiary folding for the U107-A5 P4–P6 derivative (16). Here, we sought evidence that the fluorescence responses for new pyrene-labeled P4–P6 RNAs are due to tertiary folding. One control experiment is to incorporate the pyrene-labeled 15-mer or 24-mer RNA not into the entire 160-nt foldable P4–P6 RNA but instead into only a small portion of P4–P6. In the latter case, the fluorescence response assigned to P4–P6 tertiary folding should not be observed. For labeled RNAs prepared using the 24-mer oligonucleotide (nt 238–261), pyrene can be placed within the P6 region, which is the lower right portion of P4–P6 as depicted in the secondary structure of Figure 2. As an example, we annealed the 24-mer A246-T8 RNA to the RNA oligonucleotide corresponding to P4–P6 nt 215–237, thereby reconstituting just the P6 region. Titration of the resulting RNA complex with Mg2+ led to essentially no increase in fluorescence, which is a qualitatively different outcome than for the full A246-T8 P4–P6 RNA (Figure 6). This is compelling evidence that the response of the full 160-nt A246-T8 RNA is due to tertiary folding and not to local structural changes, because the reconstituted P6 region provides the same local environment as the full P4–P6. In contrast, the reconstituted C240-T8 P6 region showed a response similar to that of the full 160-nt P4–P6 (see Supplementary Data). On this basis, we cannot safely conclude that the C240-T8 fluorescence response is due to tertiary folding.
Figure 6.
Control experiments with reconstituted P6 region to show that A246-T8 pyrene fluorescence reports on P4–P6 tertiary folding. See text for details.
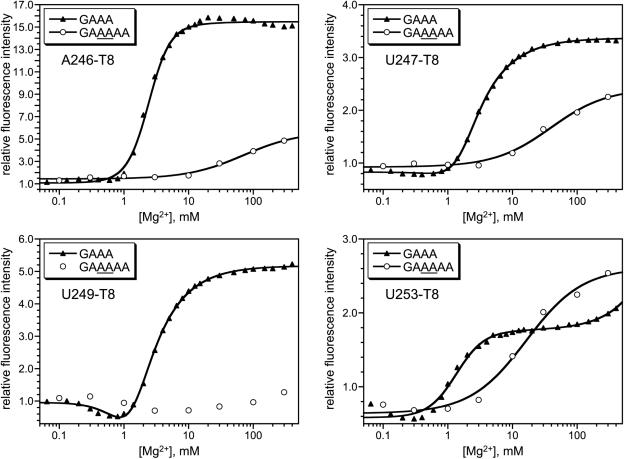
A different type of experiment can support the relationship between pyrene response and tertiary folding for most of the pyrene-labeled RNAs. Key evidence originally obtained in support of U107 pyrene RNA reporting on P4–P6 tertiary folding was that the [Mg2+]1/2 values of the fluorescence responses shift upon RNA sequence modification in accord with predictions based on native gel electrophoresis (16). We recently described nucleotide insertions in the P4–P6 GAAA tetraloop, which is remote in primary sequence from the P6 region (Figure 2). These insertions induce an increased Mg2+ dependence for P4–P6 tertiary folding according to native gels and also according to U107 pyrene fluorescence (19). If pyrene placed within the P6 region reports on tertiary folding similarly to pyrene placed at U107, then tetraloop modifications should shift the fluorescence titration curves along the [Mg2+] axis. In contrast, if pyrene within P6 is not reporting on tertiary folding, then tetraloop modifications should have little or no impact on the Mg2+ dependence of fluorescence. We performed the necessary experiments for the T8 derivatives at A246, U247, U249 and U253. In each case, the full P4–P6 RNA was prepared according to path 2 of Figure 3 by annealing the appropriate pyrene-labeled 24-nt RNA with a 136-nt transcript. The latter incorporated either an unmodified GAAA tetraloop (as a control) or a modified tetraloop of sequence GAAAAA, where the additional two AA nucleotides shift the [Mg2+]1/2 from 0.7 to 6.9 mM according to native gels (19). In all four cases, the tetraloop sequence modification had a sizeable effect upon the fluorescence titration curve (Figure 7). For A246-T8 and U247-T8, the titration curve was shifted substantially to the right, with diminution of fluorescence increase. For U249-T8, the fluorescence increase was suppressed almost completely at the accessible Mg2+ concentrations. Finally, for U253-T8, the [Mg2+]1/2 increased about 10-fold and the fluorescence increase was approximately unchanged. In contrast to all of these data, analogous experiments with tetraloop-modified C240-T8 RNAs (prepared either by the ligation or annealing pathway of Figure 3) showed no compelling change in Mg2+ dependence of the fluorescence titration (see Supplementary Data). These findings are consistent with the above experiments using reconstituted P6 regions, which showed that A246-T8 fluorescence responses are due to tertiary folding but C240-T8 responses are not due to tertiary folding.
Figure 7.
Experiments with wild-type (GAAA) or modified (GAAAAA) tetraloops to show that pyrene fluorescence reports on P4–P6 tertiary folding for the T8 tether at A246, U247, U249 and U253. See text for details.
Nondenaturing (native) gel electrophoresis experiments
The fluorescence titration data (Figures 4 and 5) demonstrate that for certain combinations of nucleotide position and tether identity, the fluorescence response of pyrene-labeled P4–P6 is poor upon Mg2+ addition, with almost no fluorescence change in some cases. In principle, a low fluorescence response could arise because the RNA simply does not fold upon Mg2+ addition, due to structural perturbation from pyrene and the appended tether. Alternatively, a low fluorescence response could derive from indifference of the chromophore to environmental change induced by Mg2+-dependent RNA tertiary folding, even though folding itself proceeds normally. The ability of P4–P6 to fold—without considering fluorescence intensity changes—may be examined using nondenaturing (native) gel electrophoresis, which directly interrogates the folded state of the RNA. Additionally, from the magnitude of any shift in the Mg2+ dependence, these data allow us to quantify the energetic penalties imposed on P4–P6 folding by appending the various tethers, independent of the fluorescence assays (28).
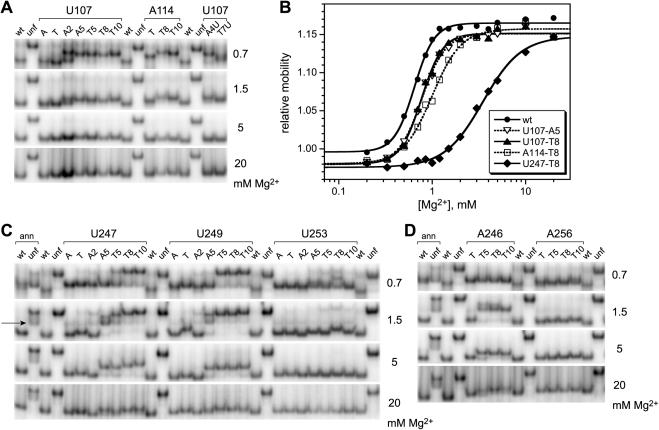
For the P4–P6 RNAs derivatized at U107, we used native gels to examine the representative A2, A5, T5, T8 and T10 pyrene derivatives, each of which showed good fluorescence response upon Mg2+ addition (Figure 4). Each of these derivatives showed conventional Mg2+-dependent native gel behavior, in that the mobility of the pyrene-labeled RNA relative to an unfolded control mutant (30) increased as the Mg2+ concentration was raised (Figure 8A). The free energy destabilizations (ΔΔG°′ values) of each pyrene-labeled P4–P6 RNA relative to unmodified P4–P6 were modest (<0.5 kcal/mol), as evidenced by only slight rightward shifts in the Mg2+ midpoints of the titration curves (Figure 8B, see Supplementary Data for computation of ΔΔG°′ values) (28). The A114-T8 and A114-T10 derivatives showed similar behavior, with ΔΔG°′ ≃ 1.0 kcal/mol (Figure 8A); the other A114 derivatives were not tested. Interestingly, the U107-A4U and U107-T7U derivatives that showed no fluorescence responses (Figure 4) folded normally according to native gel analysis (Figure 8B). This indicates that the lack of fluorescence response for these RNAs is not due to structural disruption.
Figure 8.
Nondenaturing (native) polyacrylamide gel electrophoresis of pyrene-labeled P4–P6 RNAs. The Mg2+ concentration used throughout each gel is shown on the right side of each image. Wt and unf denote unmodified wild-type P4–P6 and the unfolded control mutant (30). (A) Representative data for U107 and A114 derivatives. The lane labels A and T denote P4–P6 derivatives with 2′-amino or 2′-tethered amino groups but lacking pyrene. (B) Representative equilibrium titration curves derived from the gel data. From these curves, the energetic consequences of pyrene derivatization may be computed (28). The ΔΔG°′ values ranged from ∼0.4 kcal/mol (U107-A5 and T8) to ∼3–4 kcal/mol (U247-T8). For additional titration curves and ΔΔG°′ values, see Supplementary Data. (C) Data for U247, U249 and U253 derivatives. The wt and unf samples additionally labeled ‘ann’ were prepared by annealing the 24-mer RNA oligonucleotide to the 136-nt remainder of P4–P6 as in Figure 3C. The arrow on the left indicates the band corresponding to the unannealed 136-nt RNA that is particularly prominent at low Mg2+ concentrations (see Materials and Methods). (D) Data for A246 and A256 derivatives.
In the fluorescence studies described in this report, all of the other P4–P6 derivatives were assembled by annealing a pyrene-labeled 24-mer oligoribonucleotide to the 136-nt remainder of P4–P6, according to path 2 of Figure 3. For the native gel analyses, a 32P-radiolabel was required on the 136-mer in addition to the pyrene on the 24-mer, which was added in at least 4-fold excess (to ensure that all radiolabels were incorporated into full-length P4–P6 assemblies). The surplus pyrene-labeled 24-mer is inconsequential because fluorescence detection is not used with the native gels. Using RNAs prepared by this annealing approach, we comprehensively examined the A2, A5, T5, T8 and T10 derivatives at nucleotide positions U249, U247 and U253 (Figure 8C), which had shown varying fluorescence responses (Figure 5). At all three of these modification sites, the RNA was fully folded at high Mg2+ concentration (e.g. 20 mM) for all tested pyrene derivatives, including those derivatives that gave no fluorescence response. The ΔΔG°′ values were as high as 3–4 kcal/mol, with the longer tethers exhibiting generally larger energetic perturbations (e.g. compare U247-A2 though U247-T10 in Figure 8C). Similarly, normal folding at high Mg2+ concentration was observed for P4–P6 RNAs modified with pyrene at A246 and A256 (Figure 8D), as well as pyrene at C240 (data not shown).
Correlating native gels with fluorescence
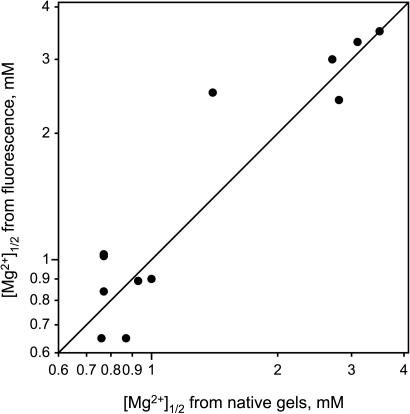
As established above, the attachment of a pyrene chromophore can modestly perturb the RNA tertiary folding, as assessed by its Mg2+ dependence. Both the fluorescence and native gel approaches allow the determination of precise [Mg2+]1/2 values (Figures 4, 5 and 8; see Supplementary Data for tabulations). When these two types of data are compared for those pyrene derivatives that were examined using both techniques, the quantitative correlation is very good (Figure 9). This outcome is additional evidence that pyrene fluorescence reports on RNA tertiary folding.
Figure 9.
Correlation between [Mg2+]1/2 values for individual pyrene-labeled P4-P6 derivatives as determined by native gels and by fluorescence. The diagonal line has a slope of exactly 1.0. Each of the 12 data points in the plot represents a different combination of pyrene site and tether. Wild-type P4-P6 without an attached pyrene has [Mg2+]1/2 of 0.64 mM in this set of experiments. The native gel [Mg2+]1/2 values represented in this plot correspond to ΔΔG°′ values between ∼0.3 and ∼6 kcal/mol (see Supplementary Data for tabulation of data). Values of [Mg2+]1/2 are reproducible with <5% error between independent determinations. Because ΔG°′ = nRT•ln[Mg2+]1/2, where n is the Mg2+ Hill coefficient (16,28), the [Mg2+]1/2 values are plotted on logarithmic axes.
DISCUSSION
The two main goals of this study were both focused on increasing our understanding of pyrene fluorescence as a probe of RNA folding. First, we sought to examine how the tether identity affects the fluorescence response. Second, we sought to explore the generality of pyrene fluorescence by placing the chromophore at various positions throughout P4–P6 (as a representative large RNA). Via the reported experiments, we have achieved both of these goals. Our data help to define the scope and limitations of using covalently attached pyrene as a probe of RNA tertiary folding.
Pyrene fluorescence changes reflect tertiary folding at many RNA positions
A primary issue underlying all of our interpretations is that the pyrene fluorescence responses must in fact arise from RNA tertiary folding. For all eight nucleotide positions examined in P4–P6 (Figure 2), pyrene labeling led to substantial Mg2+-dependent fluorescence increases for at least some tethers (Figures 4 and 5). At least three lines of evidence indicate that these fluorescence changes are indeed due to RNA tertiary folding in most—but not all—cases. First, the experiments in Figure 6 show that local RNA structure is not responsible for the fluorescence changes in the case of A246-T8; this is similar to tests previously performed for pyrene labeling at U107 (16). Second, the experiments in Figure 7 show that the previously studied GAAAAA tetraloop modification (19) shifts the dependence of the fluorescence response to higher Mg2+ concentration for all four tested labeling sites (A246, U247, U249 and U253). The diminished magnitude of fluorescence response with the modified RNA in three of the four experiments makes it difficult to interpret the [Mg2+]1/2 shift. In the case of U253-T8, where the magnitude of response is approximately unchanged by the tetraloop modification, the shift in Mg2+ dependence is quantitatively as expected from the native gels (i.e. increase in [Mg2+]1/2 from 0.7 to 6.9 mM by native gels, and from 1.4 to 16 mM by fluorescence). Only in the case of C240 pyrene labeling do the analogous experiments indicate that RNA tertiary folding is probably not responsible for the Mg2+-dependent fluorescence changes. Additional comments on the implications of this latter result for the general applicability of pyrene fluorescence are given below.
Third, the very good correlation between native gel and fluorescence [Mg2+]1/2 values (Figure 9) is most simply explained by assigning the shifts in fluorescence [Mg2+]1/2 to tertiary folding, as is already known to be the case for shifts in native gel [Mg2+]1/2 values (28). Any imperfections in this correlation are likely due to subtle effects that shift the [Mg2+]1/2 of a fluorescence titration in an unpredictable fashion, superimposed on the overall shift due to tertiary folding perturbation. Of course, ideally the plot of Figure 9 would have no dispersion at all along the x-axis; merely appending pyrene would have no effect on tertiary folding. However, appending pyrene is itself a chemical modification with quantifiable RNA folding consequences (similar to how inserting a tryptophan often requires protein mutation). In this context, the good correlation of Figure 9 not only shows that pyrene fluorescence reports on tertiary folding but also that fluorescence is capable by itself of providing meaningful data on energetic perturbations. This ability may prove useful in other RNA systems for which native gel electrophoresis is not as quantitative as for P4–P6 itself.
Tether identity is important: particularly useful tethers for pyrene-labeled RNA
At the outset of this study, we knew that pyrene is a faithful reporter of RNA tertiary folding when incorporated at the particular U107 nucleotide position within P4–P6 (16–19). Furthermore, the fluorescence response had been examined largely with only a single tether, termed A5 in this work (Figure 1). When pyrene is placed at many nucleotide positions within P4–P6 (Figure 2) by an appropriate labeling strategy (Figure 3), the fluorescence data (Figures 4 and 5) indicate that the longer tethers T8 and T10 are particularly useful for joining RNA to pyrene, whereas the shorter tethers sometimes lead to little fluorescence response. The precise break point at which a shorter tether does not lead to a good fluorescence response depends on the nucleotide position. For example, tethers at U107 ranging from A3 to T10 all report well on P4–P6 folding, whereas the longer T8 and T10 tethers report much better for U253. Therefore, the first main goal of this study—to examine dependence of fluorescence response on tether identity—has therefore been reached, with the overall finding that longer tethers such as T8 and T10 provide a more reliable fluorescence response.
Pyrene labeling at many positions leads to useful fluorescence responses: implications for general applicability of pyrene labeling
The utility of the pyrene fluorescence approach for reporting upon RNA folding at sites other than nucleotide U107 of P4–P6 was almost entirely unaddressed at the outset of this study. Here, we have demonstrated that fluorescence is a useful probe for RNA tertiary folding when pyrene is placed at many positions throughout P4–P6. Because the successfully modified nucleotides (all sites except C240 in Figure 2) are scattered throughout P4–P6 at locations tens of Å apart, pyrene fluorescence is useful for monitoring RNA folding when the chromophore is placed in a variety of RNA contexts. Our findings suggest that the folding of large RNAs that are unrelated to P4–P6 can also be studied using pyrene; this should be tested with other RNAs in further experiments that are beyond the scope of the current study. It is encouraging to note that a recent independent study with the HIV-1 TAR RNA and aminoglycoside ligands used our general pyrene methodology to obtain useful RNA folding data (38).
The inability of C240 pyrene labeling to report convincingly on P4–P6 tertiary folding is likely a consequence of C240's peripheral location on the folded RNA molecule (Figure 2). Ultimately, the available data do not permit us to state with certainty why the C240 response does not reflect tertiary folding. As a practical guideline, we suggest that future attempts at pyrene labeling of RNA should focus on placing the pyrene probe at nucleotides located internally within the RNA of interest, rather than on the periphery. We also note that it would be unreasonable to expect pyrene fluorescence to give a useful tertiary folding response in absolutely every RNA position that is examined. For calibration, tryptophan in proteins does not always give a useful response and neither does 2-aminopurine in nucleic acid studies, yet these probes have undeniably found widespread practical utility.
Because our labeling approach attaches pyrene to RNA via a ribose 2′-position, it is useful to examine other large RNAs to determine whether they have a significant fraction of their 2′-positions available for modification according to high-resolution structural data. We have checked this for two other large RNAs (see Supplementary Data). In the HDV ribozyme (39), at least 50 out of 72 positions (∼69%) meet this criterion, as assessed by manual inspection. Similarly, in the 5S RNA within the large ribosomal subunit (40), at least two-thirds of the 122 2′-positions appear accessible for derivatization, even when considering contacts between RNA and the other components of the ribosome.
RNA structure consequences of pyrene labeling
Our data allow us to infer some of the structural consequences of labeling P4–P6 RNA with pyrene. Several specific combinations of nucleotide position and tether resulted in no useful fluorescence response (Figures 4 and 5; see U107-A4U and T7U; U247-A2 and A5; U253-A2, A5 and T5). These poor responses could mean either that each RNA does not fold, or that folding occurs but leads to little fluorescence change. These possibilities are distinguished using the native gel assays of Figure 8, which report directly on the RNA's folded state. The rightward shifts in [Mg2+]1/2 shown in Figure 8B correspond to 0.4–4 kcal/mol of free energy penalty (ΔΔG°′) as a consequence of the pyrene modification (16). Even for those pyrene-labeled RNAs that have a free energy penalty on the large side of this range, the native gel titration data indicate that the overall structural perturbation is minimal, because the shape and the mobility limits of the titration curves are similar to those of wild-type (unmodified) P4–P6. Furthermore, there is no obvious correlation between magnitude of fluorescence response upon Mg2+ titration and ΔΔG°′ (e.g. A246-T8 and A246-T10 in Figure 5). Of course, the local rather than global effects of pyrene incorporation are difficult to assess without high-resolution structural information. An important finding from the native gel studies is that none of the pyrene-labeled RNAs fails to fold upon addition of sufficient Mg2+. Therefore, the lack of useful fluorescence response from certain combinations of labeling site and tether does not reflect failure to fold but instead demonstrates indifference of the chromophores to the RNA folding event.
With respect to the extent of structural perturbation due to appending pyrene, the situation faced here is similar to that encountered during a typical study of protein folding in which tryptophan is introduced by mutagenesis [e.g. (7–10)].As is the case for proteins, functional assays should be useful for demonstrating that any specific RNA retains its proper structure (and therefore function) upon pyrene labeling. Native gel electrophoresis serves this role for P4–P6 (28). The ability to incorporate pyrene independently at several nucleotide positions should alleviate concern over structural artifacts that may arise when the chromophore is incorporated at just one site.
Physical basis of the fluorescence response
In a typical fluorescence experiment to study protein folding, tryptophan emission intensity serves as an empirical correlate of protein folding. These data are often interpreted without a full (or even partial) understanding of the physical origin of the change in signal. Nevertheless, this lack of understanding does not diminish the practical utility of the fluorescence response. Similarly, we and others have previously demonstrated that pyrene fluorescence is useful for monitoring RNA folding, despite our incomplete knowledge of the underlying photophysical explanation (16–21). Here, our data allow three statements about the physical basis of the pyrene fluorescence response upon RNA folding:
The unresponsiveness of the unsaturated A4U and T7U tethers as compared with their more flexible A4 and T7 counterparts (Figure 4) suggests that the chromophore must not be held rigidly away from the RNA itself if the fluorescence signal is to change upon Mg2+-induced folding. This implies some direct interaction between the RNA and chromophore in either the unfolded or folded state (or both) for the responsive tethers. The data do not allow us to distinguish in which state such interaction occurs, or if interaction occurs in both states. An alternative and not necessarily exclusive contribution to the explanation is that the conjugated chromophores may simply be less responsive to changes in nearby RNA structure.
Similarly, the break points in tether length after which fluorescence response is gained imply that a particularly short tether does not allow the chromophore to interact with the RNA in the same way as pyrene on a longer tether. The variation of this break point with nucleotide position indicates that the RNA–pyrene interaction which changes the fluorescence intensity likely involves specific contacts. Higher-resolution information is needed to assess the structural possibilities in greater detail.
The direct comparisons of the 5-, 6- and 7-atom tethers (A5–A7 versus T5–T7; Figures 4 and 5) indicate that the tether composition and not just length affects the fluorescence response. For U247 in particular, the equal-length A5 and T5 tethers provide very different fluorescence responses (Figure 5). This demonstrates explicitly that the tether composition itself can contribute substantially to the fluorescence response, although the other data indicate that tether composition does not always contribute substantially.
A more comprehensive understanding of the relationships among derivatization site, tether identity and fluorescence response is clearly an interesting biophysical objective. In practice, the utility of pyrene fluorescence (16–21) is not hampered by our incomplete mechanistic understanding of the origin of the fluorescence changes, however desirable such understanding is from the perspective of biophysical chemistry. Similarly, tryptophan is a useful probe for protein folding even when the mechanistic explanation for its fluorescence change is unknown, which is the case in nearly all experimental applications of tryptophan fluorescence (5–10) and for other chromophores as well (41). Our data provide guidelines for future experiments in which covalently attached pyrene is used to study RNA tertiary folding.
Practical synthesis of pyrene-labeled RNA
During these experiments, we used two pyrene labeling strategies: the derivatization-ligation and derivatization-annealing approaches (Figure 3). The ligation approach (Figure 3, path 1) is required for incorporating modifications at P4–P6 nucleotides U107 and A114; in general, this approach is appropriate when the RNA backbone must be unbroken in the vicinity of the pyrene substitution. The alternative annealing approach (Figure 3, path 2) was used for the other six pyrene-modified P4–P6 nucleotides, which were chosen in part because they are amenable to this streamlined procedure. Others have used similar tactics to examine structure–function correlations for various ribozymes (42), and recently the annealing of fluorescently labeled probe oligos to large RNAs has been reported (43,44). Although the annealing approach is useful for P4–P6, the covalent integrity of the RNA backbone is probably required in many cases for meaningful fluorescence folding data to be obtained. RNA ligation strategies using either protein enzymes (34–37,45) or deoxyribozymes (46) will be valuable for such experiments.
The relatively broad applicability of the T8 tether (Figures 4 and 5) has several side benefits in terms of practical RNA labeling. The requisite organic reagent pyr3-STP is commercially available and also readily synthesized (see Supplementary Data). The precursor to the RNA portion of T8, a 2′-(2-aminoethoxy) group (Figure 1C), is relatively reactive as a nucleophile. The derivatization of this ‘tethered’ primary amino functional group proceeds rapidly, requiring only 30 min incubation at 25°C. This is particularly fast in comparison with a 2′-amino group, which reacts much more slowly (e.g., requiring 24 h incubation at 25°C) and therefore leads to more side products. The increased reactivity for the tethered amine relative to 2′-amino is presumably a combination of steric and pKa effects. The 2′-amino group is sterically hindered and substantially less basic (and therefore less nucleophilic) than a 2′-tethered amine, as revealed by nondenaturing gel electrophoresis (see Supplementary Data). At present, the tethered amine nucleosides are not commercially available within RNA oligonucleotides, but we have reported efficient synthetic procedures for each of the corresponding nucleoside phosphoramidites (33). Although the T10 derivative had a larger fluorescence response than T8 in several cases, the corresponding pyr5-STP is not as soluble as pyr3-STP under the typical incubation conditions for RNA derivatization, which apparently contributes to lower derivatization yields (see Supplementary Data). We therefore tend to favor T8 over T10 on a practical basis.
CONCLUSION
By covalently attaching a pyrene moiety onto the P4-P6 RNA with a variety of tethers and at multiple nucleotide positions, we have explored the general applicability of pyrene fluorescence as a probe to monitor RNA tertiary folding. The results reveal that the tether length and composition can substantially affect the fluorescence response observed upon Mg2+-dependent RNA folding. The longer T8 and T10 tethers have relatively large fluorescence increases upon RNA folding and therefore are most useful. In contrast, shorter tethers sometimes have diminished fluorescence response, and tethers that have a rigid spacer element between RNA and pyrene show almost no fluorescence change upon folding. The T8 and T10 tethers lead to a productive fluorescence response when placed at many—but not all—of the tested nucleotide positions dispersed throughout P4–P6. This suggests that pyrene fluorescence will be useful for studying the tertiary folding of RNAs other than P4-P6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the National Institutes of Health (GM-65966), the March of Dimes Birth Defects Foundation (Research Grant No. 5-FY02-271) and the UIUC Department of Chemistry (all to S.K.S.). S.K.S. is the recipient of a fellowship from The David and Lucile Packard Foundation. The authors thank Brian C. Smith for assistance with synthesis of the pyr2 compounds. The authors thank members of the Silverman laboratory for discussions, in particular Chandra Miduturu for advice on organic synthesis and native gel electrophoresis. The authors also thank Chris McGinley for assistance with diazomethane reactions. The Open Access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mollova E.T. Single-molecule fluorescence of nucleic acids. Curr. Opin. Chem. Biol. 2002;6:823–828. doi: 10.1016/s1367-5931(02)00394-0. [DOI] [PubMed] [Google Scholar]
- 2.Whitaker M. Fluorescent tags of protein function in living cells. BioEssays. 2000;22:180–187. doi: 10.1002/(SICI)1521-1878(200002)22:2<180::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Beechem J.M., Brand L. Time-resolved fluorescence of proteins. Annu. Rev. Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 4.Eftink M. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- 5.Munro I., Pecht I., Stryer L. Subnanosecond motions of tryptophan residues in proteins. Proc. Natl Acad. Sci. USA. 1979;76:56–60. doi: 10.1073/pnas.76.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Barkley M.D. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 7.Kwan T., Loughrey H., Brault M., Gruenheid S., Urbatsch I.L., Senior A.E., Gros P. Functional analysis of a tryptophan-less P-glycoprotein: a tool for tryptophan insertion and fluorescence spectroscopy. Mol. Pharmacol. 2000;58:37–47. doi: 10.1124/mol.58.1.37. [DOI] [PubMed] [Google Scholar]
- 8.Ding H., Mukerji I., Oliver D. Lipid and signal peptide-induced conformational changes within the C-domain of Escherichia coli SecA protein. Biochemistry. 2001;40:1835–1843. doi: 10.1021/bi002058w. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay S., Valder C.R., Huynh H.G., Ren H., Allison W.S. The βG156C substitution in the F1-ATPase from the thermophilic Bacillus PS3 affects catalytic site cooperativity by destabilizing the closed conformation of the catalytic site. Biochemistry. 2002;41:14421–14429. doi: 10.1021/bi026243g. [DOI] [PubMed] [Google Scholar]
- 10.Hagan R.M., Worner-Gibbs J., Wilton D.C. Tryptophan insertion mutagenesis of liver fatty acid-binding protein: L28W mutant provides important insights into ligand binding and physiological function. J. Biol. Chem. 2005;280:1782–1789. doi: 10.1074/jbc.M407131200. [DOI] [PubMed] [Google Scholar]
- 11.Menger M., Tuschl T., Eckstein F., Porschke D. Mg2+-dependent conformational changes in the hammerhead ribozyme. Biochemistry. 1996;35:14710–14716. doi: 10.1021/bi960440w. [DOI] [PubMed] [Google Scholar]
- 12.Millar D.P. Fluorescence studies of DNA and RNA structure and dynamics. Curr. Opin. Struct. Biol. 1996;6:322–326. doi: 10.1016/s0959-440x(96)80050-9. [DOI] [PubMed] [Google Scholar]
- 13.Jean J.M., Hall K.B. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C., Martin C.T. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 15.Berry D.A., Jung K.-Y., Wise D.S., Sercel A.D., Pearson W.H., Mackie H., Randolph J.B., Somers R.L. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides. Tetrahedron Lett. 2004;45:2457–2461. [Google Scholar]
- 16.Silverman S.K., Cech T.R. RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry. 1999;38:14224–14237. doi: 10.1021/bi991333f. [DOI] [PubMed] [Google Scholar]
- 17.Silverman S.K., Deras M.L., Woodson S.A., Scaringe S.A., Cech T.R. Multiple folding pathways for the P4–P6 RNA domain. Biochemistry. 2000;39:12465–12475. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- 18.Silverman S.K., Cech T.R. An early transition state for folding of the P4–P6 RNA domain. RNA. 2001;7:161–166. doi: 10.1017/s1355838201001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young B.T., Silverman S.K. The GAAA tetraloop–receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4–P6 RNA domain. Biochemistry. 2002;41:12271–12276. doi: 10.1021/bi0264869. [DOI] [PubMed] [Google Scholar]
- 20.Bevilacqua P.C., Kierzek R., Johnson K.A., Turner D.H. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 21.Kierzek R., Li Y., Turner D.H., Bevilacqua P.C. 5′-Amino pyrene provides a sensitive, nonperturbing fluorescent probe of RNA secondary and tertiary structure formation. J. Am. Chem. Soc. 1993;115:4985–4992. [Google Scholar]
- 22.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 23.Juneau K., Podell E., Harrington D.J., Cech T.R. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 24.Tinoco I., Jr, , Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 25.Golden B.L., Gooding A.R., Podell E.R., Cech T.R. X-ray crystallography of large RNAs: heavy-atom derivatives by RNA engineering. RNA. 1996;2:1295–1305. [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn-Charlebois A., Wang Y., Prior T.K., Rashid I., Hoadley K.A., Coppins R.L., Wolf A.C., Silverman S.K. Deoxyribozymes with 2′–5′ RNA ligase activity. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Silverman S.K. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 28.Silverman S.K., Cech T.R. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 29.Gee K.R., Archer E.A., Kang H.C. 4-Sulfotetrafluorophenyl (STP) esters: new water-soluble amine-reactive reagents for labeling biomolecules. Tetrahedron Lett. 1999;40:1471–1474. [Google Scholar]
- 30.Murphy F.L., Cech T.R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 31.Murphy F.L., Cech T.R. GAAA tetralooop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 32.Cech T.R. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 33.Jin S., Miduturu C.V., McKinney D.C., Silverman S.K. Synthesis of amine- and thiol-modified nucleoside phosphoramidites for site-specific introduction of biophysical probes into RNA. J. Org. Chem. 2005;70:4284–4299. doi: 10.1021/jo050061l. [DOI] [PubMed] [Google Scholar]
- 34.Moore M.J., Sharp P.A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice site. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 35.Moore M.J., Query C.C. RNA–Protein Interactions: A Practical Approach. In: Smith C.W.J., editor. Oxford: Oxford University Press; 1998. pp. 75–108. [Google Scholar]
- 36.Moore M.J. Joining RNA molecules with T4 DNA ligase. Methods Mol. Biol. 1999;118:11–19. doi: 10.1385/1-59259-676-2:11. [DOI] [PubMed] [Google Scholar]
- 37.Moore M.J., Query C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 38.Blount K.F., Tor Y. Using pyrene-labeled HIV-1 TAR to measure RNA-small molecule binding. Nucleic Acids Res. 2003;31:5490–5500. doi: 10.1093/nar/gkg755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferré-D' Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 40.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 41.Schönbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc. Natl Acad. Sci. USA. 2000;97:6345–6349. doi: 10.1073/pnas.120120397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira M.J., Harris D.A., Rueda D., Walter N.G. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 43.Dorywalska M., Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., Puglisi J.D. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith G.J., Sosnick T.R., Scherer N.F., Pan T. Efficient fluorescence labeling of a large RNA through oligonucleotide hybridization. RNA. 2005;11:234–239. doi: 10.1261/rna.7180305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bain J.D., Switzer C. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 1992;20:4372. doi: 10.1093/nar/20.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purtha W.E., Coppins R.L., Smalley M.K., Silverman S.K. General deoxyribozyme-catalyzed synthesis of native 3′–5′ RNA linkages. J. Am. Chem. Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.