Abstract
Many restriction enzymes require binding of two copies of a recognition sequence for DNA cleavage, thereby introducing a loop in the DNA. We investigated looping dynamics of Type IIE restriction enzymes NaeI and NarI by tracking the Brownian motion of single tethered DNA molecules. DNA containing two endonuclease recognition sites spaced a few 100 bp apart connect small polystyrene beads to a glass surface. The position of a bead is tracked through video microscopy. Protein-mediated looping and unlooping is then observed as a sudden specific change in Brownian motion of the bead. With this method we are able to directly follow DNA looping kinetics of single protein–DNA complexes to obtain loop stability and loop formation times. We show that, in the absence of divalent cations, NaeI induces DNA loops of specific size. In contrast, under these conditions NarI mainly creates non-specific loops, resulting in effective DNA compaction for higher enzyme concentrations. Addition of Ca2+ increases the NaeI-DNA loop lifetime by two orders of magnitude and stimulates specific binding by NarI. Finally, for both enzymes we observe exponentially distributed loop formation times, indicating that looping is dominated by (re)binding the second recognition site.
INTRODUCTION
In many genetic processes, proteins bind simultaneously to two separate DNA sites large distances apart, creating a DNA loop. These processes include DNA replication and repair (1,2), site-specific recombination (3,4) and transcription regulation (5–7). Recently, it has become increasingly apparent that many restriction enzymes also need to interact with two identical copies of the recognition sequence before cleavage can take place (8–11). In vivo, Type II restriction endonucleases constitute an important defense mechanism of bacteria against viral attacks. They do so by catalyzing double-stranded DNA (dsDNA) breakage at specific recognition sites using Mg2+ as co-factor. Other divalent cations such as Mn2+ or Co2+ can function as analogues for this metal ion. Interestingly, Ca2+ enhances binding to the specific sequence as well, but at the same time inhibits DNA cleavage (12,13). In contrast, non-specific binding is generally not affected by the presence of divalent metal ions (13–15).
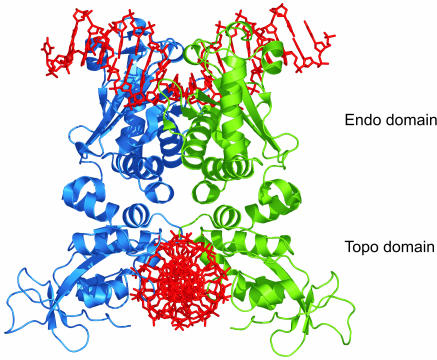
DNA loop formation by Type II restriction enzymes has been demonstrated in several ways (11,16–21). However, in these types of studies, looping could only be demonstrated indirectly and little insight in the underlying loop kinetics was gained. Here we apply a tethered particle motion (TPM) assay (22–26), to follow in real-time the loop formation by restriction enzymes within single dsDNA molecules. In particular we study the dynamics of two similar Type IIE enzymes, NaeI (from Nocardia aerocolonigenes) and NarI (from Nocardia argentinensis). Both proteins, presumably functional as dimers, are known to interact with two DNA sites before cleavage. Of these two proteins, NaeI is the best studied. The crystal structure is solved and the enzyme is considered a prototype Type IIE restriction enzyme (Figure 1). The dimeric protein has two different DNA-binding domains. Only one of these domains, the endonuclease or ‘Endo’ domain, is capable of cleaving substrate DNA, provided that a second copy of the recognition site, the activator DNA, is bound to the ‘Topo’ domain (27,28).
Figure 1.
Dimeric structure of NaeI in complex with two cognate DNA sites [(48), PDB ID: 1IAW]. The protein comprises two structurally different DNA-binding domains. Binding of cognate activator DNA to the ‘Topo’ domain functions as allosteric effector for binding and DNA cleavage by the ‘Endo’ domain (28,48). The two DNAs, shown in red, are bound under a 90° angle.
Single molecule observations have recently provided considerable new insights in the mechanochemistry of restriction enzymes (29–31). In this study we unambiguously show loop formation and disruption by these remarkable enzymes. Moreover, we compare looping kinetics under different conditions (with Mg2+, Ca2+ or no divalent cations) and reveal unexpected large differences in the stability and specificity of the loop formation and disruption between the two proteins.
MATERIALS AND METHODS
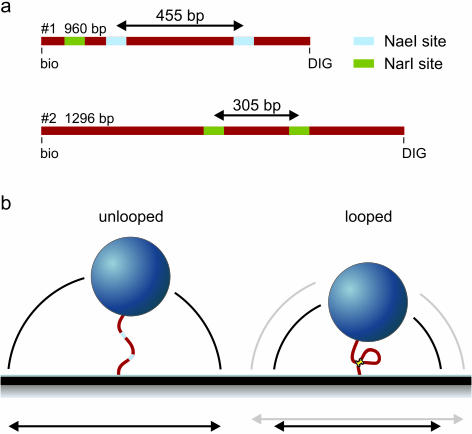
Looping kinetics were studied for restriction enzymes NaeI (Amersham Biosciences) and NarI (Roche Applied Science) in a buffer containing 33 mM Tris-acetate, 66 mM K-acetate, 1 mM DTT and 100 µg/ml α-casein (pH 7.0), with or without any divalent metals. In the latter case 0.1 mM EGTA and 0.1 mM EDTA were present in the buffer. Two different DNA substrates were used in the experiments (Figure 2a). DNA #1, a 960 bp substrate prepared by PCR using pCco5 as template, contained two NaeI recognition sites (GCCGGC) at a distance 455 bp from each other and one site for NarI. It has been shown for NaeI that the two DNA-binding domains (see Figure 1) have different affinities for the recognition sequence, depending on flanking sequence (28). In DNA substrate #1, the two NaeI sites differ in flanking sequence, but both contain an AT-rich side and a GC-rich side: CAATGCCGGCGCCG and TGATGCCGGCCTGG. However, neither of these sites very closely resemble any of the sequences tested (28). We cannot tell if there is any preference in binding of the Endo or Topo domain to either of the sites used here. DNA substrate #2, created by PCR from pRW490 (32), was slightly longer (1296 bp) and harbored two NarI sites (GGCGCC) spaced at 305 bp and no NaeI sites. For both substrates the primers (MWG Biotech) were labeled with biotin and digoxigenin (DIG).
Figure 2.
(a) DNA templates used in the experiments. Template #1 is 960 bp in length and has two NaeI recognition sites (and one NarI site). Template #2 is slightly longer, 1296 bp and harbors two NarI sites. In the looped state template #1 is 505 bp and template #2 991 bp in length. (b) Schematic representation of the experiment. Small beads are tethered with the DNA molecule in question to the glass slide. By tracking the x- and y-positions of the bead the magnitude of the Brownian motion is monitored, which is a measure for the tether length. Upon DNA loop formation by a restriction enzyme the Brownian motion of the bead suddenly decreases. This allows following DNA looping kinetics in real-time.
Single DNA molecules labeled with DIG on one end and biotin on the other end were attached to a glass surface (DIG–anti-DIG binding) and to a 440 nm diameter streptavidin-coated polystyrene bead (Indicia Biotechnology). Such constructs were assembled in a flow cell consisting of a microscope slide and a perpendicularly placed (plasma cleaned) cover slip, using double stick tape as spacer. Silicon grease was used to create reservoirs for ∼100 µl buffer on both sides of the sample chamber, thereby avoiding evaporation of water in the chamber. The volume of the flow chamber itself was ∼20 µl. After assembly the chamber was incubated for 20 min with 20 µg/ml anti-DIG (Roche Applied Science). Then the flow chamber was washed extensively with buffer solution, which included α-casein to passivate glass surfaces. Next, the labeled DNA, diluted to a few 100 ng/ml, was flown in. After incubation of 1 h, the chamber was again washed with several hundreds of microliters of restriction enzyme buffer before being studied.
Brownian motion of a bead was monitored by tracking the x and y (in-plane) coordinates of the centroid of the particle with video microscopy (50 Hz) in a bright-field microscope. The amplitude of Brownian motion is a direct measure for the DNA tether length (22,24,26,33). Recorded is the root mean square (RMS) motion 〈R〉 = {[[(x − xm)2 + (y − ym)2]/2}1/2, with xm and ym the mean values of x and y averaged over 100 frames. Doing so automatically corrects the data for thermal drift of the microscope stage. When a protein bridges two sites on the DNA a loop is introduced in the DNA. This effectively reduces the DNA tether length and restricts the movement of the bead (22,23,34) (Figure 2b). The motion of a specific bead was typically measured for 1 h continuously. The recorded x and y values were smoothed using a Gaussian filter with σ = 1.0 s (NaeI data) and σ = 2.0 s (NarI data) and analyzed using a half-amplitude threshold method (35) to obtain dwell times. This method allowed us to reliably detect DNA loops having lifetimes as short as 1.4 s (NaeI) and 2.7 s (NarI). Events shorter than these limits were omitted. The NaeI data traces display a better signal-to-noise ratio than the traces of NarI, because NaeI looping shortens the DNA by about 50% while NarI only reduces the tether length by 25%. The less significant length change between unlooped and looped state for NarI results in a smaller difference in Brownian motion.
Sometimes a bead momentarily sticks to the cover slip surface, leading to a ‘fake’ looping transition. Such events are easily recognized, since in these cases the registered Brownian motion <R> reduces to a very small value (typically <20 nm) before returning to its original value. Another possible cause of fraudulent looping transitions is the transient adsorption of the DNA tether itself to the glass (either by itself or via a bound enzyme). Whether an observed transition truly represents DNA looping was tested by evaluating the individual x- and y-coordinate traces. False transitions are generally characterized by a shift in the anchor point of the tether (36). The amount of such fake transitions in the analyzed NaeI and NarI data caused by this effect was found to be negligible with respect to the number of real events. In control experiments without proteins in the solution, we did not observe any transitions except for occasional bead sticking.
RESULTS
DNA cleavage
To examine whether NaeI and NarI restriction enzymes are active in the TPM assay, we carried out cleavage control experiments. With Mg2+ present in solution, the DNA is expected to be cleaved within several seconds after the protein has induced a DNA loop by binding to both specific sites. For these experiments we marked the positions of many tethered beads (∼20) per sample chamber. Next, enzymes in buffer containing Mg2+ (2–5 mM) were introduced by flow. This was done for NaeI and NarI on their specific DNA substrates. Directly after solution exchange (typically 10–20 s), most of the previously located beads had already disappeared, while some of the remaining beads were seen to be released into solution. These experiments were repeated with DNA tethers that were first incubated with enzymes in a buffer containing Ca2+, before the introduction of Mg2+. In this case we observed that all DNA molecules that were looped by NaeI before solution exchange were cleaved within seconds after Mg2+ was introduced (interestingly, some of the unlooped tethers survived for more than 10 min). This result indicates that divalent metal ions are exchanged quickly in the specific enzyme–DNA complexes. Exact turnover rates were not obtained, because most of the cleavage events take place faster than the dead-time in this particular assay (>10 s). These experiments demonstrate that both NaeI and NarI are active in this tethered particle assay and that the DNA templates are appropriate for their specific enzymes.
DNA looping
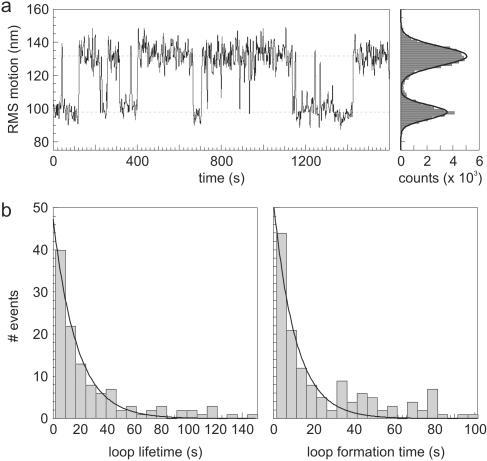
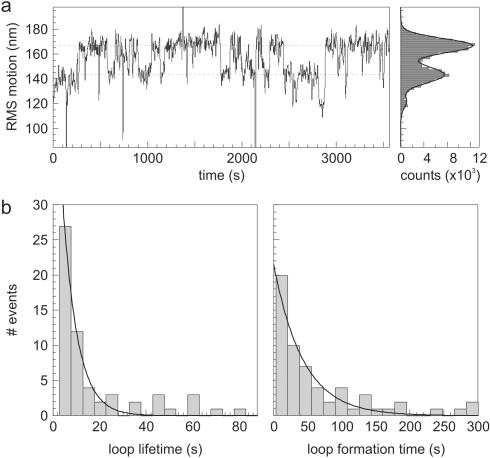
In order to observe loop formation and disruption by the restriction enzymes, the length of the DNA tether needs to be tracked with no Mg2+ in the solution. We measured the motion of beads, tethered by DNA template #1, in the presence of 2 U/ml NaeI (estimated 10–100 pM), but in the absence of divalent metal ions. A typical data trace is given in Figure 3a. We observed transitions between two distinct amplitudes of RMS Brownian motion <R>, which we attribute to DNA loops of specific size formed by NaeI. The obtained <R> in the unlooped state for both DNA substrates corresponds very well to predicted values (theory: 149 nm and 165 nm, experiment: 147 ± 4 nm and 168 ± 5, respectively; D. Segall and R. Phillips, private communication). The observed value for the looped state of the NaeI–DNA complex is slightly lower than expected (theory: 117 nm, experiment: 108 ± 4 nm). This apparent discrepancy presumably is caused by the fact that NaeI binds its two target sites under a 90° angle (Figure 1). The ‘unlooped part’ of the looped tether thus comprises a kink, which reduces its average end-to-end distance (37), resulting in a lower observed magnitude of Brownian motion.
Figure 3.
DNA looping by NaeI in the absence of divalent cations. (a) Typical data trace showing specific looping by NaeI (2 U/ml). Two distinct levels in the root mean square (RMS) amplitude of Brownian motion <R> can be recognized. The actual magnitudes of Brownian diffusion for the two states are estimated for each trace by fitting the histogram of <R> (shown on the right) to a double Gaussian, which agrees very well with the observed distribution. (b) Histograms of measured looped and unlooped state durations of NaeI (2 U/ml). As a result of the Gaussian filtering of the data with σ = 1.0 s, DNA loops with a lifetime shorter than 1.4 s cannot be reliably detected and are not taken into account. Left: NaeI–DNA looped complex lifetime. The data fits a single exponential (normalized χ2 = 1.0) with lifetime τoff = 18 ± 2 s. Right: NaeI-DNA loop formation. These data are reasonably well fitted by a single exponential (normalized χ2 = 2.2), yielding a time constant τformation of 11 ± 2 s.
In the looped configuration there are two possible binding modes for NaeI: the Topo domain is bound to site 1 and the Endo to site 2, or the reverse. Most likely the stabilities of the two differ, resulting in a double exponential distribution for the loop lifetime. However, the data (Figure 3b, left side) shows only a single exponential, indicating that one of the two possible loop structures is preferred over the other, or that the two have equal loop lifetimes. Fitting a double exponential does not give a better fit. The fitted mean lifetime of the looped state τoff is found to be 18 ± 2 s (Figure 3b), corresponding to a mean loop disruption rate koff of 0.057 ± 0.007 s−1. This value is 10 times faster than found for the Type IIF restriction enzyme SfiI [∼240 s without Ca2+ (19)] and approximately comparable to the Lac repressor (23). The same value for the lifetime of the looped state was found for a 5-fold higher NaeI concentration (20 ± 4 s), as would be expected for the unimolecular process of loop breakdown.
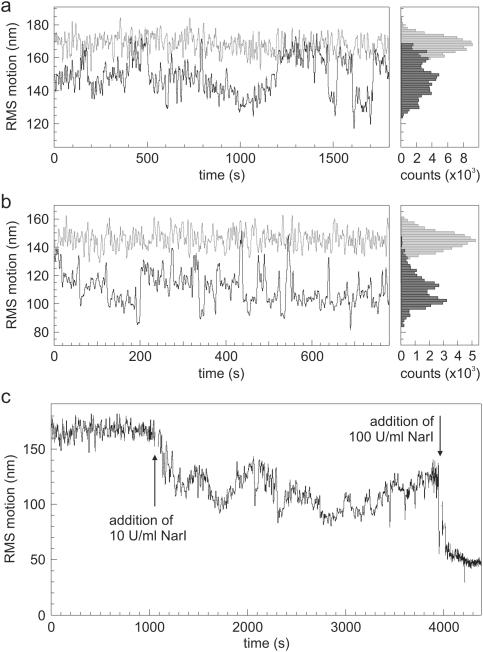
Figure 4a shows a typical data trace of looping of DNA substrate #2 by NarI (2 U/ml), again in the absence of divalent ions. In contrast to the findings for NaeI, here no discrete Brownian motion levels that relate to the expected 〈R〉 values for the unlooped and looped state can be distinguished. Instead, the signal displays a large variation with jumps of arbitrary size. The measured DNA tether length thus fluctuates heavily in time. A likely explanation for this effect is that NarI also binds to non-specific or non-cognate DNA sites, creating loops of random size. If specific DNA loops are formed in these experiments, they are clouded by many non-specific events. These findings are strengthened by the result that is obtained when the same NarI concentration is added to DNA substrate #1 tethers, containing only one recognition sequence: the same non-specific looping behavior is observed (Figure 4b). In contrast, when DNA #2 was used as template for control experiments with NaeI (no recognition sites), 2 U/ml NaeI did not cause any change in the magnitude of Brownian motion over several hours (∼20 tethers measured; data not shown), either in the absence or presence of Ca2+. NaeI thus binds its recognition sequence highly specifically, whereas NarI displays non-specific looping regardless whether one or two sites are present on the DNA.
Figure 4.
DNA looping by NarI in the absence of divalent cations. (a) Black trace: looping of DNA template #2 (two recognition sites) by NarI (2 U/ml). Grey trace: signal without protein. The histogram on the right clarifies even more that, in contrast to NaeI (Figure 3), it is not possible to distinguish two discrete levels of Brownian motion. (b) Black trace: non-specific looping by NarI (2 U/ml) on DNA template #1 (one site). The signal is similar to the trace observed with DNA template #2. Grey trace: signal without protein. (c) Effect of higher NarI concentrations. Addition of 10 U/ml NarI to DNA substrate #2 tethers results in a drop of Brownian motion from 170 nm to ∼110 nm over several minutes, presumably due to non-specific looping of multiple enzymes. After this initial process the average RMS motion of the bead fluctuates around this last value and never recovers toward the initial value. Upon addition of a high NarI concentration (100 U/ml), the Brownian motion is quickly reduced to a very low level due to non-specific DNA looping of many enzymes. This happens regardless of whether the DNA substrate has one or two NarI sites.
Loop formation
The duration of the unlooped state of NaeI represents the time between subsequent looping events. The data, displayed in Figure 3b (right panel) for 2 U/ml NaeI, is reasonably well fitted by a single exponential, resulting in a mean loop formation time τformation of 11 ± 2 s. A similar value is found for a NaeI concentration of 10 U/ml (16 ± 5 s). Still higher enzyme concentrations, however, repress the number of observed looping occurrences: at 100 U/ml NaeI almost no DNA tethers show sign of looping, leaving the molecules permanently in the unlooped state. This result is most likely caused by both sites being occupied by different enzymes and is described in more detail in the discussion.
Higher concentrations of NarI result in totally different behavior (Figure 4c). Upon addition of 10 U/ml NarI to the tethered DNA molecules (in the absence of divalent metals) the amplitude of Brownian motion on average decreases by 30–40%. Unlike in the 2 U/ml case, fewer jumps to higher tether lengths are observed. A further increase in NarI concentration to 100 U/ml reduces the effective tether length to a mere ∼100 bp in less than a minute. The same results were obtained when DNA #1 was used, containing only a single copy of the NarI recognition sequence. From these results we infer that at these concentrations increasingly more non-specific DNA loops are created by multiple enzymes, effectively resulting in a ‘compaction’ of the DNA.
Effect of calcium
In the mechanism of DNA-binding by restriction enzymes, Ca2+ can function as an analogue for Mg2+. However, it fails to support DNA cleavage (12–15). In our experiments, the presence of 2 mM Ca2+ in the buffer results in very stable specific NaeI-DNA loops. Under these conditions all observed looped molecules (3 slides, 10–20 tethers per slide) remained in the looped state for a very long time (more than 100-fold longer than without divalent ions). This durable looping is in agreement with numerous binding affinity studies on NaeI (38), SfiI (19), BamHI (13) and EcoRV (14,15,39). For the DNA looping Type IIF restriction enzyme SfiI (19), it was also shown that Ca2+ stabilizes specific DNA loops by more than two orders of magnitude. Interestingly, we observe that NarI reacts differently in the presence of 2 mM Ca2+. Instead of the indiscriminate looping seen without Ca2+, now 2 U/ml NarI does induce specific loops in the DNA (Figure 5a). These characteristics of binding resemble those of EcoRV, which binds all DNA sequences with similar affinity without divalent metal ions present in solution (40–43). However, Ca2+ greatly enhances specific DNA-binding. EcoRV binds 104-fold more tightly to the recognition site, while non-specific binding remains unaffected (14,15,40,44).
Figure 5.
DNA looping by NarI in the presence of Ca2+ ions. (a) Data trace showing specific DNA looping by NarI in the presence of 2 mM Ca2+. The histogram on the right shows two levels of Brownian motion corresponding to the expected values for specific DNA looping. The two Gaussians are less separated than for NaeI, reflecting the smaller length of the specific NarI-loop. At times along the trace non-specific loops are formed. The small third peak at 120 nm presumably represents binding to a non-cognate site on this DNA template. This extra state was observed for several tethers. From time to time the bead transiently sticks to the surface (the Brownian motion drops to almost zero). (b) Histograms of measured looped and unlooped state durations of NarI (2 U/ml). Data are filtered with σ = 2.0 s, giving rise to a loop detection limit of 2.7 s. Left: distribution of the measured lifetimes of specific NarI DNA loops in the presence of Ca2+. The exponential fit gives a mean lifetime of 6 ± 1 s. (normalized χ2 = 1.3) Right: distribution of the measured NarI DNA unlooped state durations in the presence of Ca2+. Again, the data fits a single exponential (normalized χ2 = 0.8), yielding a mean loop formation time τformation of 43 ± 8 s.
The histograms for the looped state disruption and formation times of NarI in the presence of calcium (Figure 5b) are again fitted with single exponentials, resulting in a mean lifetime τoff of 6 ± 1 s for the looped state duration, while the loop formation time τformation is found to be 43 ± 8 s. These values are more or less similar as observed for NaeI without calcium. The trace displayed in Figure 5a, compared to Figure 4a and b demonstrates that Ca2+ favors specific looping by NarI rather than non-specific looping. However, occasionally an extra ‘lower state’ of Brownian motion is observed. The system spends a few percent of the time in this third state. This behavior was seen for several traces on different tethers and is presumed to be binding to a (preferred) non-cognate site on the DNA. Which site on the target DNA exactly this might be remains unclear. There are several plausible candidate sequences on DNA substrate #2 that differ 1 bp from the recognition sequence. No less than five of these sequences are located in such a way that looping would induce the observed tether length.
Ca2+ thus promotes specific binding, but its effect on non-specific binding is presumably much weaker. In any case, high NarI concentrations (100 U/ml) in the presence of Ca2+ still lead to a large reduction in Brownian motion, similar to the results obtained without Ca2+, indicating that non-specific binding is at least not vastly diminished. We show here that Ca2+ reduces the relative amount of non-specific looping events by NarI. The stability of the specific loop in the presence of Ca2+ is, however, still two orders of magnitude weaker than observed for NaeI (corresponding to about 5 kbT or 3 kcal/mol binding energy)
DISCUSSION
Many biochemical studies regarding proteins that interact with two DNA sites have been carried out, greatly enhancing the knowledge of enzyme-mediated looping. The Type IIE and IIF restriction enzymes are often considered ideal examples. DNA cleavage can be readily detected and is only possible when two specific sites are brought together.
One recent study revealed that NarI, though interacting with two DNA sites, cuts only one phosphodiester bond before dissociating from the DNA, leaving the DNA nicked. The second bond is then cut in a separate, slower reaction (11). This kinetic scheme differs from the common reaction pathway of Type IIE and Type IIF restriction enzymes (including NaeI), where two strands of the same DNA are cleaved concertedly in one binding event, creating a double-stranded break (10,18,20,45). We now begin to appreciate the result that, even in the presence of divalent metal ions, NarI forms short-lived loops of only a few seconds. It might very well be that the reason why NarI acts differently is purely due to this short-lived binding: the available time for the hydrolysis reaction is so short that at most one bond is cleaved within one binding event. Turnover rates of restriction enzymes on long DNA substrates are generally rate-limited by product release: both strands are cleaved very fast compared to enzyme dissociation from the product (on long DNA substrates and at physiological pH) (38,46). The short binding time of NarI indicates that for this particular enzyme product release is not the rate-limiting step.
In another nice study on DNA looping by restriction enzymes, Milsom et al. (19) ingeniously exploited site-specific recombination by resolvase (of a plasmid into catenanes) so as to detect enzyme-mediated looped complexes. DNA looping by a restriction enzyme segregated the two resolvase sites into isolated topological domains, which inhibited the recombination reaction. The degree of recombination reflects the fraction of DNA looping in equilibrium. Although this method proved to be very successful for Cfr10I and SfiI, DNA looping in the presence of Ca2+ could not be detected for either NaeI or NgoMIV. The authors argue that the resolvase method only functions for DNA loops that are stable for more than 30 s. It was therefore concluded that NaeI and NgoMIV must produce very short-lived DNA loops, even in the presence of Ca2+. Our results for NaeI, obtained in a buffer with equal pH, are in contrast with these observations. In the more direct TPM method utilized here, loops in the presence of Ca2+ have a lifetime on the order of thousands of seconds, comparable to the loop lifetime Milsom et al. found for the tetrameric Type IIF restriction enzyme SfiI. Possibly the recombination reaction by resolvase was not entirely inhibited by NaeI induced DNA looping.
In DNA cleavage tests with NaeI, we observed some tethers that stayed unlooped for 10 min or more, whereas most tethers were cleaved within a few seconds. Such impeded loop formation also showed up in experiment in the presence of Ca2+. In a test with high NaeI concentration (100 U/ml) and 2 mM Ca2+, just 1 out of 15 tethers became looped directly after flowing in the enzymes. This looped complex then remained for more than 1 h, while the other tethers all stayed unlooped for this time period. When a buffer containing 2 mM Mg2+ and NaeI was flown in afterwards, only the previously looped DNA was cleaved within the dead-time of buffer exchange (∼20 s). None of the remaining DNA tethers, however, got released in 25 min. A feasible explanation for this resistance of NaeI against looping (and thus cleavage) at higher concentrations is that two individual NaeI dimers bind to the two specific sites, blocking the formation of the loop that is required for cleavage. Diminished cleavage at high enzyme concentrations was also shown earlier for SfiI (20,47).
We also tested the cleavage of DNA tethers by the Type IIP restriction enzyme EcoRV (50 nM) on DNA template #2, which contained one recognition sequence. In contrast to NaeI and NarI, EcoRV does not need to interact with two recognition sites. In this case all tethered beads were released within seconds after introducing the enzymes.
There are four possible pathways that can lead to specific loop formation by NaeI on DNA substrate #1 (ignoring a potential difference between a positively and negatively twisted loop). In principle, the initial association to the first site can occur via either the Topo or the Endo domain of NaeI, at site 1 or site 2. Binding of the second site will then be to the remaining unfilled domain. Nonetheless, the distribution of the loop formation time (time in between consecutive DNA loops), in the absence of divalent metal ions for NaeI and with Ca2+ for NarI, is found to fit a single exponential for both NaeI and NarI (Figures 3b and 5b). This result implies that the process of repetitive loop formation is rate-limited by a single reaction step. Two plausible candidates come to mind.
The rate-limiting step is the association of a protein out of solution to one of the DNA sites. Once this complex is formed it binds very fast to the second site, producing the loop. When the complex dissociates, there is a high probability that NaeI is released from both sites before a new loop is created.
After loop disruption the enzyme remains bound to one of the sites. Further DNA looping/unlooping then occurs as a unimolecular process, where the rate-limiting step is the association of the protein–DNA complex to the second site. In this case the rate of finding the second site is required to be much slower than in scheme a.
If scheme a is true, the loop formation time τformation should decrease with enzyme concentration for non-saturating conditions, whereas in scheme b it should be independent. We measured DNA looping by NaeI at 2 U/ml, 10 U/ml and 100 U/ml concentrations. The two lowest concentrations both resulted in an exponentially distributed loop formation time with similar time constants (11 ± 2 s and 16 ± 5 s, respectively, see Figure 3b (2 U/ml)). The DNA looping at 100 U/ml was heavily suppressed and yielded too few events to reliably measure the loop formation time (presumably because the DNA is saturated with enzymes). Moreover, for the lowest concentration, 2 U/ml, we observed that after some time, free (unlooped) tethers suddenly started displaying looping dynamics, which then generally continued for a number of transitions, until finally the DNA remained unlooped for a much longer time. This suggests that a single NaeI molecule is responsible for making multiple consecutive DNA loops. Based on these results we conclude that what occurs upon loop disruption is the unbinding of the enzyme from one cognate DNA site, while the other binding domain of the protein stays bound to the second site (scheme b). The distribution of the loop formation time we observe thus represents the association of the protein–DNA complex to this second site. This concurs with earlier studies of NaeI (28), where it was found that the two binding domains of NaeI can have different affinities (up to 14-fold) for the DNA recognition site (depending on the context flanking the cognate site). The crystal structure of NaeI (Figure 1) (48) shows that the two DNA-binding domains, although they recognize the same nucleotide sequence, are in fact structurally different. By means of structural comparison between the free and DNA bound NaeI structures, Huai et al. (48) put forward that initial DNA-binding occurs via the Topo domain, which then triggers a conformational change that facilitates binding of substrate DNA to the Endo domain. The same model was also speculated earlier by Colandene and Topal, based on the characteristics of the separate DNA-binding domains (49). Our results further strengthen these assumptions.
The association of restriction enzymes to their substrates usually is very efficient and limited by diffusion. If the same is true for NaeI and NarI, the time between consecutive DNA looping events represents the time it takes to bring the two DNA sites together (via 3D diffusion). In other words, after the initial binding of the protein, loop formation becomes entirely dependent on the dynamics of the DNA molecule. It is interesting to check whether the values we find are realistic for such a process. Earlier Brownian dynamics simulations revealed that the typical time for juxtaposition within 10 nm of two specific DNA sites spaced at ∼400 bp for relaxed DNA is less than a second (50,51) (for supercoiled DNA this process is 100-fold faster). However, the DNA tethers in the experiments are not entirely free to move: one end is fixed to an infinite glass surface and the other end to a 440 nm bead. These steric constraints give rise to a volume-exclusion effect, inducing a small entropic stretching force on the DNA. Due to this force the rate of juxtaposition of the two recognition sites decreases. The rate of loop formation changes accordingly. For our experimental parameters the stretching force is predicted to be 50–60 fN, resulting in an ∼10-fold increase in average loop formation time (D. Segall and R. Phillips, private communication). Given all the theoretical uncertainties, the values we obtain for τformation, 11 ± 2 s for NaeI and 43 ± 8 s for NarI, seem to correspond to the expected time for juxtaposition of the two sites in our tethered particle measurements. This result is in line with our conjecture posed above about the process of loop formation.
The formation of a relatively short DNA loop, such as demonstrated here, introduces a certain amount of mechanical strain to be stored in the DNA. The accompanied bending energy has to be paid by the protein that produces the loop: the binding energy is weakened by the strain in the DNA, affecting the lifetime of the looped complex. In previous experiments with the Lac repressor, however, the looped complex seemed unperturbed by the DNA bending energy (23). In our particular case, the loop produced by NaeI is 455 bp, or about three persistence lengths. However, NarI has to induce a loop of only two persistence lengths long (305 bp). In the presence of Ca2+ we observe a 100 to 1000-fold difference in looped state lifetime between NaeI and NarI. Although the better part of this difference in stability most likely originates from a weaker protein–DNA interaction for NarI, the smaller loop size may contribute to the observed smaller loop lifetime (estimated max. 10-fold). Even so, it cannot explain the observed non-specific looping by NarI. Future experiments could investigate the loop lifetime as a function of loop length. Single molecule experiments using DNA tension as a variable can explore the binding strength of NaeI by pulling on the DNA loop, as well as test the process of loop formation under tension.
In this paper we have demonstrated that TPM analysis can be a simple and effective technique to study the looping kinetics of DNA molecules by Type IIE restriction enzymes. We discovered a fundamental difference in the way NaeI and NarI bind and bridge two copies of their specific target sites. In the absence of divalent metal ions, NaeI produces only specific loops, while NarI mostly displays non-specific looping. Higher NarI concentrations effectively result in a ‘condensation’ of the DNA. We show that, in contrast to previous studies (19), in the presence of Ca2+ NaeI produces very stable DNA loops. Ca2+ also notably stimulates specific looping by NarI, although the observed loop lifetime is much shorter than that of NaeI. The exponential distribution found for the loop formation time by NaeI and the concentration-independence of the rate of this process imply that the enzyme remains bound to one of the DNA recognition sites upon loop disruption. Finally, real-time measurements of DNA looping as demonstrated here have so far only been carried out on specific gene regulators, such as Lac repressor (23) and Gal repressor (52). Therefore, the presented results provide an opportunity to compare mechanisms of DNA looping proteins that are very different in structure and function.
Acknowledgments
We thank Darren Segall, Seth Blumberg and Rob Phillips for sharing results prior to publication, useful comments and stimulating discussions. B.v.d.B. performed and conceived research, analyzed data and wrote the paper, F.V. designed research and analysis tools, D.N. performed research in the initial stages, F.S.P. designed research, G.J.L.W. conceived the experiment and wrote the paper. This work was supported by a Marie Curie fellowship, (human potential, PHD20) (to F.S.P.), an NWO Vernieuwingsimpuls grant and a grant from the Dutch Foundation of Fundamental Research on Matter (FOM) (to G.J.L.W.). Funding to pay the Open Access publication charges for this article was provided by FOM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dodson M., Roberts J., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc. Natl Acad. Sci. USA. 1985;82:4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen D.J., Makhov A., Grilley M., Taylor J., Thresher R., Modrich P., Griffith J.D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. Nature. 1987;325:401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- 4.Moitoso de Vargas L., Pargellis C.A., Hasan N.M., Bushman E.W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 5.Mastrangelo I.A., Courey A.J., Wall J.S., Jackson S.P., Hough P.V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl Acad. Sci. USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis M., Chang G., Horton N.C., Kercher M.A., Pace H.C., Schumacher M.A., Brennan R.G., Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd G., Landini P., Busby S. Activation and repression of transcription initiation in bacteria. Essays Biochem. 2001;37:17–31. doi: 10.1042/bse0370017. [DOI] [PubMed] [Google Scholar]
- 8.Halford S.E., Bilcock D.T., Stanford N.P., Williams S.A., Milsom S.E., Gormley N.A., Watson M.A., Bath A.J., Embleton M.L., Gowers D.M., et al. Restriction endonuclease reactions requiring two recognition sites. Biochem. Soc. Trans. 1999;27:696–699. doi: 10.1042/bst0270696. [DOI] [PubMed] [Google Scholar]
- 9.Deibert M., Grazulis S., Sasnauskas G., Siksnys V., Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 10.Bath A.J., Milsom S.E., Gormley N.A., Halford S.E. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 11.Gowers D.M., Bellamy S.R.W., Halford S.E. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagunavicius A., Grazulis S., Balciunaite E., Vainius D., Siksnys V. DNA binding specificity of MunI restriction endonuclease is controlled by pH and calcium ions: involvement of active site carboxylate residues. Biochemistry. 1997;36:11093–11099. doi: 10.1021/bi963126a. [DOI] [PubMed] [Google Scholar]
- 13.Engler L.E., Sapienza P., Dorner L.F., Kucera R., Schildkraut I., Jen-Jacobson L. The energetics of the interaction of BamHI endonuclease with its recognition site GGATCC. J. Mol. Biol. 2001;307:619–636. doi: 10.1006/jmbi.2000.4428. [DOI] [PubMed] [Google Scholar]
- 14.Engler L.E., Welch K.K., Jen-Jacobson L. Specific binding by EcoRV endonuclease to its DNA recognition site GATATC. J. Mol. Biol. 1997;269:82–101. doi: 10.1006/jmbi.1997.1027. [DOI] [PubMed] [Google Scholar]
- 15.Vipond I.B., Halford S.E. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 16.Topal M.D., Thresher R.J., Conrad M., Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 17.Wentzell L.M., Halford S.E. DNA looping by the Sfi I restriction endonuclease. J. Mol. Biol. 1998;281:433–444. doi: 10.1006/jmbi.1998.1967. [DOI] [PubMed] [Google Scholar]
- 18.Siksnys V., Skirgaila R., Sasnauskas G., Urbanke C., Cherny D., Grazulis S., Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 19.Milsom S.E., Halford S.E., Embleton M.L., Szczelkun M.D. Analysis of DNA looping interactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol. 2001;311:515–527. doi: 10.1006/jmbi.2001.4893. [DOI] [PubMed] [Google Scholar]
- 20.Embleton M.L., Siksnys V., Halford S.E. DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol. 2001;311:503–514. doi: 10.1006/jmbi.2001.4892. [DOI] [PubMed] [Google Scholar]
- 21.Katiliene Z., Katilius E., Woodbury N.W. Single molecule detection of DNA looping by NgoMIV restriction endonuclease. Biophys J. 2003;84:4053–4061. doi: 10.1016/S0006-3495(03)75131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin H., Landick R., Gelles J. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys J. 1994;67:2468–2478. doi: 10.1016/S0006-3495(94)80735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finzi L., Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 24.Qian H., Elson E.L. Quantitative study of polymer conformation and dynamics by single-particle tracking. Biophys J. 1999;76:1598–1605. doi: 10.1016/S0006-3495(99)77319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian H. A mathematical analysis for the Brownian dynamics of a DNA tether. J. Math. Biol. 2000;41:331–340. doi: 10.1007/s002850000055. [DOI] [PubMed] [Google Scholar]
- 26.Pouget N., Dennis C., Turlan C., Grigoriev M., Chandler M., Salome L. Single-particle tracking for DNA tether length monitoring. Nucleic Acids Res. 2004;32:e73. doi: 10.1093/nar/gnh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad M., Topal M.D. DNA and spermidine provide a switch mechanism to regulate the activity of restriction enzyme Nae I. Proc. Natl Acad. Sci. USA. 1989;86:9707–9711. doi: 10.1073/pnas.86.24.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C.C., Topal M.D. Nonidentical DNA-binding sites of endonuclease NaeI recognize different families of sequences flanking the recognition site. Biochemistry. 1992;31:9657–9664. doi: 10.1021/bi00155a019. [DOI] [PubMed] [Google Scholar]
- 29.Seidel R., van Noort J., van der Scheer C., Bloom J.G., Dekker N.H., Dutta C.F., Blundell A., Robinson T., Firman K., Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nature Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 30.van den Broek B., Noom M.C., Wuite G.J. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J., Skoko D., Marko J.F. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E. Stat. Nonlin Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh W.T., Whitson P.A., Matthews K.S., Wells R.D. Influence of sequence and distance between two operators on interaction with the lac repressor. J. Biol. Chem. 1987;262:14583–14591. [PubMed] [Google Scholar]
- 33.Schafer D.A., Gelles J., Sheetz M.P., Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 34.Tolic-Norrelykke S.F., Engh A.M., Landick R., Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 35.Colquhoun D., Sigworth F.J. Fitting and statistical analysis of single-channel records. In: Sakmann B., Neher E., editors. Single Channel Recording. NY and London: Plenum Press; 1983. pp. 191–263. [Google Scholar]
- 36.Blumberg S., Gajraj A., Pennington M.W., Meiners J.C. Three-dimensional characterization of tethered microspheres by total internal reflection fluorescence microscopy. Biophys J. 2005;89:1272–1281. doi: 10.1529/biophysj.105.061242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dame R.T., van Mameren J., Luijsterburg M.S., Mysiak M.E., Janicijevic A., Pazdzior G., van der Vliet P.C., Wyman C., Wuite G.J. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Res. 2005;33:e68. doi: 10.1093/nar/gni073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C.C., Baxter B.K., Topal M.D. DNA cleavage by Naei: protein-purification, rate-limiting step, and accuracy. Biochemistry. 1994;33:14918–14925. doi: 10.1021/bi00253a031. [DOI] [PubMed] [Google Scholar]
- 39.Vipond I.B., Baldwin G.S., Halford S.E. Divalent metal ions at the active sites of the EcoRV and EcoRI restriction endonucleases. Biochemistry. 1995;34:697–704. doi: 10.1021/bi00002a037. [DOI] [PubMed] [Google Scholar]
- 40.Taylor J.D., Badcoe I.G., Clarke A.R., Halford S.E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991;30:8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- 41.Alves J., Selent U., Wolfes H. Accuracy of the EcoRV restriction endonuclease: binding and cleavage studies with oligodeoxynucleotide substrates containing degenerate recognition sequences. Biochemistry. 1995;34:11191–11197. doi: 10.1021/bi00035a026. [DOI] [PubMed] [Google Scholar]
- 42.Szczelkun M.D., Connolly B.A. Sequence-specific binding of DNA by the EcoRV restriction and modification enzymes with nucleic acid and cofactor analogues. Biochemistry. 1995;34:10724–10733. doi: 10.1021/bi00034a004. [DOI] [PubMed] [Google Scholar]
- 43.Erskine S.G., Halford S.E. Reactions of the eco RV restriction endonuclease with fluorescent oligodeoxynucleotides: identical equilibrium constants for binding to specific and non-specific DNA. J. Mol. Biol. 1998;275:759–772. doi: 10.1006/jmbi.1997.1517. [DOI] [PubMed] [Google Scholar]
- 44.Hiller D.A., Fogg J.M., Martin A.M., Beechem J.M., Reich N.O., Perona J.J. Simultaneous DNA binding and bending by EcoRV endonuclease observed by real-time fluorescence. Biochemistry. 2003;42:14375–14385. doi: 10.1021/bi035520w. [DOI] [PubMed] [Google Scholar]
- 45.Wentzell L.M., Nobbs T.J., Halford S.E. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 46.Halford S.E., Goodall A.J. Modes of DNA cleavage by the EcoRV restriction endonuclease. Biochemistry. 1988;27:1771–1777. doi: 10.1021/bi00405a058. [DOI] [PubMed] [Google Scholar]
- 47.Szczelkun M.D., Halford S.E. Recombination by resolvase to analyse DNA communications by the SfiI restriction endonuclease. EMBO J. 1996;15:1460–1469. [PMC free article] [PubMed] [Google Scholar]
- 48.Huai Q., Colandene J.D., Topal M.D., Ke H. Structure of NaeI-DNA complex reveals dual-mode DNA recognition and complete dimer rearrangement. Nature Struct. Biol. 2001;8:665–669. doi: 10.1038/90366. [DOI] [PubMed] [Google Scholar]
- 49.Colandene J.D., Topal M.D. The domain organization of NaeI endonuclease: separation of binding and catalysis. Proc. Natl Acad. Sci. USA. 1998;95:3531–3536. doi: 10.1073/pnas.95.7.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jian H., Schlick T., Vologodskii A. Internal motion of supercoiled DNA: brownian dynamics simulations of site juxtaposition. J. Mol. Biol. 1998;284:287–296. doi: 10.1006/jmbi.1998.2170. [DOI] [PubMed] [Google Scholar]
- 51.Vologodskii A., Cozzarelli N.R. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lia G., Bensimon D., Croquette V., Allemand J.F., Dunlap D., Lewis D.E., Adhya S., Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]