Abstract
The development of high-throughput technologies has generated the need for bioinformatics approaches to assess the biological relevance of gene networks. Although several tools have been proposed for analysing the enrichment of functional categories in a set of genes, none of them is suitable for evaluating the biological relevance of the gene network. We propose a procedure and develop a web-based resource (BIOREL) to estimate the functional bias (biological relevance) of any given genetic network by integrating different sources of biological information. The weights of the edges in the network may be either binary or continuous. These essential features make our web tool unique among many similar services. BIOREL provides standardized estimations of the network biases extracted from independent data. By the analyses of real data we demonstrate that the potential application of BIOREL ranges from various benchmarking purposes to systematic analysis of the network biology.
INTRODUCTION
The post-genomic era has introduced several high-throughput methodologies. A wide range of possibilities was opened for exploring the dynamics of biological processes. An exciting prospective that emerged from high-throughput techniques is extracting gene functional interactions in the context of genetic networks. Recently, a massive amount of experimental data was generated (1–8) and a number of computational approaches (9–13) were proposed to infer gene networks. Systematic exploration of the network biology, including systematic exploration of the biases introduced by gene associations in the network, is very important for understanding perspectives and limitation of different high-throughput technologies and computational methods. Moreover, knowledge of biases of different high-throughput technologies may allow the user to select an optimal one that is most suitable (i.e. less noisy, provides more relevant data) for the purposes of the particular investigation, before running a possibly long experimental study. Therefore, the development of tools for the functional analysis of gene networks is currently of particular interest.
A bias in the network related to some category can be defined as an enrichment (compared with null hypotheses) of associations in the network between genes sharing such category. We understand the term ‘category’ in a very broad sense. It relates to any gene property, such as functional category, domain composition or sequence similarity to another gene. The biologically unrelated characteristics such as gene length or geometric distance between gene probes on the chip (microarray technology) can be considered as category as well. The strong network biases associated with such biologically unrelated categories indicate shortcomings and limitations of the technology used to extract networks (14). On the contrary, the biological relevance can be defined as a bias related only to gene biological activities/properties categories. If quantified, the network biological relevance allows estimating the potential of high-throughput techniques to discover gene relations.
The information related to gene biological activities/properties can be retrieved from heterogeneous widely distributed public databases, such as FunCat (15), GO (16), Swiss-Prot (17), PFAM (18) and KEGG (19). Recently, a lot of tools such as DAVID (20), GFINDer (21), GOToolBox (22), FatiGO (23), GoMiner (24), MAPPFinder (25) and GOTM (26) have been proposed that use gene annotations provided through the Gene Ontology (GO) to detect the GO categories more relevant for a given set of genes. All these publicly available sources are focused towards the analysis of an unordered gene set rather a gene network structure.
In this study, we propose a procedure and develop a web-based resource (BIOREL) for evaluating the quantitative value of the overall network bias (http://mips.gsf.de/proj/biorel). The weights of the edges in the network may be either binary or continuous. To our knowledge, this is a first procedure of this kind. In general, the network bias of any nature can be evaluated by BIOREL. The web version integrates several sources of information, which allow one to estimate biologically related biases in the network. BIOREL provides a standardized estimation of the network bias extracted from independent data. The potential application of the BIOREL system ranges from various benchmarking purposes to systematic analysis of the network biology.
METHODS
A gene network structure can be formalized in matrix form. Each element of a matrix quantifies the edge weight between a corresponding pair of genes. Each column of the matrix reflects the associations (edge weights) of a particular gene. The whole gene network structure can be decomposed into small sub networks (further referred to as elementary networks). For the purpose of our study we will decompose the network so that each elementary network reflects associations of one particular gene. Therefore, the elementary network is formalized mathematically as a vector, namely the column of the corresponding network matrix.
The networks that are extracted from biological knowledge databases or from other reliable sources will be referred to as reference networks. These networks represent current knowledge about gene functional associations. The gene network whose biological relevance one should quantify will be referred to as target networks.
To avoid the ambiguity we give strict definitions to employed terms and concepts. We define the overall network bias (relevance) score as the proportion of genes in the target network with significantly biased associations. A gene is defined to be biased if its associations in the network significantly enriched with some categories (the null distribution is estimated based on statistics from random networks). We define the term ‘category’ as any principle to generate gene pairwise similarity matrix (further referred as reference network). For example, the category ‘metabolism’ [protein functional classification category from FunCat (15)] generates binary similarity matrix (a pair of genes that share this category get similarity score 1 and 0 otherwise). The category ‘sequence similarity’ generates corresponding similarity matrix (a pair of genes get similarity score proportional to their sequence similarity). To quantify the bias introduced by associations of individual gene, in the case when both target and reference network are binary, one can use standard statistical techniques (hypergeometric distribution). However, in other cases (the weights of the edges in the target or reference network are not binary) such techniques are inapplicable. For this reason to quantify the bias of each gene we use the regression analysis. The term ‘relevance’ we use only in relation to bias related to FunCat (15) categories.
For each gene X from the target network the following procedure is applied. The information from knowledge databases is formalized in a reference matrix . The element of the matrix quantifies the association of gene i (index i runs over all genes from the target network) with gene X in the reference network k (index k runs over all reference networks, e.g. categories, selected for analysis). On the other hand, the association of gene i with gene X in the target network is formalized by the element yi. In the next step the vector y is regressed against the matrix . The multiple correlation coefficient R is a quantitative measure of correlation between the reference matrix and vector y. The R-value is used to estimate the bias (related to employed reference networks) introduced by associations of gene X in the target network. The value of R can vary from 1 (perfect match between target network and reference networks) to 0 (the absence of any correlation). The corresponding P-value reflects the statistical significance of R and represents the probability to get the same correlation between the elementary target network and reference matrix by chance assuming as a null hypothesis that both and yi were generated randomly. In reality, this assumption is not true and statistical significance of R should be estimated based on statistics from random networks. For this purpose, random vector z (random analogue of vector y, represents associations of gene X in the random network) is generated and Rz (multiple correlation coefficient between random target network (vector z) and reference matrix) is estimated. The procedure is repeated an appropriate number of times (in respect to chosen significance level) to gain statistics of Rz-value for random networks. Based on Rz statistics we estimate the significance of R-value. Therefore, we classify the associations of gene X as biased/nonbiased at given significance level. The overall network bias is defined as the proportion of genes in the network with significantly biased associations.
Two options are realized to generate the random networks. In both cases, the topology of the target network is preserved and only genes in the nodes are permutated. In the first case, genes are permutated only from the target network. In the second case, the permutation process involves the whole set of genes from the analysed organism. The difference between random models allows evaluating the bias introduced by the target network set of genes.
Along with the bias of the target network the set of genes with significantly biased associations is identified. For each gene from the set of categories that make major contributions in an explanation of its associations in the target network are inferred. In other words these categories were significantly over- or under-represented among gene associations in the network. The overall statistics of such categories in the network provides information on the kind of gene interactions that prevail in the target network. This information can be used as a basis for a deeper insight into the network biology/biases.
Knowledge modules and principles to extract reference networks
The reference networks can be extracted from principally different sources of biological knowledge. As a core of our system, we employ the MIPS functional catalogue (27). Gene sequence similarity, Gene Neighborhood, Protein/Protein Interaction data (28) and InterPro domains data (29) were employed as additional independent knowledge data sources. Our system is very flexible in use. Each knowledge module can be switched on/off depending on the purpose of the study. There is an option, which allows the user to upload his/her own knowledge modules in the specified format.
Functional catalogue module (FunCat module)
The FunCat (27) is an annotation scheme for the functional description of proteins. Taking into account the broad and highly diverse spectrum of known protein functions, the FunCat consists of 28 main functional categories (or branches) that cover general fields, such as cellular transport, metabolism and cellular communication/signal transduction. The main branches exhibit a hierarchical, tree-like structure with up to six levels of increasing specificity. In total, the FunCat includes 1307 functional categories.
Each of the functional categories is assigned to a unique two-digit number. The upward context of the hierarchical tree consists of the prefix of the preceding nodes, located in the upper levels in the hierarchy. The levels of categories are separated by dots, e.g. 01 metabolism is a representative of the highest level, and 01.01.03.02.01 biosynthesis of glutamate belongs to the most specific level of FunCat.
According to the total number of different functional categories (1307) one can extract the same number of different networks. Each network corresponds to one category. The extraction procedure is very simple. If two genes have the same category then they are connected in the corresponding network. The hierarchical tree-like structure of FunCat presumes a hierarchical organization of the extracted networks. The networks generated by very specific categories (e.g. 01.01.03.02.01 biosynthesis of glutamate) are subnetworks of the networks generated by corresponding unspecific ones (e.g. 01 metabolism).
Sequence similarity (SS) module
The base information used by the module is a pairwise similarity score between the amino acid sequences of two genes. The FASTA pairwise scores were retrieved from the SIMAP database (15). The input values were calculated as −log10 (E-value). Pairwise scores with E-value > 0.1 were excluded from the analysis. The edge weight between two genes is proportional to the similarity score.
There are several reasons to include sequence similarity (SS) module to the BIOREL system. First of all it reflects any bias in the network that can be attributed to the genes sequence similarity. This module, for example, may be very helpful for analyses of gene expression data to estimate unspecific cross-hybridization effects. Any systematic bias towards similarity in expression profiles of genes with similar sequences will be detected.
InterPro domain (IPD) module
The core information used in this module is the protein domain composition provided by the InterPro database (29). The number of different networks extracted by this module corresponds to the number of domains. Each domain generates a network. The extraction procedure creates an edge between two genes if their proteins both have the corresponding domain. Any systematic bias in the network due to similar domain composition of interacting genes will be estimated by this module.
Gene neighborhood (GNH) module
The base information used in this module is the physical distance between two genes on the chromosome. The weight of the edge between two genes is inversely proportional to the distance separating them physically on the chromosome. Two options are implemented. The distance is measured in a number of genes or in a number of nucleotides. Any systematic bias in the gene interactions reflected in the network due to gene neighborhood on the chromosome will be estimated by this module.
Protein–protein interaction (PPI) module
There are several databases on PPI in yeast (28). Among them most reliable information comes from manually curated catalogues of known protein complexes (28,30). In addition, data from high-throughput experiments such as two hybrid experiments (1), genetic interactions (31), etc. are available. In all cases, the same network extraction procedure can be used. An edge of the binary network is constructed if two proteins are involved in an interaction according to the database record. The BIOREL system as available in the web configuration employs only manually curated catalogues of known protein complexes.
User-defined knowledge modules
This option can be flexibly used for various purposes. New sources of information considering genes from different biological perspectives may not yet have been included in our set of knowledge modules. Thus, we allow the user to add new knowledge for analysis. Second, the networks extracted from high-throughput data (or in some other way) may be significantly biased due to shortcomings of the technology or another reason. Therefore, the user can evaluate any bias in his network by uploading corresponding data.
RESULTS
Benchmarking BIOREL on synthetic data (Saccharomyces cerevisiae)
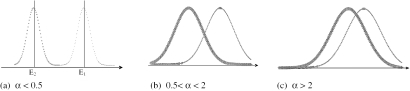
To benchmark the potential of BIOREL, we initially assessed the data with known and varied biases. For this purpose, we generated synthetic target gene networks with built-in bias related to FunCat (15) categories and evaluated them by BIOREL. The FunCat module was used to generate reference networks (see Methods). The edge weights in the synthetic gene networks were generated such that they follow two normal distributions: EdgeWeight1 ∼ N1(E1,SD1) and EdgeWeight2 ∼ N2(E2,SD2) where E1 and E2 correspond to the two means and SD1 and SD2 correspond to the standard deviations. We used the first distribution to generate the edge weights between gene pairs sharing the same FunCat category. In the current context, only these relations were considered as ‘relevant associations’. The second distribution was used to generate the edge weights for all other gene pairs. We name these relations as ‘irrelevant associations’. Several synthetic networks corresponding to different values of normal distributions parameters were generated. Parameters E1 and E2 (E1 > E2) were fixed and parameters SD1 = SD2 = SD = α(E1 − E2) were varied (α = 0.2, 0.5, 1, 1.2, 1.4, 1.5, 2, 3). In the generated networks, the signal-to-noise ratio was controlled by the value I = (E1 − E2)/SD = 1/α. The difference between edge weights of relevant/irrelevant associations ranged (Figure 1) from complete separation (α < 0.5, I > 2) to almost inseparable (α > 2, I < 0.5). In the completely separable case (i) the weight for any relevant association is greater than for any irrelevant association. For the partially separable case (ii) there are some irrelevant edges with weights greater than some relevant ones. The proportion of such edges depends on parameter α. For the inseparable case (iii) there is no difference between the edge weights of relevant/irrelevant associations in the synthetic network. Thus, the built-in bias in the generated data varied from maximal possible to zero.
Figure 1.
Statistical model used to generate synthetic data. The edge weight is increasing along horizontal axis and the probability (density) to generate such weight for relevant (left curve on each plot)/irrelevant (right curve on each plot) association in the synthetic network is shown.
The bias of each generated dataset was assessed by BIOREL. In Figure 2, the bias score is plotted against the value of parameter α.
Figure 2.
The bias score (computed by BIOREL) in the synthetic data is plotted against parameter α. The bias score is a proportion of genes in the network with significantly (P < 0.01) biased associations.
At low noise levels (α < 0.5, I > 2) the relevance score converges approximately to 0.7. This value is the share of annotated genes for S.cerevisiae genome in FunCat and represents the maximal possible bias (all annotated genes had significantly biased associations in the corresponding synthetic network). From Figure 2 we can also derive that if the relevance score of the network is >0.5 then we expect the value of α to be <1. The relevance score between 0.1 and 0.4 means that 1 < α < 2. For α > 2 (I < 0.5) we expect no difference between relevant/irrelevant edge weights and, therefore, no bias is found in the data.
The results clearly demonstrate that BIOREL correctly ranks the datasets according to the value of built-in bias. Thus, it can be used for systematic benchmarking purposes to evaluate the potential of various data sources to reveal biological relations between genes as well as to detect systematic biases related to the limitations and shortcomings of high-throughput methodologies.
Analyses of high-throughput technologies for inferring PPI
At the moment there are two major alternative methodologies to discover PPIs on the whole genome scale. Without taking into acount minor technological variations they can be classified as yeast two-hybrid systems and mass spectrometric protein complex identification (MS-PCI). Previous comparison of the MS-PCI dataset with interactions reported in the literature revealed an average 3-fold higher success rate in the detection of known complexes compared with large-scale two-hybrid studies (2). In this example, we systematically benchmarked the biological relevance and biases of gene networks extracted from independent sources of PPI data.
PPI two-hybrid data
We analysed gene networks extracted from PPI data yielded in two-hybrid high-throughput experiments for S.cerevisiae genome. We analysed two publicly available datasets produced by different groups. It is widely accepted that interactions, which are detected simultaneously by independent experimental groups are more reliable. We investigated the bias of the overlapped network constructed from interaction pairs that were detected in both analysed sets. The details of the benchmark procedure can be found in Table 1.
Table 1.
BIOREL evaluation of PPI two hybrid data
| Experiment | Network bias tested by BIOREL | BIOREL modules | Bias scorea | ||
|---|---|---|---|---|---|
| Ito | Uetz | Overlapped, Ito+Uetz | |||
| 1 | Interacting proteins share the same function | FunCat module | 0.1 | 0.18 | 0.30 |
| 2 | Interacting proteins share sequence similarity (or partial similarity) | Sequence Similarity and InterPro Domain modules | 0.05 | 0.10 | 0.15 |
| 3 | Interacting proteins have the same length | Gene length module | ∼0.01b | ∼0.01b | ∼0.01b |
aThe bias score is a proportion of genes in the network with significantly (P < 0.01) biased associations.
bThe biases were not found to be significant compared with random networks.
The PPI data of (1) contain information about 4549 putative two-hybrid interactions among 3276 genes (referred to as Ito). The PPI data of (3) contain information about 957 putative interactions involving 1004 genes (referred to as Uetz). The overlapped network contained 343 interactions involving 268 genes. The standard output of the BIOREL system for all three cases is available on the web site (see Examples section). The bias score at the significance level (P < 0.01) for (1) and (3) networks was 10 and 18%, respectively, while the overlapped network bias was ∼30% (Table 1). These results suggest that the increased bias detected is reflective of the likely increased reliability in the overlapped network. If this is true, then the BIOREL analysis of the PPI networks not only supports the view that the intersection of both datasets is more reliable but also provides a quantitative estimation of the effect.
Along with biological relevance we evaluated the bias in PPI data related to sequence similarity or partial similarity (domains) of interacting proteins. For this reason we repeated the evaluation with only two knowledge modules: SS and IPD (see Methods). We found a strong bias in the data. At the significance level (P < 0.01) the bias score for (1), (3) and the overlapped network was 5, 10 and 15%, respectively. These results partially reflect the higher than expected number of interacting pairs of paralogous proteins in PPI networks previously reported in (4).
We also investigated whether or not there is a bias that relates to the length of interacting proteins. For this purpose, we construct a corresponding reference network. The edge of the network between two genes was proportional to the relative difference between the lengths of two corresponding proteins. Thus, we test the hypothesis that two interacting genes in PPI data have approximately equal length. There was no such bias detected in the data.
PPI MS-PCI data
The MS-PCI technology does not directly measure PPIs. Rather, the protein composition of purified (putative) protein complexes is determined. Not all proteins involved in a complex necessarily physically interact. Nevertheless, we assume that there is ‘functional association’ between every pair of proteins in such a complex. If a complex of 10 proteins is purified, we suppose that there are 10(10 − 1)/2 = 45 pairwise interactions.
We analysed two dataset yielded by MS-PCI technology. The PPI data from (2) contain information ∼30 000 pairwise interactions among ∼1350 genes (referred to as Gavin). The PPI data from (5) contain information ∼30 000 pairwise interactions among ∼1570 genes (referred to as Ho). We tested the same spectrum of biases by BIOREL as we did for PPI two hybrid data. The results are summarized in the Table 2.
Table 2.
BIOREL evaluation of PPI MS-PCI data
| Experiment | Network bias tested by BIOREL | BIOREL modules | Bias scorea | |
|---|---|---|---|---|
| Gavin | Ho | |||
| 1 | Interacting proteins share the same function | FunCat module | 0.64 | 0.29 |
| 2 | Interacting proteins share sequence similarity (or partial similarity) | Sequence Similarity and InterPro Domain modules | 0.19 | 0.09 |
| 3 | Interacting proteins have the same length | Gene length module | ∼0.04 | ∼0.02 |
aThe bias score is a proportion of genes in the network with significantly (P < 0.01) biased associations.
The BIOREL results suggest that the bias in MS-PCI data related to the associations between functionally similar genes is substantially higher than for PPI two-hybrid data. Therefore, as expected, the MS-PCI technology is preferable for the identification of functional relations between genes. At the same time, we would like to point out to extremely significant sequence similarity bias related to MS-PCI data and the presence of slight bias related to the length of sequences of interacting proteins. AASAs been already mentioned this bias reflects the higher than expected number of interacting pairs of paralogous proteins in PPI networks (4).
Microarray expression data
Among other high-throughput methodologies microarray technology became a routine in many laboratories. Many expression datasets were generated by various microarray platforms. A number of complex network extracting procedures have been employed. However, there is no common understanding of the limitation and reliability of expression data. For instance, normalization artifacts or unspecific cross-hybridization effects may cause a systematic bias. The quality of different publicly available datasets produced by different generations of microarray chips and by different laboratories varies considerably. Thus, a systematic evaluation of the relevance and biases of genetic networks extracted from different expression datasets is an essential need.
In this example, we assessed using BIOREL expression data from S.cerevisiae generated by different platforms (oligonucleotide, spotted cDNA arrays). An abundance of cDNA data is publicly available from Stanford Microarray Database (SMD). We downloaded two datasets (referred to as Gasch and Spellman) related to studies of (6,7). These datasets were widely used in studies, which proposed new methods for extracting gene functional associations from expression data. The last analysed set (referred to as Causton) was generated by Affymetrix platform (8).
A variety of methods were proposed to extract gene functional networks from expression data. Most of them consider gene expression profiles as vectors in multidimensional space and attempt to explore geometrical relations between them. The Pearson correlation coefficient reflects the linear statistical relation and reciprocal geometric position in a vector space between two expression profiles. The correlation matrix collects such information for all pairs of genes and was a primary data source used for many network extraction methods. To benchmark relevance and biases of expression data we analysed the relevance and biases of information from the correlation (Pearson) matrix.
For each expression dataset, we apply the same benchmark procedure. Initially, we preselect four sets of the 1000, 2000, 3000 and 4000 most highly expressed genes (average value across available measurements). For the selected genes, the Pearson correlation coefficient was computed. Absolute correlation values <0.5 were considered as insignificant and ignored. The weight of an edge in the network between two genes was proportional to the absolute value of correlation (>0.5).
We estimated the biological relevance of the expression networks as well as functional bias, which can be attributed to unspecific cross-hybridization. For this reason, we estimated the network relevance twice by different sets of knowledge modules. First, we used FunCat module. Thus, we evaluated the network bias due to associations between genes related to the same functional groups. In the second run, we used sequence similarity and domain (reflects strong partial sequence similarity) modules only. Therefore, the network bias due to associations between genes sharing strong sequence similarity was estimated. The bias value can serve as an indicator of unspecific cross-hybridization effects (the cross-hybridization between probes related to different genes that share sequence similarity).
The results are summarized in the Table 3. A brief analysis of the table indicates that the relevance of the gene networks inferred from Affymetrix expression data with the threshold 0.5 is remarkably higher than the relevance of the gene networks inferred from cDNA data with the same threshold value. At the same time the bias attributed to sequence similarity is almost equal in all networks however slightly lower for Affymetrix data. Therefore, the rate of co-expressed genes which simultaneously share the same function and strong sequence similarity is approximately twice as high for cDNA data compared with Affymetrix data. The 2-folds bias clearly points out that the unspecific cross-hybridization effect is higher for cDNA platform. The result is easily explained by the technical differences between two technologies: cDNA chips use full-length gene transcripts attached to the slide while Affymetrix use several oligonucleotides. Among many other reasons that may explain this effect we point out that the conditions of hybridization (due to the technological differences) for one probe are more stringent for Affymetrix than for cDNA platforms.
Table 3.
BIOREL evaluation of expression datasets
| Dataset | Platform and number of measurements | Biological relevance (functional bias scorea) | Sequence similarity bias scorea | Bias scores ratio | Number of selected genes |
|---|---|---|---|---|---|
| Gasch | cDNA, 53 | 0.24 | 0.08 | 3.0 | 1000 |
| Gasch | cDNA, 53 | 0.21 | 0.07 | 3.0 | 2000 |
| Gasch | cDNA, 53 | 0.18 | 0.07 | 2.6 | 3000 |
| Gasch | cDNA, 53 | 0.19 | 0.08 | 2.4 | 4000 |
| Spellman | cDNA, 148 | 0.16 | 0.07 | 2.3 | 1000 |
| Spellman | cDNA, 148 | 0.20 | 0.09 | 2.2 | 2000 |
| Spellman | cDNA, 148 | 0.18 | 0.08 | 2.3 | 3000 |
| Spellman | cDNA, 148 | 0.17 | 0.08 | 2.1 | 4000 |
| Causton | Affy, 45 | 0.35 | 0.06 | 6.0 | 1000 |
| Causton | Affy, 45 | 0.43 | 0.06 | 7.1 | 2000 |
| Causton | Affy, 45 | 0.52 | 0.07 | 7.5 | 3000 |
| Causton | Affy, 45 | 0.50 | 0.06 | 8.3 | 4000 |
aThe bias score is a proportion of genes in the network with significantly (P < 0.01) biased associations.
Gene neighborhood networks
In the next example using BIOREL we estimated the relevance of gene networks extracted based on gene neighborhood information for different model organisms. The principle to construct the neighborhood network was very simple. The weight of the edge between two genes was inversely proportional to the number of genes separating them physically on the chromosome. The weight of the edge between two consecutive genes was set to 1. The weight of the edge between genes separated only by 1 gene was set to 0.5, by two genes 0.33 and so on [the edge weight equal to 1/(n + 1), where n is a number of separating genes]. We estimated the functional bias of such networks for eight model organisms: Arabidopsis thaliana, Bacillus subtilis, Helicobacter pylori, Listeria monocytogenes, Listeria innocua, Thermoplasma acidophilum, Saccharomyces cerevisiae and Neurospora crassa. The standard output of the BIOREL system for all cases is available at the web site (see Examples section). As it was expected (see Table 4) the functional bias of neighborhood network for bacteria species was much stronger (∼20%) than the functional bias for eukaryotes [4–7%, except A.thaliana (20%)].
Table 4.
BIOREL evaluation of gene neighborhood networks in eu- and prokaryote genomes
| Genome | Network bias tested by BIOREL | BIOREL modules | Bias scorea | Top enriched categories |
|---|---|---|---|---|
| A.thaliana | Genes located closely on the chromosome have the same function or have similar sequences | FunCat module, Sequence Similarity and InterPro Domain modules | 0.20 | Sequence similarity (>90%) |
| B.subtilis | 0.22 | FunCat categories (>70%) | ||
| H.pylori | 0.18 | FunCat categories (>80%) | ||
| L.monocytogenes | 0.19 | FunCat categories (>70%) | ||
| T.acidophilum | 0.14 | FunCat categories (>50%) | ||
| S.cerevisiae | 0.04 | Sequence similarity (>60%) | ||
| N.crassa | 0.07 | Sequence similarity (>90%) |
aThe bias score is a proportion of genes in the network with significantly (P < 0.01) biased associations.
The statistical analyses of categories enriched in the network for bacteria and eukaryotes species reveal the principal difference in the roots of both effects. The number of cases for eukaryotes when sequence similarity was only one category, which explains the associations of genes classified as relevant was strikingly higher (∼80% of cases) than for bacteria species (∼15–20% of cases) where in most cases (∼70% of cases) the associations were mainly explained by FunCat categories. Thus, the functional bias of the neighborhood network for eukaryotes mainly can be attributed to gene duplication events while for bacteria the neighborhood network bias is accounted for the operon genome structure as genes within the same operon does not necessarily share strong sequence similarity but are involved in the same biological function.
We would like to point out that the functional bias of gene neighborhood network for bacteria species is less than the share of genes in the genome expected to be organized in operon structures (50–80%). The gene neighborhood network as it was constructed in the example reflects operon structure only partially. For instance, genes at the operon boarders functionally unrelated but are connected in the gene neighborhood network. On the other hand, functionally related operons are sometimes separated physically on the chromosome and thus a lot of relevant edges are absent in the gene neighborhood network.
BIOREL web server
Using the BIOREL web server (http://mips.gsf.de/proj/biorel), the typical analysis steps are (i) upload the gene network structure, (ii) select the knowledge modules that should be used for bias evaluation and (iii) receive output results.
The BIOREL output file consists of three sections. The first section contains information on the uploaded network (the number of genes and nonzero edges). The second section reports the functional bias of the supplied network at different significance levels. This information is summarized in a table and a graph. The third section specifies in the table format the genes with significantly biased associations in the network along with the categories, which mostly explain these associations. Therefore, the user gets systematic information about the bias of the supplied network. Currently, the BIOREL provides tools for analysis of eight genomes, including H.pylori, L.innocua, L.monocytogenes, B.subtilis, T.acidophilum, A.thaliana, N.crassa and S.cerevisiae. The functional annotation data for these genomes are described in (15,27,32,33).
It has been shown in the above examples that BIOREL system can help the user in analysis of the genetic network at least in two ways: it identifies the quantitative value of the network biological (technical) relevance (biases) as well as supplies detailed information about functional associations of each gene (along with functional categories, which play major role in explanation of gene interactions in the network). It is obvious that a reliable classification of the genes for any set of genes submitted to BIOREL analysis is a prerequisite for the accuracy of the relevance factor.
DISCUSSION
The development of high-throughput technologies has generated the need for bioinformatics approaches to assess the biological relevance of gene network structures. Although several tools have been proposed for analysing the enrichment of functional categories in a list of genes, none of them is applicable for evaluating the biological relevance of any specific gene network. Such evaluation is related to the analysis of the functional bias introduced by the gene associations in the network. Unlike most similar services on the web the BIOREL is able to analyse not just a list of genes but a network structure. The weights of the edges in the network may be either binary or continuous. This essential feature makes our web tool unique among many similar services.
A systematic way to integrate different and biologically independent sources of information is particularly useful for the interpretation of genetic network. It can lead to a better understanding of the biological nature of the associations in the network. The examples shown above demonstrate the potential of BIOREL. As one can see it can be successfully used for variety of goals and supply statistical basis for interesting biological conclusions.
The BIOREL is the first system on the web, which automatically infers the biological relevance of any genetic network for several model organisms. The potential application of the BIOREL system ranges from various benchmarking purposes to systematic analysis of the network biology. BIOREL can be flexibly extended to incorporate any other type of information relevant for functional interaction such as literature data, protein complex information and the like.
At the end we would like to summarize that BIOREL can be used for several goals:
To discover new information and generate new hypotheses in a completely automated mode. An interpretation of the hypotheses can provide new knowledge. Examples of such analyses were considered in the article.
To guide a high-throughput experiment, as demonstrated by the PPI data example.
Acknowledgments
This work was supported by grants 031U212C BFAM (BMFB) to H.W.M. and TE/308/1-1 (DFG) to I.V.T. and H.W.M. The authors would like to thank Philip Pagel, Philip Wong, Michael Mader and Martin Munsterkotter for fruitful discussions and anonymous reviewers for their helpful remarks. Funding to pay the Open Access publication charges for this article was provided by 031U212C BFAM (BMFB) grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 3.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 4.Ispolatov I., Yuryev A., Mazo I., Maslov S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tornow S., Mewes H.W. Functional modules by relating protein interaction networks and gene expression. Nucleic Acids Res. 2003;31:6283–6289. doi: 10.1093/nar/gkg838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart C.E., Sharenbroich L., Bornstein B.J., Trout D., King B., Mjolsness E., Wold B.J. A mathematical and computational framework for quantitative comparison and integration of large-scale gene expression data. Nucleic Acids Res. 2005;33:2580–2594. doi: 10.1093/nar/gki536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C.F., Wai K.M., Patterton H.G. Calculating the statistical significance of physical clusters of co-regulated genes in the genome: the role of chromatin in domain-wide gene regulation. Nucleic Acids Res. 2004;32:1798–1807. doi: 10.1093/nar/gkh507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Xu D. Global protein function annotation through mining genome-scale data in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:6414–6424. doi: 10.1093/nar/gkh978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonov A.V., Tetko I.V., Kosykh D., Surmeli D., Mewes H.W. Exploiting scale-free information from expression data for cancer classification. Comput. Biol. Chem. 2005;29:288–293. doi: 10.1016/j.compbiolchem.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Ioannidis J.P. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mewes H.W., Amid C., Arnold R., Frishman D., Guldener U., Mannhaupt G., Munsterkotter M., Pagel P., Strack N., Stumpflen V., et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnhammer E.L., Eddy S.R., Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Masseroli M., Martucci D., Pinciroli F. GFINDer: Genome Function INtegrated Discoverer through dynamic annotation, statistical analysis, and mining. Nucleic Acids Res. 2004;32:W293–W300. doi: 10.1093/nar/gkh432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin D., Brun C., Remy E., Mouren P., Thieffry D., Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 24.Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doniger S.W., Salomonis N., Dahlquist K.D., Vranizan K., Lawlor S.C., Conklin B.R. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B., Schmoyer D., Kirov S., Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Guldener U., Mannhaupt G., Munsterkotter M., et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guldener U., Munsterkotter M., Kastenmuller G., Strack N., van Helden J., Lemer C., Richelles J., Wodak S.J., Garcia-Martinez J., Perez-Ortin J.E., et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csank C., Costanzo M.C., Hirschman J., Hodges P., Kranz J.E., Mangan M., O'Neill K., Robertson L.S., Skrzypek M.S., Brooks J., et al. Three yeast proteome databases: YPD, PombePD, and CalPD (MycoPathPD) Methods Enzymol. 2002;350:347–373. doi: 10.1016/s0076-6879(02)50973-3. [DOI] [PubMed] [Google Scholar]
- 31.Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 32.Riley M.L., Schmidt T., Wagner C., Mewes H.W., Frishman D. The PEDANT genome database in 2005. Nucleic Acids Res. 2005;33:D308–D310. doi: 10.1093/nar/gki019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tetko I.V., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Fobo G., Ruepp A., Antonov A.V., Surmeli D., Mewes H.W. MIPS bacterial genomes functional annotation benchmark dataset. Bioinformatics. 2005;21:2520–2521. doi: 10.1093/bioinformatics/bti380. [DOI] [PubMed] [Google Scholar]