Abstract
5-fluorouracil (5-FU) is a widely used anticancer drug that disrupts pyrimidine nucleotide pool balances and leads to uracil incorporation in DNA, which is then recognized and removed by the uracil base excision repair (BER) pathway. Using complementary biochemical and genetic approaches we have examined the role of uracil BER in the cell killing mechanism of 5-FU. A yeast strain lacking the enzyme uracil DNA glycosylase (Ung1), which excises uracil from the DNA backbone leaving an abasic site, showed significant protection against the toxic effects of 5-FU, a G1/S cell cycle arrest phenotype, and accumulated massive amounts of U/A base pairs in its genome (∼4% of T/A pairs were now U/A). A strain lacking the major abasic site endonuclease of Saccharomyces cerevisiae (Apn1) showed significantly increased sensitivity to 5-FU with G2/M arrest. Thus, efficient processing of abasic sites by this enzyme is protective against the toxic effects of 5-FU. However, contrary to expectations, the Apn1 deficient strain did not accumulate intact abasic sites, indicating that another repair pathway attempts to process these sites in the absence Apn1, but that this process has catastrophic effects on genome integrity. These findings suggest that new strategies for chemical intervention targeting BER could enhance the effectiveness of this widely used anticancer drug.
INTRODUCTION
The prodrug 5-fluorouracil (5-FU) (Figure 1A) has historically been used to treat varied types of malignancies including colorectal, breast, and head and neck cancers. In the year 2002, 5-FU was given to over 2 million patients worldwide making it one of the most widely used anticancer drugs (1,2). The mechanism of 5-FU involves enzymatic conversion to the active metabolite 5-fluorodeoxyuridine monophosphate (FdUMP) which covalently inhibits thymidylate synthase (TS), an essential enzyme responsible for synthesizing deoxythymidine monophosphate (dTMP) from deoxyuridine monophosphate (dUMP) (Figure 1A) (3). This is the major de novo pathway by which the cell produces thymidine precursors for DNA replication, and it is widely accepted that depletion of thymidine nucleotides for DNA synthesis following 5-FU treatment directly results in 5-FU cell killing (‘thymineless death’) (4).
Figure 1.
Possible biochemical linkages between 5-FU toxicity and damage to RNA and DNA. (A) The 5-fluorouracil may interfere with nucleic acid structure and function through inhibition of TS and disruption of nucleotide pool balance or the direct incorporation of 5-fluoronucleotides into DNA and RNA. (B) Although it has been hypothesized that uracil BER plays an important role in mediating 5-FU toxicity, the lethal intermediate along this pathway remains to be identified. Possible intermediates moving clockwise around the cycle are the mutagenic 5-FU base itself, the abasic product of the Ung1 reaction, or various incised products derived from the intact abasic site (see text).
Although the thymineless death mechanism is attractive in its simplicity, recent evidence suggests a more complex scenario involving pyrimidine nucleotide balances (5,6), DNA repair pathways and disruptions in RNA metabolism (4,7–9). For example, consider that inhibition of TS by 5-FU not only depletes the dTTP pool within the cell, but also increases the pools of dUTP and FdUTP that can be incorporated during DNA replication (Figure 1A). Although under normal conditions uracil is excluded from DNA by maintaining a low cellular dUTP pool through the action of deoxyuridine triphosphate nucleotidohydrolase (dUTPase), upon TS inhibition large amounts of dUTP and FdUTP accumulate which overwhelms the dUTPase activity. Thus, dUMP and FdUMP are incorporated into genomic DNA, which may directly lead to cytotoxicity either by causing mutations and resulting protein miscoding (10), or by triggering apoptosis (11). Alternatively, the presence of uracil and 5-FU in DNA may result in ‘futile cycling’ of uracil base excision repair (BER) because of the depleted dTTP pool (Figure 1B). In this case, the unwanted uracil base is removed by uracil DNA glycosylase (Ung1), the DNA backbone is nicked by an abasic endonuclease, the blocking 5′ deoxyribose phosphate is removed by a flap endonuclease (Rad27) and the resulting gap is filled in using another dUMP or FdUMP residue through the action of a repair DNA polymerase (pol ɛ) and DNA ligase (cdc9) (Figure 1B). During this cycling, there may be an accumulation of toxic intermediates such as abasic sites, single-strand breaks (SSBs) or double-strand breaks (DSBs) in the DNA, any of which may promote cell death (Figure 1B) (12). In addition to these DNA-based routes that promote 5-FU toxicity, at very high concentrations FUTP is also extensively incorporated into RNA, thereby inhibiting processing of pre-rRNA (13–15), post-transcriptional modification of tRNAs (16,17), and polyadenylation and splicing of mRNA (18,19). Despite its use in the clinic for over 40 years, the relative importance of these potential mechanisms for 5-FU-mediated cell killing is not clearly established. Understanding the salient mechanisms is of significant interest because novel targets for therapeutic development may be uncovered that could enhance the clinical efficacy of 5-FU and related fluoropyrimidines (20).
A potential approach to enhance the DNA-based mechanisms of 5-FU-mediated cell killing is to identify the toxic intermediates during uracil BER, and then develop small molecule inhibitors of downstream enzymes, leading to accumulation of the lethal intermediate. For instance, if accumulation of uracil or 5-FU in DNA is the trigger for cell death, then inhibition of the first step of BER catalyzed by Ung1 would be effective (Figure 1B). Alternatively, if the accumulation of abasic sites or DNA nicks is critical for toxicity, then inhibition of the subsequent steps of BER would potentially be the most productive route. Although it seems counterintuitive to inhibit processes of DNA repair, such a strategy could allow targeting of rapidly dividing cells in a sea of quiescent normal cells. Since up regulation of BER enzymes has been implicated in the resistance mechanisms and progression of various tumor cells (21–24), inhibitors of BER could also target one mechanism of drug resistance. The same inhibition strategy could be used to target pathogenic microorganisms such as the fungus Candida albicans, herpesviruses, and parasites from the Trypanosomatidae family (such as Leishmania major and Trypanosoma cruzi), all of which depend on stringent pyrimidine nucleotide pool balance for viability (25–27).
The budding yeast Saccharomyces cerevisiae is an ideal organism to investigate the importance of uracil BER in 5-FU toxicity because deletion yeast strains in most steps of this process are available (polε and cdc9 are lethal deletions), as well as other DNA repair pathways, and the yeast pathway is closely related to that in humans (28). Thus, the important intermediates and enzymes that enhance (or diminish) the potency and efficacy of 5-FU can be ascertained. Using complementary biochemical and genetic approaches we find that deletion of uracil DNA glycosylase, with a resultant massive increase in uracilated DNA, has a protective effect against 5-FU. In contrast, deletion of the major abasic site endonuclease of yeast (Apn1) results in a large increase in the potency and efficacy of 5-FU. Thus, abasic sites appear to be a major progenitor giving rise to the DNA-mediated toxic effects of 5-FU. The significance of these findings to cancer chemotherapy is considered.
MATERIALS AND METHODS
Yeast media and growth conditions
Wild-type yeast were grown in YEPD [1% yeast extract, 2% peptone (BD Biosciences), 2% dextrose (J. T. Baker)]; mutant strains in YEPD supplemented with 200 µg/ml geneticin (Gibco). Selection for fresh MATa single mutants was performed as described (29) using sporulation buffer (1% KOAc, 0.005% ZnOAc; Sigma) and MATa selective ‘magic medium’ (SC-Leu-His-Arg+L-canavanine (60 µg/ml) + geneticin; all amino acids and l-canavanine from Sigma, yeast nitrogenous base without amino acids and ammonium sulfate from Difco). The apn1 and quadruple mutants derived from FF18733 and rad27 yeast prepared using a URA3 deletion cassette were plated on the appropriate selective media (Supplementary Table S1).
Yeast strains
Table 1 details strains of S.cerevisiae used in this study. Apn1, ung1, rad27, rad2, msh2 and rad51 are isogenic to parental strain BY4741. Homozygous and convertible heterozygous mutants were supplied courtesy of Dr Jef Boeke. All mutations of strain BY4741 were confirmed via PCR using primers as described by the Saccharomyces Genome Deletion Project (30). The apn1 (strain BG1) and apn1apn2ntg1ntg2 (strain BG105) yeast and their parental strain (FF18733) were gifts of Dr Serge Boiteux and were verified for auxotrophy. The complete genotypes of these strains are reported in Supplementary Table S1.
Table 1.
S.cerevisiae mutant yeast strains used in this study
| Mutant strain | Missing enzyme activity | Potential effect of mutation after 5-FU treatment |
|---|---|---|
| ung1 | Uracil DNA glycosylase. Removes uracil and 5-FU from DNA. | Accumulation of uracil and 5-FU in DNA |
| apn1 | Abasic endonuclease. Cleaves DNA backbone 5′ to abasic sites; also 3′ phosphodiesterase activity. | Accumulation of abasic sites in DNA |
| rad27 | Flap endonuclease. Removes 5′ deoxyribose phosphate (dRP) blocking groups. | Accumulation of 5′ dRP-blocked sites in DNA |
| apn1 apn2 ntg1 ntg2 | Abasic endonucleases (apn1, apn2) and 3′ AP lyases (ntg1 and ntg2). | Accumulation of intact abasic sites in DNA |
| rad51 | ssDNA binding protein. Critical for homologous recombination. | Diminished capacity for recombinational repair of DSB's and SSB's generated from cleaved abasic sites |
| rad2 | Endonuclease required for incision on the 3′ side of a DNA lesion during NER. | Ablation of alternative pathway for repair of 5-FU induced DNA lesions |
| msh2 | Dimerizes with Msh3 or Msh6 to bind DNA mismatches and initiate MMR. | Ablation of alternative pathway for repair of 5-FU induced DNA lesions |
| apn1rad27 | Deficient in abasic site endonuclease and flap endonuclease | Increased levels of abasic sites and/or 5′ dRP-blocked sites in DNA |
5-FU survival time courses
Yeast were grown to logarithmic phase and diluted to 5 × 106 cells/ml into YEPD +/− 150 µM 5-FU (Sigma) based on a preliminary estimate of the EC50 of 5-FU for the parental BY4741 strain. At given times, cells were diluted, plated and incubated at 30°C until countable colonies had formed. Colony counting was assisted with the use of a Gel Doc imaging system (BioRad), and data were plotted using GraphPad Prism.
5-FU dose-response studies
Yeast were grown to logarithmic phase and diluted to 5 × 106 cells/ml into media containing varying concentrations of 5-FU. After 4 h of shaking at 30°C, the cells were diluted, plated and incubated until counting. Data were plotted and EC50 values were determined using GraphPad Prism.
Flow cytometry
Yeast were grown to logarithmic phase, diluted to 5 × 106 cells/ml into media +/−150 µM 5-FU, and prepared for flow cytometry as described previously (31). Briefly, at given time intervals, 5 × 106 cells were pelleted, washed and resuspended in 250 µl sterile water. Cold 100% ethanol was added to 70%, with vortexing. The cells were allowed to fix overnight at 4°C before being washed and resuspended in 100 µl of 50 mM sodium citrate, pH 7.0 (Fisher), and digested with 100 µg/ml RNase A at 37°C for 2 h. Additional buffer and propidium iodide (10 µg/ml final concentration; Sigma) were added, and samples were stored at 4°C in the dark. Flow cytometry was carried out using a FACScan cytometer (Beckton–Dickinson), and data were analyzed with Cell Quest software.
Analysis of U and 5-FU in DNA and RNA
Yeast were grown to logarithmic phase, diluted to 5 × 106 cells/ml into media +/− 150 µM 5-FU, and incubated for varying times (wt, 6 h, ung1, 6 h, apn1, 3 h) at 30°C with agitation in order to kill 70–90% of the wt and apn1 cells and achieve maximum cell killing with the ung1 yeast at this 5-FU concentration. Small aliquots were diluted and plated, and the remaining cells were spun down, lysed with lyticase (Sigma) and genomic DNA was isolated using Genomic-tips (Qiagen). Each sample (3 µg) was digested overnight at room temperature with 3 nM Escherichia coli uracil DNA glycosylase (Ung) and 3 nM human AP endonuclease (Ape1) in buffer containing 50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl and 10 mM MgCl2. Half of each reaction was run on a 0.8% agarose gel, which was stained with 1 µg/ml EtBr prior to imaging.
For GC-MS, yeast cells were grown to logarithmic phase and diluted to 5 × 106 cells/ml into 5-FU to achieve 70–90% cell killing across all samples (wt, 150 µM 5-FU for 6 h, ung1, 3 mM 5-FU for 4 h, and apn1, 150 µM 5-FU for 3 h). Aliquots were diluted and plated, and the genomic DNA was purified and digested with 10 nM Ung overnight at room temperature in buffer containing 10 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2 and 25 mM NaCl. Following digestion, aliquots of uracil-13C4, 15N2 and 5-FU-13C4, 15N2 were added as internal standards (stable isotope labeled U and 5-FU were purchased from Cambridge Isotope Laboratories). Then the DNA was precipitated with 70% EtOH, centrifuged and the supernatant and pellet fractions were separated. Ethanol was removed from supernatant fractions under vacuum in a SpeedVac at room temperature. Aqueous supernatant fractions were frozen in liquid nitrogen, lyophilized to dryness for 18 h, and then trimethylsilylated and analyzed by GC-MS as described (32–34). For identification and quantification, selected-ion monitoring was used to monitor the characteristic ions of the trimethylsilyl derivatives of uracil (m/z 256 and m/z 241), uracil-14C4,15N2 (m/z 262 and m/z 247), 5-FU (m/z 274 and m/z 259) and 5-FU-13C4,15N2 (m/z 280 and m/z 265) during GC/MS analysis. (In each case, the first ion is the molecular ion and the second one is the ion that results from the loss of methyl radical from the molecular ion.)
For RNA 5-FU incorporation analysis, wt yeast were grown to logarithmic phase, diluted to 5 × 106 cells/ml into YEPD +/− one EC50 of 5-FU, and shaken at 30°C for 6 h. Cellular RNA was then isolated using the RNeasy Kit (Qiagen). RNA (10 µg) was digested to nucleosides using mung bean nuclease (10 U) and calf intestinal phosphatase (10 U) overnight at 37°C in buffer containing 10 mM Tris–HCl, pH 7.9, 10 mM MgCl2, 50 mM NaCl and 1 mM DTT (all reagents from New England Biolabs). High-performance liquid chromatography (HPLC) was carried out using an analytical Aqua reversed-phase C18 column (Phenomenex) and isocratic elution with 3% acetonitrile in aqueous 0.1 M TEAA, pH 7.0, at a flow rate of 1 ml/min.
Aldehyde reactive probe slot-blot (ASB) assay
Yeast were grown to logarithmic phase, diluted to 5 × 106 cells/ml into YEPD +/− one EC50 of 5-FU, and shaken for 6 h at 30°C. The DNA was isolated using Genomic-tips (Qiagen) and subjected to the ASB assay as described (35–37). Briefly, 4 µg of each DNA sample was digested with E.coli exonuclease III (145 U; New England Biolabs) for 1 min at 37°C, 100 mM putrescine at 37°C for 30 min (Acros Organics), both exonuclease III and putrescine, or left undigested. All samples were precipitated with EtOH, resuspended in phosphate-buffered saline, and incubated with 1 mM aldehyde reactive probe (ARP; Dojindo Laboratories) for 10 min at 37°C. Following a second precipitation, the DNA was resuspended in TE buffer (10 mM Tris–HCl, pH 7.4 and 1 mM EDTA), quantified by ethidium bromide spotting, and 0.5 µg of each sample was diluted to a volume of 100 µl in TE, heat denatured and mixed with an equal volume of 20× SSC (3 M NaCl and 0.3 M sodium citrate). The denatured DNA was slot-blotted onto Hybond-C Super nitrocellulose (Amersham Biosciences) using a Minifold II slot-blot manifold (Schleicher & Schuell). The membrane was soaked in 5× SSC for 15 min at 37°C, dried, baked at 75°C under vacuum for 2 h, and then rewet and blocked in 10 ml of preincubation buffer (20 mM Tris–HCl, pH 7.5, 0.1 M NaCl, 1 mM EDTA, 0.5% casein, 0.25% BSA and 0.1% Tween 20) for 45 min at room temperature, with shaking. An additional 10 ml of prehybridization buffer containing a 1:500 dilution of streptavidin conjugated–horseradish peroxidase (HRP) was added to the membrane, and following an additional 40 min of shaking at room temperature, the membrane was reacted with enhanced chemiluminescence (ECL) reagents and imaged using Hyperfilm (streptavidin-HRP, ECL reagents and film from Amersham Biosciences). The films were analyzed using a Gel Doc System (Bio-Rad), and data were plotted using GraphPad Prism. Quantification was based on abasic site DNA standards purchased from Dojindo Laboratories.
RESULTS
General strategy
Many cancer chemotherapeutic strategies act by damaging DNA in various ways and therefore elicit a cellular DNA damage response that either repairs the damage, or alternatively, generates toxic repair intermediates that may be the true progenitors of drug-induced lethality. Our general strategy to examine the role of uracil BER in the mechanism of 5-FU toxicity is to use yeast strains deficient in various enzymatic steps of the pathway, thereby unmasking repair intermediates that may be progenitors of cell death (Figure 1B and Table 1). In addition, we sought to support the genetic results with quantitative analyses of uracil and 5-FU in DNA to determine the load of uracilated DNA in wild-type and the Ung1 and Apn1 deficient yeast, both in the absence and presence of5-FU. In a final analytical approach, we determine the number of intact and cleaved abasic sites in the presence and absence of 5-FU treatment for wild-type, ung1 and apn1 yeast. To investigate the possibility of alternative processing of abasic sites by other AP endonucleases or lyases, we also examined the yeast strain apn1apn2ntg1ntg2, which is defective in all pathways for endonucleolytic cleavage of abasic sites opposite to adenine bases (Table 1). Although the focus of this study is on uracil BER, the findings also necessitated an examination of potential backup roles for DSB repair, nucleotide excision repair (NER) and mismatch repair pathways, and an assessment of 5-FU incorporation into RNA. Combining both genetic and biochemical approaches allows assessment of the discrete molecular events that are most strongly correlated with the lethal effects of 5-FU treatment.
5-FU survival time courses for wild-type and BER deficient yeast
To investigate the influence of uracil BER enzyme activities on sensitivity to 5-FU, the survival curves of these yeast strains were measured in the absence and presence of the drug. In the absence of 5-FU, all uracil BER deficient strains exhibited indistinguishable doubling times over a 6 h growth period (not shown). Exceptions were the rad51 and rad27 strains that had doubling times about 1.5 times longer. In the presence of 150 µM 5-FU, a concentration similar to the EC50 for the wild-type parent yeast strain (see below), marked differences in survival were observed (Figure 2A). The ung1 strain, which is deficient in the removal of uracil and 5-FU, showed marked protection against 5-FU as compared with the wild-type strain, and it was not possible to achieve any greater than about 60% killing even after 54 h of treatment using this concentration of 5-FU. In contrast, the apn1 strain that is deficient in cleavage of the 5′ phosphodiester linkage of the abasic site product of the Ung1 reaction showed rapid and complete cell killing (Figure 2A). The log linear survival time course without lags for this strain suggests the rapid formation of a toxic species from 5-FU. For the rad27 strain, which is deficient in the 5′-dRPase activity required to process the 5′-incised abasic sites generated from Apn1 action, a strong protection against 5-FU was observed similar to ung1 (Figure 2A).
Figure 2.
Time courses for killing of wild-type and repair-deficient S.cerevisiae by 5-FU. Yeast were shaken in 150 µM 5-FU for given times and then diluted and plated. (A) Single mutants lacking enzymes along the uracil BER pathway have distinct 5-FU survival curves. Ung1 and rad27 yeast are protected from the effects of 5-FU while apn1 yeast are more sensitive to the drug relative to wild type. (B) Apn1apn2ntg1ntg2 yeast is more sensitive to 5-FU relative to wild-type and the apn1 strain. (C) HR deficient rad51 yeast are modestly protected against 5-FU as compared with wild-type yeast. (D) The NER and mismatch repair-deficient rad2 and msh2 yeast show no increased sensitivity relative to wild type at this concentration of 5-FU.
It should be noted that this protective rad27 phenotype was only observed for a haploid strain that was freshly prepared from a convertible heterozygote (29). In contrast, we found that an older rad27 strain showed similar sensitivity to 5-FU as compared with the wild type (data not shown). Our results with the older strain are similar to a recent report in which antifolate drugs were used to deprive yeast of thymidine (i.e. modest or no effect of the rad27 deletion on survival was observed) (38). These results emphasize that care must be taken when dealing with deletion yeast strains that show genetic instability and slow growth due to defects in DNA repair. Since rad27 acts downstream of apn1 in BER (Figure 1B), it would be expected that the phenotype of the apn1rad27 double mutant would recapitulate the 5-FU sensitivity of apn1 alone. Surprisingly, this was not the case, as apn1rad27 showed the same protective phenotype as rad27 (not shown).
The above findings suggest the following conclusions (i) the protective effect of the ung1 deletion indicates that accumulation of uracil or 5-FU in DNA is not responsible for the toxic effects of 5-FU, (ii) the increased 5-FU sensitivity of the apn1 strain indicates that efficient 5′ endonucleolytic processing of abasic sites plays an important role in cell survival in the presence of 5-FU, and (iii) the protective effect of deleting the 5′-dRPase activity provided by Rad27 suggests that single-nucleotide gapped DNA generated by this enzyme is more toxic than the 5′-dRp group itself, or alternatively, that another toxic species is derived from this product. Of course, an unknown effect of deleting Rad27 that is unrelated to its enzymatic activity cannot be excluded from these studies.
Role of alternative repair pathways
Although Apn1 is known to be the major abasic site endonuclease in S.cerevisiae (39), it was possible that other enzymes capable of catalyzing strand breaks at abasic sites were acting as surrogates to initiate repair of abasic sites in the absence of Apn1. To investigate this possibility, the survival curve for the apn1apn2ntg1ntg2 quadruple mutant which lacks all known enzymes capable of nicking the DNA backbone 5′ or 3′ to abasic sites opposite to adenine bases was measured (Figure 2B and also Table 1). Since this quadruple deletion mutant was constructed using a different wild-type strain that is less sensitive to 5-FU (FF18733; Figure 2B), we also measured the survival curve for the apn1 mutant in this genetic background. The FF18733 wild-type strain showed 70% survival at 24 h incubation with 150 µM 5-FU, while the apn1 and apn1 apn2ntg1ntg2 strains showed increasing sensitivities (40 and 2% survival, respectively). Thus, these results support the proposal that processing of abasic sites by Apn1 is important for initiating productive repair of 5-FU generated abasic sites, and that Apn2 and/or Ntg1 and Ntg2 can play a backup role.
To investigate a role for homologous recombination (HR) in ameliorating the toxicity of 5-FU, the survival curve for the rad51 strain was also measured (Figure 2C). Rad51 is the yeast homolog for the bacterial RecA protein, which is critical for HR, but this deletion had no effect on the time dependence and only a small effect on 5-FU efficacy at 8 h as compared with the wild-type strain. However, after 24 h incubation with 5-FU the rad51 strain showed about 10-fold greater survival than BY4741. This increasing protection as a function of time, with a pronounced lag phase (Figure 2C), suggests that HR plays a modest role in drug-induced lethality, but that the mechanism requires time-dependent accumulation of recombinogenic intermediates. This time course differs considerably from apn1 that shows log linear killing with no lags (Figure 2A).
A role for the NER pathway was examined using the rad2 yeast strain (Figure 2D). Rad2 is a single-stranded DNA endonuclease that nicks the DNA backbone 3′ to a damaged site, and is the yeast homolog of the human XPG protein (40,41). In contrast, with the BER deletion strains, rad2 showed an indistinguishable survival curve from that of the wild-type strain BY4741 as did the mismatch repair-deficient strain msh2 (Figure 2D). We conclude that deletions in the BER pathway have the largest effects on 5-FU toxicity.
5-FU dose-response studies
The above survival time courses indicated that the ung1, apn1 and apn1apn2ntg1ntg2 deletion strains had significant effects on 5-FU induced cell death. We therefore examined the dose-response curves for the wild-type strain and these mutants after 4 h of growth in 5-FU containing media (Supplementary Data). The BY4741 wild-type parent strain showed an EC50 of 110 ± 20 µM, which may be compared with the EC50 values of 110 ± 37 µM and 22 ± 4 µM for the ung1 and apn1 deletion strains, respectively. A key aspect of these dose-response curves is that 100% killing is never achieved for the wild-type or ung1 strains, but that the apn1 mutant shows both complete killing and a 5-fold decrease in the EC50 value for 5-FU as compared with wild-type. Similar findings were observed with another yeast strain (FF18733) and its apn1 and apn1apn2ntg1ntg2 deletion mutants. In this case, 60% killing was observed for the wild-type strain (EC50 = 400 ± 150 µM), while complete killing was observed for the single apn1 mutant and the quadruple mutant (EC50 = 200 ± 30 µM and 66 ± 8 µM, respectively). Thus, singular deletion of the major abasic site endonuclease of yeast (Apn1) significantly increases both the potency and efficacy of 5-FU, and further deletion of the backup endonuclease (Apn2) and two AP lyases (Ntg1, Ntg2) further increases potency.
To further examine possible roles for NER and mismatch repair in 5-FU toxicity we examined the dose-response curves for the rad2 and msh2 strains. In accord with their survival curves (Figure 2D), rad2 and msh2 showed only a slightly lower EC50 values as compared with BY4741, providing no evidence for a significant role of these pathways in repair of 5-FU induced lesions.
Cell cycle responses to 5-FU
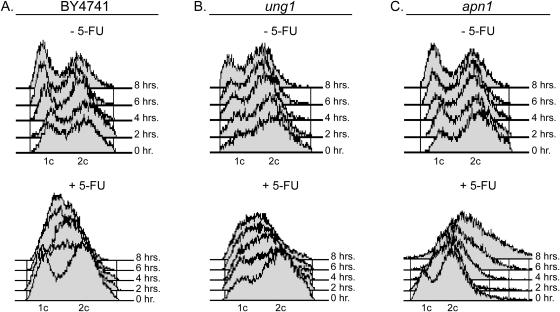
Flow cytometry was used to examine the cell cycle distribution of wild-type and the ung1 and apn1 yeast strains (Figure 3). In the absence of 5-FU, the wild-type and the uracil BER deficient strains showed no significant time-dependent changes in their cell cycle distribution over 8 h, with the exception of a slight increase in the G1 population at the later time points (upper panels, Figure 3A–C). In addition, the ung1 strain showed a somewhat larger steady-state distribution of cells in the G2 phase of the cell cycle as compared with the other two strains (Figure 3B). Upon treatment with 150 µM 5-FU, most of the wild-type cells arrested early in S phase as indicated by an intermediate content of DNA (lower panel, Figure 3A). The ung1 strain showed similar S phase arrest as wild-type, although the mechanism may differ (lower panel, Figure 3B). A dramatically different profile was observed for the apn1 strain upon 5-FU treatment (lower panel, Figure 3C). Unlike wild type or ung1, S phase arrest was bypassed, and apn1 arrested in the G2 phase of the cycle, with significant polyploidy and/or dead cells present. These findings indicate a significant change in the cell killing mechanism of 5-FU in the absence of Apn1 involving activation of a G2/M checkpoint.
Figure 3.
Cell cycle profiles for S.cerevisiae treated with 5-FU. Yeast were incubated in media with or without 150 µM 5-FU, fixed at given times with ethanol, and stained with propidium iodide. (A) Wild-type yeast treated with 5-FU arrest in the G1/S phase of the cell cycle. (B) Ung1 yeast arrest in G1/S phase of the cell cycle. (C) Apn1 yeast arrest in the G2/M phase of the cell cycle.
Uracil and 5-FU accumulation in DNA and RNA
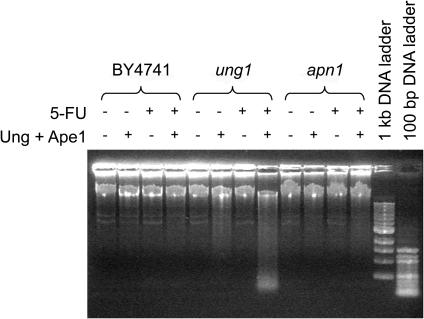
Unlike humans, yeast have no other known enzymatic activity that can remove uracil (or 5-FU) from DNA, and therefore, deletion of Ung1 should completely block the removal of these bases. Thus, the predicted outcome of the ung1 deletion is the accumulation of uracil and/or 5-FU in DNA. In a preliminary experiment to examine this prediction, we isolated genomic DNA from the wild-type, ung1 and apn1 yeast after 6 h growth in media containing 0 or 150 µM 5-FU. The DNA was then analyzed by agarose gel electrophoresis before and after treatment of the DNA with purified E.coli Ung and human AP endonuclease (Ape1) (Figure 4). If a significant amount of uracil had accumulated in the DNA of these samples, then combined digestion with Ung/Ape1 would result in significant DNA fragmentation, appearing as low molecular weight DNA after electrophoretic analysis. This analysis revealed that the wild-type and apn1 strains grown in the absence and presence of 5-FU had no detectable low molecular weight DNA fragments before or after Ung/Ape1 treatment. In contrast, the ung1 strain showed a detectable amount of lower molecular weight bands in the absence of 5-FU, and a large amount of lower molecular weight products in the presence of 5-FU. Indeed, under these 5-FU growth conditions about 50% of the DNA migrated with fragment sizes <4 kb. These results suggest a very high level of uracil or 5-FU incorporation in the ung1 strain.
Figure 4.
Uracil, 5-FU and abasic site accumulation in yeast genomic DNA. DNA from yeast grown in media containing 150 µM 5-FU or its absence was digested with E.coli UNG1 and human APE1 and then fractionated by electrophoresis using an agarose gel. DSBs resulting from the enzymatic removal of U and 5-FU and cleavage of abasic sites appear as a smear of lower molecular weight fragments.
To more accurately quantify the amounts of uracil and 5-FU incorporated into the genomes of these strains in the absence and presence of 5-FU, we performed GC-MS analysis of the uracil and 5-FU contents (Figure 5). All of the strains exhibited similar levels of uracil when growth was performed in the absence of 5-FU, suggesting that the dUTPase activity is sufficient to keep dUTP levels low enough to preclude its incorporation into DNA (Figure 5A). In the presence of 5-FU, the wild-type and apn1 strains were still capable of keeping genomic uracil levels as low as in the absence of 5-FU, exemplifying the high efficiency of uracil base excision by Ung1. However, as suggested from the electrophoretic assay for DNA fragmentation, the ung1 strain showed a 36-fold increase in the genomic uracil content in the presence of 5-FU (Figure 5A), indicating that about 1 in every 25 thymidine bases (4%) is substituted with deoxyuridine under these conditions. GC-MS analysis for 5-FU incorporation established that in the absence of a uracil DNA glycosylase activity this base is present at about 1/100 the levels of uracil (Figure 5B). Thus, proposals that direct incorporation of 5-FU into DNA is responsible for its toxic effects are not supported by these findings.
Figure 5.
GC-MS quantification of U and 5-FU incorporation into the genomic DNA of yeast grown in the absence and presence of 5-FU. Cells were incubated in media containing 5-FU such that 70–90% cell killing was achieved. Genomic DNA was then isolated and digested with E.coli Ung to release U and 5-FU. (A) Uracil levels detected in the presence and absence of 5-FU. (B) 5-FU levels detected in the presence and absence of 5-FU.
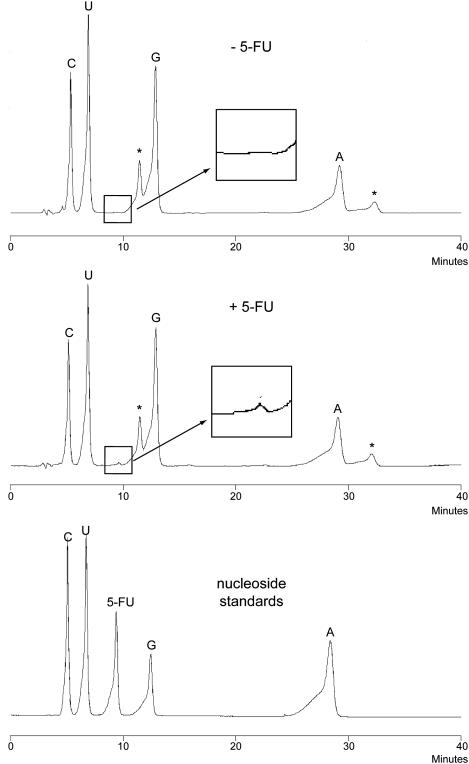
Another possible outcome of 5-FU treatment is the incorporation of 5-FUTP into RNA (see Figure 1A). To examine the extent of incorporation, total cell RNA was isolated and subjected to extensive enzymatic digestion with mung bean nuclease and calf intestinal phosphatase followed by reversed-phase HPLC analysis of the resulting nucleosides (Figure 6). We reproducibly detected a very small amount 5-FUrd in RNA corresponding to ∼3% of the uridine that was present. In other words, about 1 in every 30 uridine nucleotides in RNA is replaced with 5-FUrd. This much higher level of 5-FU in RNA as compared with DNA may reflect the higher pool of cellular 5-FUTP as compared to FdUTP.
Figure 6.
5-FU incorporation into RNA. Wild-type yeast were grown in media in the absence and presence of 5-FU and cellular RNA was isolated and digested to its constituent nucleosides using mung bean nuclease and calf intestinal phosphatase. A small peak corresponding to 5-fluorouridine is visible in the HPLC spectrum of the digested RNA from the drug-treated cells (middle panel, inset). This peak is not seen with yeast grown in the absence of 5-FU (upper panel, inset). The lower panel shows the elution pattern for authentic nucleoside standards.
Analysis of abasic sites and DNA strand breaks
The large increase in potency and efficacy of 5-FU for the apn1 and apn1apn2ntg1ntg2 strains suggests that abasic sites, or some processed form of abasic sites, may potentiate the lethal effects of 5-FU. To investigate this further, the total number of genomic abasic sites as well the number of 5′ and 3′ nicked forms were measured after growth for 6 h in the absence and presence of 5-FU (Figure 7). This analysis was performed using the ASB assay which detects the ring open aldehyde form of abasic sites by forming a covalent Schiff base linkage with a reactive semicarbazide that is covalently linked to biotin (35,42,43). Thus, sensitive detection is afforded by chemiluminescence methods after covalently derivatizing the DNA, fixing it to a nitrocellulose membrane, and then probing with strepavidin-conjugated HRP. This assay may also be combined with enzymatic analyses to ascertain the fraction of the total abasic sites which are nicked on the 5′ and 3′ phosphodiester linkages. For instance, 3′ nicks can be detected using a 5′ AP endonuclease (ExoIII) which completely severs the site from the DNA, precluding its binding to nitrocellulose and chemiluminescence detection. Similarly, 5′ nicks can be inferred by chemical cleavage of the 3′ phosphodiester linkage using putrescine (35). It should be noted that if rapid processing of sites occurs, then this assay will grossly underestimate the flux of total abasic sites. In other words, the assay is most informative if the rate-limiting process is a step or steps involved in endonucleolytic turnover of these sites.
Figure 7.
ASB assay for intact and nicked abasic sites. (A) Wild-type yeast grown in the absence and presence of 5-FU. (B) ung1 yeast, same as (A). (C) apn1 yeast, same as (A).
ASB analysis of genomic DNA isolated from the BY4741 wild-type strain showed about 12 abasic sites per 106 bp in the absence of 5-FU. Essentially all of these sites were nicked at either the 5′ or 3′ sides, indicating that the initial abasic site product of the Ung1 reaction is efficiently processed to these nicked forms (upper panel, Figure 7A). The genomic DNA of the ung1 strain showed ∼5-fold lower levels of abasic sites as compared with the wild-type strain in the absence of 5-FU, and essentially all of these were in the 5′ nicked form (upper panel, Figure 7B). An overall decrease in the steady-state load of abasic sites is consistent with the expectation that abasic sites generated from the excision of uracil or 5-FU would be diminished in the absence of Ung1, because it is the only known enzyme in yeast capable of processing uracil lesions into abasic sites. The remaining sites that are detected in the absence of Ung1 must therefore arise from the action of other glycosylases, or alternatively, from spontaneous hydrolysis of the glycosidic linkage of DNA nucleotides. From comparing the steady-state number of abasic sites in the wild-type and ung1 strain under growth conditions in the absence of 5-FU, we estimate that ∼80% of the total sites detected are derived from the excision of uracil. Finally, the apn1 strain showed similar levels of all abasic site forms as the wild-type strain in the absence of 5-FU (upper panel Figure 7C), consistent with the existence of effective alternative pathways for processing these sites.
In the presence of 5-FU only modest changes in the levels of intact and cleaved abasic sites were observed for the wild-type, ung1 and apn1 strains: (i) the wild-type strain showed at most a small decrease in the total number of abasic sites, all of which were found in the two nicked forms (lower panel, Figure 7A), (ii) the ung1 strain showed a modest 3-fold increase in the number of abasic sites in the presence of 5-FU (lower panel, Figure 7B), and (iii) the apn1 strain showed no significant change in abasic site levels as compared with the absence of 5-FU (lower panel, Figure 7C). Compared with the large flux of uracil through DNA as indicated from the GC-MS and DNA fragmentation studies (Figures 4 and 5), these data provide no evidence for a large accumulation of abasic sites arising from growth in the presence of 5-FU. An inescapable conclusion is that the vast majority of these sites are efficiently processed to strand gaps or DSBs irregardless of the presence or absence of apn1.
DISCUSSION
Effects of 5-FU on genomic DNA
Despite the widespread use of 5-FU in cancer therapy, previous studies of BER in yeast and mammalian cell culture have predominantly used the alkylating agent methylmethanesulfonate (MMS) to introduce base damage, primarily in the form of 3-methyladenine and 7-methylguanine (44–46). Consequently, the biological and biochemical effects of 5-FU treatment have not been well characterized. Proposals involving mechanisms as diverse as incorporation of 5-FU into RNA and DNA, or its disruption of nucleotide pool imbalances have been invoked, with little data to support which effects may give rise to its useful cytotoxicity (Figure 1A). Thus, the studies performed here have tracked both the biochemical consequences of growth in the presence 5-FU, and the DNA repair mechanisms that are used to repair 5-FU induced lesions.
The primary outcome of 5-FU treatment in yeast, using concentrations relevant to the observed EC50 values for cell killing and growth times equivalent to two doublings, is the flux of large amounts of uracil (but not 5-FU) through DNA. In addition, there is a significant incorporation of 5-FUTP into RNA (see below and Figures 5 and 6). The flux through DNA is revealed by the Ung1 deletion strain, which accumulates uracil to a level corresponding to substitution of about 1 in every 25 thymidines in the genome (Figure 5A). This contrasts with the wild-type and Apn1 deficient yeast, which show no significant increases in the uracil content of their DNA as a consequence of growth in 5-FU containing media. Taken together, these findings indicate that processing of uracil sites is extremely efficient in the presence of Ung1, resulting in no detectable increases in the uracil content even after 5-FU treatment. In the absence of Ung1, uracil accumulates in DNA, but this damage is less toxic to cells than the subsequent processed forms arising from BER or other repair pathways that may be operative in the wild-type strain (Figure 2A and see also below).
Incorporation of 5-FU into RNA
A recent genome-wide screen of yeast heterozygotes revealed 5-FU induced perturbation of rRNA processing suggesting a specific mechanism for its inhibition of cell growth that is independent of its incorporation and removal from DNA (47). We have measured a level for 5-FU in RNA of ∼1 in 30 U bases, which is similar to the ratio of U to T found in DNA. Previous studies of 5-FU toxicity using brewers yeast found much more extensive incorporation into RNA (∼75% of the uracil residues were substituted) (48,49), a difference that likely arises from the much higher concentrations of 5-FU that were used in the previous work. With respect to mRNA incorporation, the biological effects are likely to be of minor consequence because translation of 5-FU containing mRNA is apparently unhampered by this substitution, at least as reported for a reticulocyte lysate translation system (50). In conclusion, an rRNA-dependent pathway for 5-FU induced cell growth inhibition may be important, but it should be considered in parallel with the DNA repair path elucidated here.
Repair and toxicity of 5-FU induced DNA lesions
The high uracil flux through DNA in the presence of 5-FU implies an equal flux of abasic sites, as well as 3′ and 5′ incised abasic sites, but high steady-state levels of these lesions are not observed, even in the apn1 strain (Figure 7). Given the protective effect of the Ung1 deletion against 5-FU toxicity, it seems inescapable that a downstream repair intermediate arising from Ung1-catalyzed deglycosylation is responsible for the toxic effects of this drug. In this regard, only one BER enzyme we investigated, Apn1, gave rise to an increase in 5-FU toxicity (Figures 2A and 3B). The simplest interpretation for these results is that Apn1 cleavage of the 5′ phosphodiester, either by directly acting on the intact abasic site, or by removing the 3′-dRP group generated from AP lyase cleavage of the 3′ phosphodiester, is critical for cell survival in the presence of 5-FU (28).
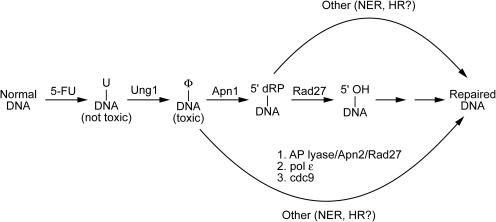
The data suggest a mechanistic basis for the increased 5-FU toxicity observed with the Apn1 deletion mutant, and possible backup pathways for repair (Figure 8). Naïve intuition would predict that removal of Apn1 would result in increased steady-state levels of intact abasic sites. However, detailed measurements of the levels of intact and cleaved abasic sites during 5-FU growth conditions revealed no detectable intact sites for the Apn1 deletion strain, and instead, an increase in both 3′ and 5′ cleaved sites was observed as compared with the wild-type strain (compare Figure 7A and C). This increase in cleaved sites could imply that in the absence of Apn1, AP lyase cleavage on the 3′ side occurs preferentially leading to a 3′-dRP group that is then inefficiently processed by the minor AP endonuclease, Apn2. Such 3′-blocked SSBs are strongly implicated in the formation of DSBs and in the collapse of replication forks, leading to activation of a RAD9-dependent G2/M checkpoint (51,52). The observed G2/M cell cycle arrest of Apn1 is consistent with this explanation.
Figure 8.
DNA repair pathways implicated in 5-FU-mediated cell killing. The model is supported by the following observations: (i) a massive amount of uracil is incorporated into DNA, but the ung1 yeast are much less sensitive to 5-FU than the wild-type strain indicating that uracilated DNA is not the mediator of 5-FU toxicity; (ii) the apn1apn2ntg1ntg2 strain that is entirely defective in processing abasic sites by a BER mechanism is more sensitive to 5-FU, indicating that intact abasic sites (or repair products derived from abasic sites) have inherent toxicity; and (iii) the rad27 and apn1rad27 yeast strains show protection against 5-FU toxicity, suggesting the presence of a toxic repair intermediate downstream of the Rad27 flap endonuclease reaction. Several backup pathways for repair of abasic sites and 5′dRp groups are indicated. The lower path involving Apn2 and other BER enzymes is important in the absence of Apn1 and accounts for the efficient removal of abasic sites in the apn1 strain. NER and HR pathways are likely to be important with the apn1apn2ntg1ntg2 and rad27 knockout strains. Consistent with this, yeast deficient in both BER and NER are not viable.
The other post-glycosylase enzyme, Rad27, that removes the 5′-dRP group resulting from the Apn1 reaction, was unexpectedly found to be strongly protective against 5-FU toxicity in both the wild-type and apn1 strains (Figure 2A). Since 5′-dRP groups block ligation of the DNA strands, and are converted into highly toxic DSBs after DNA replication, it would seem essential that another repair pathway is operative in this mutant. One bypass pathway would be cleavage on the 3′ side of the intact abasic site by Ntg1 or Ntg2, followed by removal of the 3′-dRP group by Apn1 or Apn2 (Figure 8), as implicated previously for yeast grown in the presence of MMS (53,54). Why this alternative path, or other additional backup pathways such as NER (Figure 8), would result in greater resistance to 5-FU for the Rad27 deletion strain is not obvious. The protective or null effect of the rad27 deletion to both 5-FU and antifolate drugs (38), both of which result in thymidine deprivation, differs from the MMS sensitizing effect of this deletion (55). Thus, it appears that the damage response to uracilated and methylated DNA differs even after the glycosylase step. Assuming that the 5-FU protective phenotype of the rad27 strain arises from removal of its enzymatic activity, then deletion of the next enzyme in the pathway (pol ε) could produce hypersensitivity to 5-FU if the immediate product of the Rad27 reaction is toxic. However, since pol ε is essential this could not be tested. The toxic intermediates and DNA repair pathways implicated in 5-FU-mediated cell killing are summarized in the legend to Figure 8.
Our results extend upon two previous studies that investigated the influence of uracil BER on the cytotoxicity induced by two antifolate drugs aminopterin and sulfanilamide (12,38). The first study focused largely on modulation of dUTP pools by overexpression of dUTPase (Figure 1A), an activity which was found to be strongly protective against the combined cytotoxic effects of these dihydrofolate reductase inhibitors. These previous findings suggested a pathway involving drug-induced blockage of TTP synthesis and an increase in dUTP pools leading to UNG-dependent DNA damage. The second study used the same two antifolates and an apn1 strain to find that deletion of Apn1 produced extreme sensitivity to these agents (38). However in this previous study, deletion of Ung1 was not protective in the wild-type background, suggesting that the observed cytotoxicity resulted at least in part from effects other than deoxyuridine incorporation into DNA. This is not unexpected since aminopterin and sulfanilamide have widespread effects on both purine and pyrimidine nucleotide pools, while 5-FU specifically targets TS. Nevertheless, processing of abasic sites by Apn1 was found to be important both in our work and this previous study.
Pharmacological implications
Toxicity studies in yeast now suggest a viable synergistic approach for enhancing 5-FU cancer therapy involving manipulation of both dUTP pools and DNA repair. Small molecule inhibitors of human dUTPase could dramatically increase the levels of dUTP in tumor cells, and consequently, increase uracil incorporation into DNA. This could decrease the dose of 5-FU required to give a clinical effect, and perhaps diminish unwanted side effects. In addition, after excision of uracil by Ung, inhibitors of the major human abasic endonuclease (Ape1) could efficiently block the repair of abasic sites, resulting in the massive accumulation of highly toxic lesions that would lead to apoptosis or cell death. The synergistic effect of a small molecule Ape1 inhibitor was recently demonstrated in cell culture using several base-targeting agents (56).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Drs Serge Boiteux, Jef Boeke and Xuewen Pan for providing yeast strains and for invaluable advice. We also thank Daniel J. Krosky for his helpful suggestions. This work was supported by NIH grant GM56834-10 (to J.T.S). The Open Access publication charges for this article have been waived by Oxford University Press.
Conflict of interest statement. Certain commercial equipment, instruments and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
REFERENCES
- 1.Malet-Martino M., Martino R. Clinical studies of three oral prodrugs of 5-fluorouracil (capecitabine, UFT, S-1): a review. Oncologist. 2002;7:288–323. doi: 10.1634/theoncologist.7-4-288. [DOI] [PubMed] [Google Scholar]
- 2.Rich T.A., Shepard R.C., Mosley S.T. Four decades of continuing innovation with fluorouracil: Current and future approaches to fluorouracil chemoradiation therapy. J. Clin. Oncol. 2004;22:2214–2232. doi: 10.1200/JCO.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Santi D.V., McHenry C.S., Raines R.T., Ivanetich K.M. Kinetics and thermodynamics of the interaction of 5-fluoro-2′-deoxyuridylate with thymidylate synthase. Biochemistry. 1987;26:8606–8613. doi: 10.1021/bi00400a017. [DOI] [PubMed] [Google Scholar]
- 4.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nature Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 5.Kunz B.A., Kohalmi S.E. Modulation of mutagenesis by deoxyribonucleotide levels. Annu. Rev. Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 6.Kunz B.A., Kohalmi S.E., Kunkel T.A., Mathews C.K., Mcintosh E.M., Reidy J.A. Deoxyribonucleoside triphosphate levels—a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 7.Ladner R.D. The role of dUTPase and uracil-DNA repair in cancer chemotherapy. Curr. Protein Pept. Sci. 2001;2:361–370. doi: 10.2174/1389203013380991. [DOI] [PubMed] [Google Scholar]
- 8.Fang F., Hoskins J., Butler J.S. 5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenz H.J., Manno D.J., Danenberg K.D., Danenberg P.V. Incorporation of 5-fluorouracil into U2 and U6 snRNA inhibits mRNA precursor splicing. J. Biol. Chem. 1994;269:31962–31968. [PubMed] [Google Scholar]
- 10.Rosen B., Rothman F., Weigert M.G. Miscoding caused by 5-fluorouracil. J. Mol. Biol. 1969;44:363–375. doi: 10.1016/0022-2836(69)90181-8. [DOI] [PubMed] [Google Scholar]
- 11.Morio A., Miyamoto H., Izumi H., Futagawa T., Oh T., Yamazaki A., Konno H. Enhanced induction of apoptosis in lung adenocarcinoma after preoperative chemotherapy with tegafur and uracil. Surg. Today. 2004;34:822–827. doi: 10.1007/s00595-004-2814-5. [DOI] [PubMed] [Google Scholar]
- 12.Tinkelenberg B.A., Hansbury M.J., Ladner R.D. dUTPase and uracil–DNA glycosylase are central modulators of antifolate toxicity in Saccharomyces cerevisiae. Cancer Res. 2002;62:4909–4915. [PubMed] [Google Scholar]
- 13.Ghoshal K., Jacob S.T. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994;54:632–636. [PubMed] [Google Scholar]
- 14.Kanamaru R., Kakuta H., Sato T., Ishioka C., Wakui A. The inhibitory effects of 5-fluorouracil on the metabolism of preribosomal and ribosomal RNA in l-1210 cells in vitro. Cancer Chemother. Pharmacol. 1986;17:43–46. doi: 10.1007/BF00299864. [DOI] [PubMed] [Google Scholar]
- 15.Lum P.Y., Armour C.D., Stepaniants S.B., Cavet G., Wolf M.K., Butler J.S., Hinshaw J.C., Garnier P., Prestwich G.D., Leonardson A., et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 16.Randerath K., Tseng W.C., Harris J.S., Lu L.J. Specific effects of 5-fluoropyrimidines and 5-azapyrimidines on modification of the 5 position of pyrimidines, in particular the synthesis of 5-methyluracil and 5-methylcytosine in nucleic acids. Recent Results Cancer Res. 1983;84:283–297. doi: 10.1007/978-3-642-81947-6_22. [DOI] [PubMed] [Google Scholar]
- 17.Santi D.V., Hardy L.W. Catalytic mechanism and inhibition of tRNA (uracil-5-)methyltransferase: evidence for covalent catalysis. Biochemistry. 1987;26:8599–8606. doi: 10.1021/bi00400a016. [DOI] [PubMed] [Google Scholar]
- 18.Carrico C.K., Glazer R.I. Effect of 5-fluorouracil on the synthesis and translation of polyadenylic acid-containing RNA from regenerating rat liver. Cancer Res. 1979;39:3694–3701. [PubMed] [Google Scholar]
- 19.Doong S.L., Dolnick B.J. 5-fluorouracil substitution alters pre-mRNA splicing in vitro. J. Biol. Chem. 1988;263:4467–4473. [PubMed] [Google Scholar]
- 20.de Bono J.S., Twelves C.J. The oral fluorinated pyrimidines. Invest. New Drugs. 2001;19:41–59. doi: 10.1023/a:1006404701008. [DOI] [PubMed] [Google Scholar]
- 21.Dusseau C., Murray G.I., Keenan R.A., O'Kelly T., Krokan H.E., McLeod H.L. Analysis of uracil DNA glycosylase in human colorectal cancer. Int. J. Oncol. 2001;18:393–399. doi: 10.3892/ijo.18.2.393. [DOI] [PubMed] [Google Scholar]
- 22.Pugacheva E.N., Ivanov A.V., Kravchenko J.E., Kopnin B.P., Levine A.J., Chumakov P.M. Novel gain of function activity of p53 mutants: activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene. 2002;21:4595–4600. doi: 10.1038/sj.onc.1205704. [DOI] [PubMed] [Google Scholar]
- 23.Tanner B., Grimme S., Schiffer I., Heimerdinger C., Schmidt M., Dutkowski P., Neubert S., Oesch F., Franzen A., Kolbl H., et al. Nuclear expression of apurinic/apyrimidinic endonuclease increases with progression of ovarian carcinomas. Gynecol. Oncol. 2004;92:568–577. doi: 10.1016/j.ygyno.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 24.Ladner R.D., Lynch F.J., Groshen S., Xiong Y.P., Sherrod A., Caradonna S.J., Stoehlmacher J., Lenz H.J. dUTP nucleotidohydrolase isoform expression in normal and neoplastic tissues: Association with survival and response to 5-fluorouracil in colorectal cancer. Cancer Res. 2000;60:3493–3503. [PubMed] [Google Scholar]
- 25.McIntosh E.M., Looser J., Haynes R.H., Pearlman R.E. MluI site-dependent transcriptional regulation of the Candida albicans dUTPase gene. Curr. Genet. 1994;26:415–421. doi: 10.1007/BF00309928. [DOI] [PubMed] [Google Scholar]
- 26.Studebaker A.W., Balendiran G.K., Williams M.V. The herpesvirus encoded dUTPase as a potential chemotherapeutic target. Curr. Protein Pept. Sci. 2001;2:371–379. doi: 10.2174/1389203013380946. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo-Zarco F., Gonzalez-Pazanowska D. Trypanosomal dUTPases as potential targets for drug design. Curr. Protein Pept. Sci. 2001;2:389–397. doi: 10.2174/1389203013381026. [DOI] [PubMed] [Google Scholar]
- 28.Boiteux S., Guillet M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst.) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Pan X., Yuan D.S., Xiang D., Wang X., Sookhai-Mahadeo S., Bader J.S., Hieter P., Spencer F., Boeke J.D. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 30.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 31.Sazer S., Sherwood S.W. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell. Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 32.Dizdaroglu M. Chemical determination of oxidative DNA damage by gas chromatography-mass spectrometry. Methods Enzymol. 1994;234:3–16. doi: 10.1016/0076-6879(94)34072-2. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M., Karakaya A., Jaruga P., Slupphaug G., Krokan H.E. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zastawny T.H., Doetsch P.W., Dizdaroglu M. A novel activity of E.coli uracil DNA N-glycosylase excision of isodialuric acid (5,6-dihydroxyuracil), a major product of oxidative DNA damage, from DNA. FEBS Lett. 1995;364:255–258. doi: 10.1016/0014-5793(95)00400-4. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura J., Swenberg J.A. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 36.Nakamura J., Walker V.E., Upton P.B., Chiang S.Y., Kow Y.W., Swenberg J.A. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- 37.Nakamura J., La D.K., Swenberg J.A. 5′-nicked apurinic/apyrimidinic sites are resistant to beta-elimination by beta-polymerase and are persistent in human cultured cells after oxidative stress. J. Biol. Chem. 2000;275:5323–5328. doi: 10.1074/jbc.275.8.5323. [DOI] [PubMed] [Google Scholar]
- 38.Dornfeld K., Johnson M. AP endonuclease deficiency results in extreme sensitivity to thymidine deprivation. Nucleic Acids Res. 2005;33:6644–6653. doi: 10.1093/nar/gki975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popoff S.C., Spira A.I., Johnson A.W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease—homology to Escherichia coli endonuclease-IV. Proc. Natl Acad. Sci. USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habraken Y., Sung P., Prakash L., Prakash S. Structure-specific nuclease activity in yeast nucleotide excision-repair protein Rad2. J. Biol. Chem. 1995;270:30194–30198. doi: 10.1074/jbc.270.50.30194. [DOI] [PubMed] [Google Scholar]
- 41.O'Donovan A., Davies A.A., Moggs J.G., West S.C., Wood R.D. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision-repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 42.Asaeda A., Ide H., Terato H., Takamori Y., Kubo K. Highly sensitive assay of DNA abasic sites in mammalian cells optimization of the aldehyde reactive probe method. Anal. Chim. Acta. 1998;365:35–41. [Google Scholar]
- 43.Kubo K., Asaeda A., Takamori Y., Ide H. Quantitation of abasic sites in mammalian-cells by the aldehyde reactive probe. J. Cell. Biochem. 1995:280–280. doi: 10.1080/07328319808005194. [DOI] [PubMed] [Google Scholar]
- 44.Ramotar D., Popoff S.C., Gralla E.B., Demple B. Cellular role of yeast Apn1 apurinic endonuclease 3′-diesterase—repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A.S.H., Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett R.A.O. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 1999;19:1800–1809. doi: 10.1128/mcb.19.3.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lum P.Y., Armour C.D., Stepaniants S.B., Cavet G., Wolf M.K., Butler J.S., Hinshaw J.C., Garnier P., Prestwich G.D., Leonardson A., et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 48.Strijker P.J. Effect of 5-fluorouracil on induced enzyme synthesis in yeast. Biochim. Biophys. Acta. 1969;182:262. doi: 10.1016/0005-2787(69)90545-0. [DOI] [PubMed] [Google Scholar]
- 49.Hendrick D.V., Andrean B.A.G., Dekloet S.R. Effects of cycloheximide and 5 fluorouracil on formation of low-molecular-weight ribonucleic acid in yeast. J. Bacteriol. 1969;97:743. doi: 10.1128/jb.97.2.743-748.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glazer R.I., Hartman K.D. In vitro translational activity of messenger-RNA following treatment of human-colon carcinoma-cells with sangivamycin. Mol. Pharm. 1983;24:509–512. [PubMed] [Google Scholar]
- 51.Weinert T.A., Hartwell L.H. Characterization of rad9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell-cycle arrest after DNA damage. Mol. Cell. Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinert T.A., Hartwell L.H. The RAD9 gene controls the cell-cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 53.Unk I., Haracska L., Gomes X.V., Burgers P.M., Prakash L., Prakash S. Stimulation of 3′→5′ exonuclease and 3′-phosphodiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol. Cell. Biol. 2002;22:6480–6486. doi: 10.1128/MCB.22.18.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unk I., Haracska L., Johnson R.E., Prakash S., Prakash L. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 2000;275:22427–22434. doi: 10.1074/jbc.M002845200. [DOI] [PubMed] [Google Scholar]
- 55.Wu X., Wang Z. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madhusudan S., Smart F., Shrimpton P., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.