Abstract
Human TIN2 interacts with the telomeric-DNA-binding protein TRF1, suppresses telomere elongation in telomerase-positive cells, and may control telomere length by modulating telomere structure. To test the latter idea, we developed an in vitro assay, using biotinylated telomeric DNA probes and streptavidin–agarose, to quantify the ability of TRF1 and TIN2 to stimulate interactions of telomeric DNA tracts with each other (probe clustering). This assay revealed that TRF1 alone had weak probe-clustering activity, but TIN2 stimulated activity fivefold to tenfold. A dominant-negative TIN2 mutant protein that increased telomere length in vivo disrupted probe clusters formed by TRF1 and TIN2, suggesting that the ability to stimulate telomeric DNA interactions is important for telomere-length regulation. Unlike TRF1, TIN2 did not form homodimers. We propose that TIN2 alters the conformation of TRF1, which favours a tertiary telomeric structure that hinders telomerase from gaining access to telomeres.
Introduction
Maintenance of telomere length and structure is crucial for genomic stability and for preventing cellular senescence, which in turn suppresses the development of cancer and ageing phenotypes (Campisi et al., 2001). Among the factors that regulate telomere structure and function are telomerase, several proteins that associate exclusively with telomeres,and proteins that have both telomeric and non-telomeric functions (for example, components of certain DNA repair pathways; Chan & Blackburn, 2002; Lundblad, 2000; Shore, 1997).
Two dimeric proteins, TRF1 and TRF2, bind exclusively and directly to double-stranded telomeric DNA in mammalian cells (Bilaud et al., 1997; Broccoli et al., 1997; Chong et al., 1995). A third protein, TIN2, also associates exclusively with telomeres, but does so by binding TRF1. All three proteins are thought to control telomere length by promoting a telomeric architecture that limits the ability of telomerase to access telomeres.
TRF1 contains an acidic amino terminus, a conserved homodimerization domain and a carboxyl terminus that has homology to the DNA-binding domain of Myb oncoproteins (Bilaud et al., 1997; Broccoli et al., 1997). Although a single TRF1 Myb domain can interact with telomeric DNA (Konig et al., 1998), TRF1 binds DNA predominantly as a homodimer (Bianchi et al., 1997). A flexible connection between the dimerization and Myb domains may explain how TRF1 can facilitate the pairing and looping of telomeric DNA tracts, which was detected by electron microscopy (EM; Bianchi et al., 1999; Griffith et al., 1998).
TRF1 interacts with TIN2 through a region in the TRF1 homodimerization domain (Kim et al., 1999). The crystal structure of this TRF1 domain revealed a large surface area, suitable for interactions with other proteins, which is proposed to stabilize the structure and position the two Myb domains on a single telomeric DNA repeat unit (Fairall et al., 2001). Because TIN2 binds the TRF1 homodimerization domain, TIN2 may alter the structural stability, and therefore the DNA binding characteristics, of TRF1.
Telomere-length regulation by TIN2 is telomerase dependent. Overexpression of wild-type TIN2 slightly shortened telomeres in telomerase-positive cells. By contrast, an N-terminally truncated protein (TIN2-13) increased telomere length, suggesting a dominant-negative activity (Kim et al., 1999; Rubio et al., 2002). However, neither TIN2 nor TIN2-13 altered telomere length in telomerase-negative cells or telomerase activity in vitro. In electrophoretic mobilityshift assays (EMSAs), TIN2 formed an unusually large complex with TRF1 and telomeric DNA (Kim et al., 1999). We therefore hypothesized that TIN2 controls telomere length by modulating telomere structure.
Here, we show that TIN2 facilitates the physical association of telomeric DNA tracts in a TRF1-dependent manner and that the dominant-negative TIN2 mutant protein lacks this activity. These results suggest an important role for TIN2 in organizing telomeric structure.
Results and Discussion
TIN2 does not have a homodimerization domain
TIN2 forms a large, apparently multimeric complex with TRF1 in EMSAs (Kim et al., 1999). One mechanism by which TIN2 might stimulate the formation of this complex is by forming homodimers or oligomers that create bridges among TRF1 dimers. To test this possibility, we used yeast two-hybrid analysis (Kim et al., 1999). We fused TIN2 or TRF1 to the yeast Gal4 DNA-binding (Gal4-DB) or transactivation (Gal4-AD) domains. Yeast transformed with the vectors Gal4-DB–TIN2 and Gal4-AD–TIN2 failed to grow in selective medium, despite growth in nonselective medium, but showed the expected TIN2–TRF1 interactions (Fig. 1A). These results suggest that TIN2 does not form homodimers in yeast.
Figure 1.
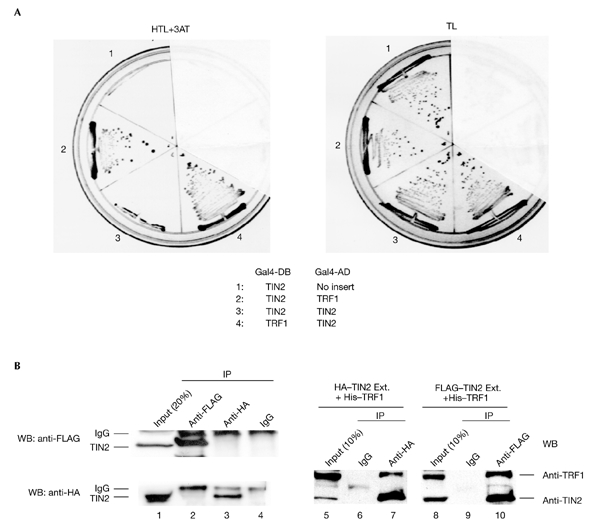
Homotypic TIN2 interactions. (A) TIN2 and TRF1 complementary DNAs were cloned into the yeast vectors pGBT-9, pGAD-10 (Clontech), pTGB-2 or pDAG-2 (Cary et al., 1998), and the indicated vector pairs (1–4) were transformed into yeast. Yeast cells were cultured on selective (HTL+3AT: −His, −Trp, −Leu, plus 10 mM 3-aminotriasol) or nonselective (TL: −Trp, −Leu) media. (B) Amino-terminal FLAG-tagged TIN2 and carboxy-terminal haemagglutinin (HA)-tagged TIN2 were cloned into the pLXSN retroviral vector, and infectious virus was produced and used to infect HT1080 cells, as described in Kim et al. (1999). Lysates alone (lanes 1–4) or mixed with purified His–TRF1 (lanes 5–10) were immunoprecipitated (IP) using anti-FLAG (Sigma) or anti-HA (Santa Cruz) antibodies. The precipitates were analysed by western blotting (WB) using polyclonal anti-HA (Santa Cruz), anti-TRF1, anti-TIN2 or anti-FLAG antibodies, respectively. Ext., cell extract; Gal4-AD, Gal4 activation domain; Gal4-DB, Gal4 DNA-binding domain.
To determine whether TIN2 interacts with itself in human cells, we overexpressed two epitope-tagged proteins, FLAG–TIN2 and haemagglutinin (HA)–TIN2, in HT1080 cells. Immuno-precipitation using anti-FLAG did not co-precipitate HA–TIN2, as detected by western blotting. Similarly, immunoprecipitation using anti-HA did not co-precipitate FLAG–TIN2 (Fig. 1B, lanes 1–4). However, FLAG–TIN2 and HA–TIN2 were able to interact with TRF1 tagged with six histidine residues (Fig. 1B, lanes 5–10). These results suggest that the formation of the multimeric complex stimulated by TIN2 does not result from TIN2 dimerization or oligomerization.
To determine the specificity and nature of the complex stimulated by TIN2 in EMSAs (the supercomplex), we purified His-tagged TIN2 and TRF1 from baculovirus-infected insect cells (Kim et al., 1999) and His–TIN2-13 and His–TIN2-12 from Escherichia coli (Fig. 2A). We confirmed protein purity using denaturing polyacrylamide gel electrophoresis (PAGE; Fig. 2B), and confirmed protein identity by western blotting (Fig. 2C) using an anti-TIN2 antibody (Kim et al., 1999).
Figure 2.
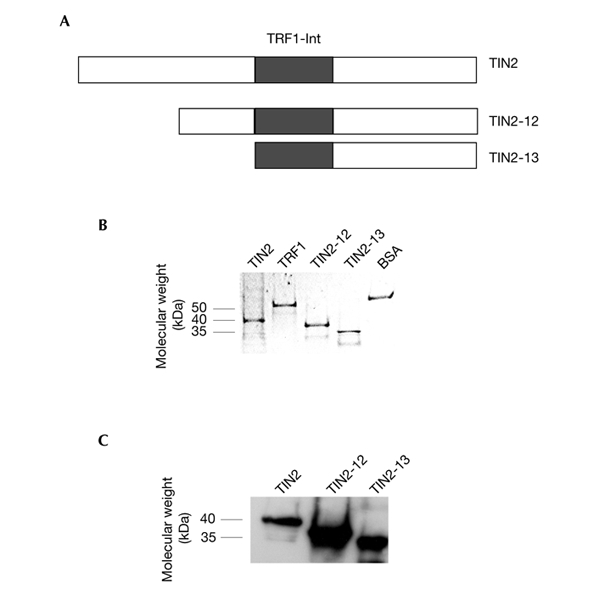
TIN2 proteins. (A) Wild-type TIN2 (amino acids 1–354), TIN2-12 (amino acids 120–354) and TIN2-13 (amino acids 196–354). (B,C) Approximately 2 µg of purified His–TRF1, His–TIN2, His–TIN2-12, His–TIN2-13 and BSA were detected by Coomassie-blue staining and identified by western blotting using an anti-TIN2 antibody. TRF1-Int, TRF1-interacting domain.
TIN2 stimulates telomeric DNA interactions in vitro
Electron microscopy showed that TRF1 promotes the pairing of telomeric DNA tracts in vitro (Griffith et al., 1998). Because EM assays are difficult and are not quantitative, we developed a biochemical assay to measure interactions between telomeric DNA tracts and to test the effects of TIN2. This assay does not distinguish between interactions of two versus several DNA tracts, and we therefore refer to it as a telomeric 'probe-clustering' assay.
In the clustering assay two probes were used, 6X-Tel (178 bp), and B-6X-Tel (126 bp), which contained two biotins at a branched 5′-end. Both contained six telomeric repeats (TTAGGG)6, but differed in length due to flanking non-telomeric sequences (Fig. 3A). We first optimized EMSAs (Fig. 3B) to detect three TRF1–probe complexes (lane 1; indicated by one, two and three asterisks), which correspond to the binding of one, two and three TRF1 dimers (indicated by the number of asterisks in Fig. 3B; Bianchi et al., 1997). Under these conditions, TRF1 alone formed a small amount of supercomplex (Fig. 3B, lanes 2 and 7), but supercomplex formation was strongly stimulated by TIN2 (Fig. 3B, lanes 4 and 9). These findings suggest that the supercomplex is formed due to the telomeric-DNA-pairing activity of TRF1, and that TIN2 stimulates this activity.
Figure 3.
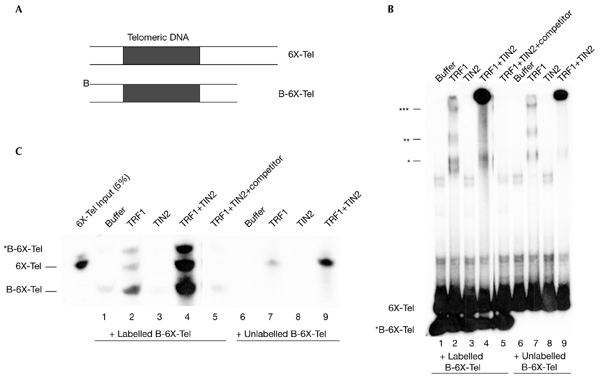
TIN2 stimulates probe clustering. (A) The telomere probes (6X-Tel and B-6X-Tel) used in the assay. (B) Reaction mixtures containing probes (10 µl) and the proteins indicated were analysed by electrophoretic mobility-shift assays. Asterisks indicate TRF1–probe complexes that correspond to the binding of one, two or three TRF1 dimers, as indicated by the number of asterisks. (C) Reaction mixtures (90 µl) in the absence ('buffer') or in the presence of TRF1 (0.25 µM), TIN2 (0.25 µM) or both were captured on streptavidin–agarose beads, released by phenol extraction, precipitated with ethanol, and analysed by native polyacrylamide gel electrophoresis. The B-6X-Tel probe was either labelled (lanes 1–5) or unlabelled (lanes 6–9). For lane 5, excess (100×) unlabelled double-stranded (TTAGGG)7 was added before the addition of proteins ('competitor').
To test this idea, we mixed labelled or unlabelled B-6X-Tel with labelled 6X-Tel, added buffer (as a control) or recombinant proteins and captured the biotinylated probes (B-6X-Tel), and associated complexes, on streptavidin–agarose beads. We then dissociated the complexes by phenol extraction. Phenol disrupts protein–protein and protein–DNA interactions, but not biotin– streptavidin binding. We identified the labelled probes that were released by PAGE and autoradiography. Because 6X-Tel is not biotinylated, it cannot associate with streptavidin unless it is bound to captured B-6X-Tel probes, which can only occur through protein–DNA interactions.
When we mixed 6X-Tel and B-6X-Tel probes with buffer (Fig. 3C, lanes 1 and 6) or TIN2 alone (Fig. 3C, lanes 3 and 8), phenol released little or no 6X-Tel. Thus, the probes did not interact with each other under these conditions. TRF1 alone increased the amount of released 6X-Tel slightly but significantly (Fig. 3C, lanes 2 and 7), consistent with its reported telomeric-DNA-pairing activity. Although the amount of labelled probe released was low when only TRF1 was present, it was dependent on TRF1 concentration (data not shown). TRF1-dependent 6X-Tel release was stimulated markedly (fivefold to tenfold) by TIN2 (Fig. 3C, lanes 4 and 9). This TIN2 activity depended on the telomeric sequence, as the signal was abolished by excess unlabelled telomeric DNA (TTAGGG)7 (Fig. 3C, lane 5). These results support the idea that TIN2 stimulates TRF1 telomeric-pairing activity.
B-6X-Tel was released from reactions that contained TRF1 because, in stimulating probe–probe interactions, TRF1 cannot distinguish biotinylated from unbiotinylated probes. Thus, TRF1-mediated probe–probe interactions occur not only between streptavidin-anchored B-6X-Tel and free 6X-Tel, but also between anchored B-6X-Tel and free B-6X-Tel. Another form of B-6X-Tel, which we refer to as *B-6X-Tel, was also released. This slowly migrating species must have been derived from B-6X-Tel because it was absent when only unlabelled B-6X-Tel was used (Fig. 3C, lanes 6–9), and was released coincidentally with B-6X-Tel (Fig. 3C, lanes 2 and 4). We failed to detect *B-6X-Tel in lanes 6–9, even when the autoradiograph was overexposed (data not shown). We do not know the precise nature of *B-6X-Tel, but speculate that the two branched biotinylated 5′-ends were modified by the probe–probe interaction reactions and by the phenol extraction, which altered a fraction of the probe and hence its migration.
Effects of probe size and mutant TIN2
To gain insights into how TRF1, TIN2 and telomeric DNA interact, we varied the TRF1 concentration and telomeric probe size and analysed the activities of TIN2 mutant proteins.
We first determined TRF1 and TIN2 activity in the clustering assay (Fig. 4A) and in EMSAs (Fig. 4B), keeping TIN2 concentration constant (1 µM), but varying TRF1 concentration (0.1 µM, 0.3 µM and 1 µM) so that one, two or three TRF1 dimers assembled on the probe (Fig. 4B; indicated by one, two and three asterisks, respectively). At all TRF1 concentrations tested, TIN2 stimulated probe clustering (Fig. 4A, lanes 5–7) and supercomplex formation (Fig. 4B, lanes 5–7). Quantification of the probes released in the clustering assay (Fig. 4C) showed that the absolute and relative amounts of probe interactions varied depending on whether one, two or three TRF1 dimers were bound (compare Fig. 4B with 4C). TIN2 was least effective at stimulating TRF1-induced probe clustering when three TRF1 dimers were bound. A control protein (BSA) did not stimulate probe clustering or supercomplex formation (Fig. 4A,B, lane 14). Quantification of the supercomplex showed similar results, supporting the idea that EMSAs and the clustering assay detect a similar complex.
Figure 4.
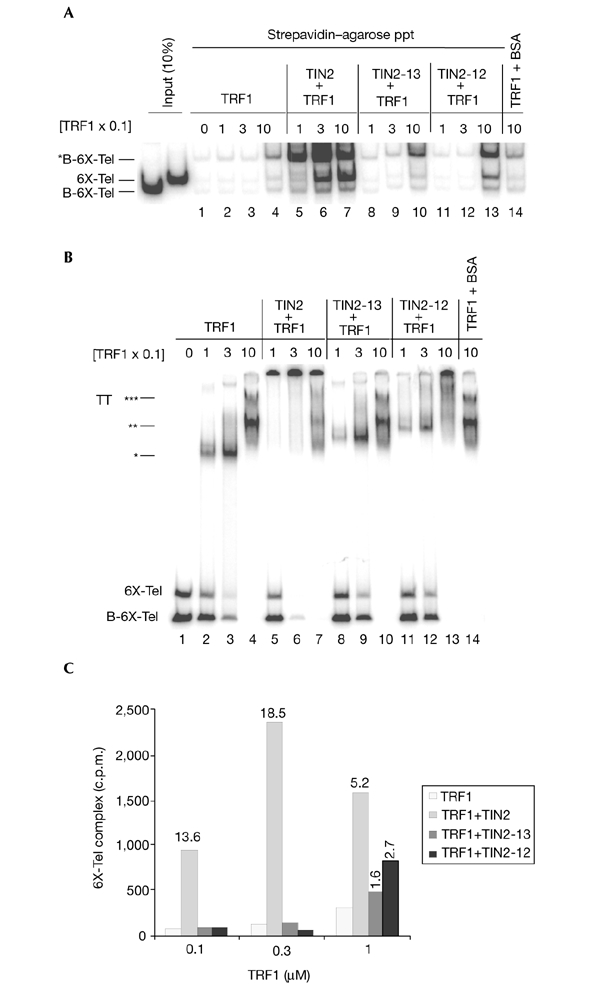
Effect of TRF1 concentration and TIN2 mutant proteins. (A) Reaction mixtures (100 µl) containing 6X-Tel and B-6X-Tel with different concentrations of TRF1 (0.1, 0.3 and 1 µM) were assembled. 1 µM each of TIN2 (lanes 5–7), TIN2-13 (lanes 8–10), TIN2-12 (lanes 11–13) and BSA (control; lane 14) were then added. The captured complexes were released by phenol. After measuring the amount of radioactivity, the released probes were precipitated and analysed by native polyacrylamide gel electrophoresis. (B) 10 µl of the reaction mixtures were analysed by electrophoretic mobility-shift assays. Asterisks indicate TRF1–probe complexes that correspond to the binding of one, two and three TRF1 dimers, as indicated by the number of asterisks. (C) We quantified the amount of probe released by phenol (c.p.m.) by scintillation counting. The amount of stimulation by TIN2 proteins was calculated by dividing the amount of radioactivity released from reactions containing TRF1 and TIN2 by that released from reactions containing only TRF1 ((TRF1+TIN2)/TRF1). Ppt, precipitate; TT, TRF1–probe complex.
Amino-terminal TIN2 truncation mutants (TIN2-12 and TIN2-13) were defective in stimulating probe interactions in both the clustering assays and in EMSAs (Fig. 4A,B, lanes 8–13). Moreover, TIN2-13 was more defective than TIN2-12, consistent with their respective strong and weak dominant-negative activities in vivo (Kim et al., 1999). Both mutants were almost inactive when a single TRF1 dimer was bound (Fig. 4A,B, lanes 8 and 11), despite being able to bind a single TRF1 dimer, as shown by their ability to supershift the TRF1–probe complex (Fig. 4B, lanes 8 and 11). Surprisingly, however, both mutants weakly stimulated probe clustering when two or three TRF1 dimers were bound to the probe (Fig. 4A,B, lanes 10 and 13; Fig. 4C).
One possible explanation for these results is shown in the supplementary information online. TRF1 can induce DNA bending when several dimers bind (Bianchi et al., 1997). TIN2 may stimulate the intra-probe (DNA bending) interactions that occur at higher TRF1 concentrations, which may in turn compete with inter-probe interactions (clustering) stimulated by TIN2. That is, bending may reduce clustering, and the ability of TIN2 to stimulate probe clustering at high TRF1 concentrations may consequently decline. Dominant-negative TIN2 mutants may reduce the ability of TIN2 to stimulate intra-probe (bending) interactions, thereby increasing its ability to stimulate inter-probe (clustering) interactions.
Indeed, the ability of TIN2 to stimulate probe clustering depended on the number of TRF1 dimers bound to the probe. To show this, we used probes containing 6 (6X-Tel) or 13 (13X-Tel) telomeric repeats, which bind a maximum of 3 or 6 TRF1 dimers (Bianchi et al., 1997, 1999). EMSAs showed that increasingly larger TRF1 telomeric DNA complexes (TT) formed as both TRF1 concentration and probe length increased (Fig. 5A). We assume that the fraction of probe bound by TRF1 in each reaction is similar, but could not estimate this directly because the stability of TRF1 complexes during EMSAs changed (increased) as the number of bound TRF1 dimers increased (Fig. 5A, compare lanes 2 and 6, 3 and 7, and 4 and 8). We mixed labelled 6X-Tel or 13X-Tel probes with unlabelled B-6X-Tel or B-13X-Tel probes, varying the TRF1 concentration but keeping the TIN2 concentration constant. Complexes were recovered on streptavidin beads and dissociated by phenol, and probes present in the complexes were quantified (Fig. 5B). As expected, increasing TRF1 promoted increasing probe clustering, and TIN2 strongly stimulated this activity. However, the stimulation by TIN2 was diminished by increasing the probe length and, hence, TRF1 loading (Fig. 5B,C). The amount of unbound TRF1 is expected to be higher in reactions with 6X-Tel compared with 13X-Tel (Fig. 5A), suggesting that unbound TRF1 does not compete with bound TRF1 for TRF2 in the 13X-Tel reactions. These results are consistent with TIN2 stimulating the bending of probes with two or more TRF1 dimers, which reduces inter-probe clustering (see supplementary information online). Probes with longer telomeric tracts have more intra-probe (bending) interactions, and hence show less stimulation of inter-probe clustering by TIN2. From a biological perspective, our results suggest that TIN2 may increase the complexity of the telomeric structure.
Figure 5.
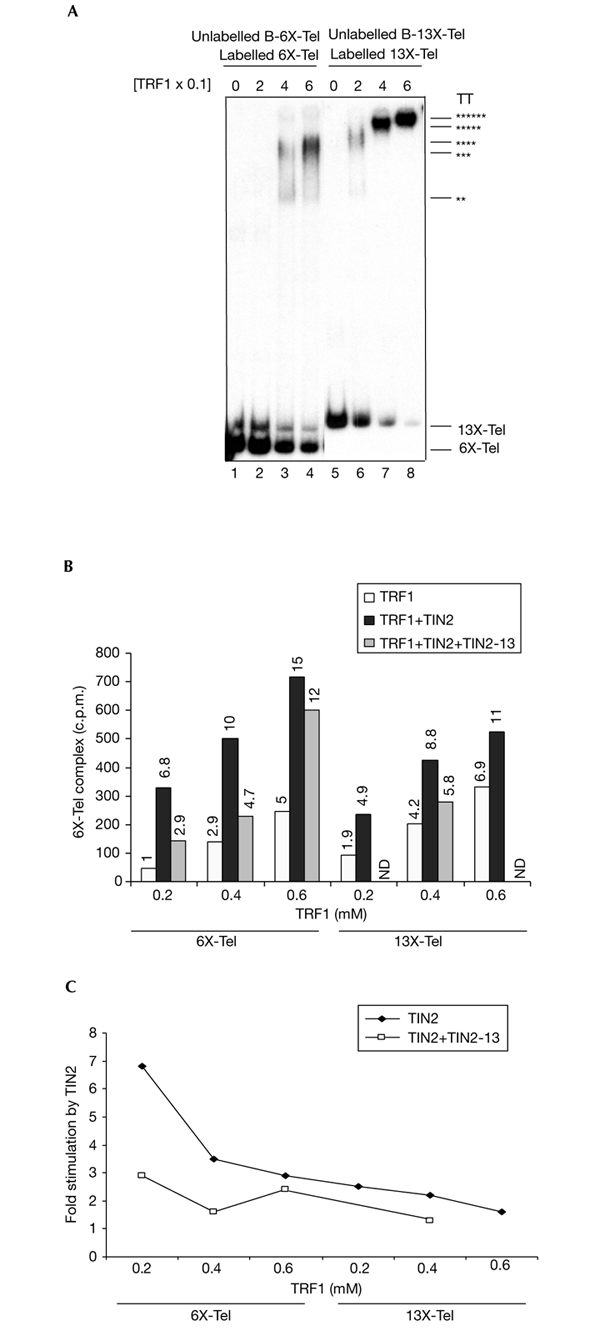
Effect of probe length. (A) Labelled 6X-Tel and unlabelled B-6X-Tel (lanes 1–4) or labelled 13X-Tel and unlabelled B-13X-Tel (lane 5–6) probes were incubated with 0, 0.2, 0.4 or 0.6 µM TRF1 and analysed by electrophoretic mobility-shift asssays to detect complexes with increasing numbers (indicated by the number of asterisks) of TRF1 dimers. (B) Labelled 6X-Tel and unlabelled B-6X-Tel or labelled 13X-Tel and unlabelled B-13X-Tel probes were incubated with 0, 0.2, 0.4 or 0.6 µM TRF1, followed by incubation with buffer (control), 0.25 µM TIN2 or 0.25 µM each of TIN2 or TIN2-13. The amount of probe released by phenol (measured as c.p.m.) was quantified by scintillation counting, and the relative change compared with 0.2 mM TRF1 is shown. (C) Stimulation induced by TIN2 or TIN2 and TIN2-13 was calculated by quantifying the radioactivity released in reactions containing both TIN2 and TRF1 and dividing this by the radioactivity released in reactions containing only TRF1 ((TRF1+TIN2) radioactivity/TRF1 radioactivity). ND, no data.
As shown in Fig. 4, TIN2-13 was unable to stimulate the clustering of probes on which one or two TRF1 dimers were bound, but did stimulate (albeit not as well as wild-type TIN2) clustering of probes with three TRF1 dimers. This result suggests that TIN2-13 was more effective at reducing intra-probe interactions (bending) than inter-probe interactions. To test this idea, we added equimolar amounts of TIN2 and TIN2-13 to 6X-Tel and 13X-Tel probes (Fig. 5B,C). TIN2-13 reduced the stimulation of probe clustering by TIN2 when 6X-Tel probes with one or two TRF1 dimers were used. However, TIN2-13 was less effective at reducing TIN2stimulated clustering when 6X-Tel or 13X-Tel probes with more than two TRF1 dimers were used. These results suggest that the ability of TIN2-13 to abolish probe clustering and its ability to abolish DNA bending can be balanced, depending on the complexity of the telomeric structure. One possibility is that TIN2 and TRF1 negatively control telomere length by increasing intra-telomeric bending, and that TIN2-13 increases telomere length by inhibiting this bending.
The hinge regions between the DNA-binding and homo-dimerization domains of TRF1 seem to be flexible (Bianchi et al., 1999). Moreover, TRF1 binds (TTAGGG)2 halfsites in direct and inverted orientations equally well, and TRF1 binding tolerates relatively large variations in half-site distances, possibly by inducing DNA loops in the intervening segments (Bianchi et al., 1999). In addition, TRF1 has a relatively high DNA-binding off-rate, which may facilitate dynamic rearrangements within telomere tracks (Bianchi et al., 1999). One possibility is that TIN2 facilitates TRF1-dependent DNA looping and bending on long telomeric tracks by changing the TRF1 off-rate, thereby increasing the complexity of the telomeric structure. EM showed that TRF1 stimulates parallel pairing between long telomeric DNA tracts (Griffith et al., 1998). It remains to be determined whether or how TIN2 induces pairing or other higher-order interactions between telomeric DNA sequences. Our results suggest that TIN2 stimulates the ability of TRF1 to induce pairing or higher-order interactions between telomeric DNA tracts by causing a conformational change in TRF1.
Methods
Telomeric probes.
We used pBluescript-SK containing 6 (6X-Tel) or 13 (13X-Tel) telomeric repeats (TTAGGG), flanked by non-telomeric sequences, to synthesize and label probes by PCR. A primer containing biotin at two branched sites at the 5′ end (BioSynthesis) was used to synthesize biotinylated probes. 32P-labelled PCR products were separated on 1.5% agarose gels, excised, extracted using a commercial kit (Qiagen) and resuspended in TE buffer at ∼6 ng µl−1 DNA or ∼500 c.p.m. µl−1.
Probe-clustering and electrophoretic mobility-shift assays.
Reaction mixtures (100 µl) containing 6X-Tel and B-6X-Tel probes, buffer and the indicated proteins (Zhong et al., 1992) were incubated for 30 min at 23 °C. 10 µl was used for EMSAs, as described in Kim et al. (1999). We diluted 90 µl of the reactions with 250 µl of binding buffer (Zhong et al., 1992) and incubated this with 50 µl of a 50% slurry of streptavidin–agarose (BRL) for 1 h at 4 °C. We washed the streptavidin–agarose complexes four times with 500 µl of binding buffer. The pellets were mixed with 200 µl of extraction solution (0.1 × SSC, 0.1% SDS) and extracted with phenol/chloroform/isoamyl alcohol. Where indicated, radioactivity in the supernatants was quantified by direct counting. Probes in the supernatants were precipitated with ethanol, solubilized, and separated on 10% polyacrylamide gels, which run in 1 × TBE. The dried gels were analysed by autoradiography or using a phosphorimager.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor872-s1.pdf).
Supplementary Material
Supplementary information
Acknowledgments
This work was supported by grants from the California Breast Cancer Research Program, the National Institutes of Health (grants AG09909 to J.C. and AG18949 to D.J.C.) and by the Ellison Medical Foundation under contract DE-AC03-76SF00098 from the US Department of Energy to the University of California.
References
- Bianchi A., Smith S., Chong L., Elias P. & de Lange T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J., 16, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D. & de Lange T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J., 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T. & Gilson E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L. & de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Campisi J., Kim S.H., Lim C.S. & Rubio M. (2001) Cellular senescence, cancer and aging: the telomere connection. Exp. Gerontol., 36, 1619–1637. [DOI] [PubMed] [Google Scholar]
- Cary R.B., Chen F., Shen Z. & Chen D.J. (1998) A central region of Ku80 mediates interaction with Ku70 in vivo. Nucleic Acids Res., 26, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W. & Blackburn E.H. (2002) New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene, 21, 553–563. [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P. & de Lange T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Fairall L., Chapman L., Moss H., de Lange T. & Rhodes D. (2001) Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell, 8, 351–361. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bianchi A. & de Lange T. (1998) TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol., 278, 79–88. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kaminker P. & Campisi J. (1999) TIN2, a new regulator of telomere length in human cells. Nature Genet., 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P., Fairall L. & Rhodes D. (1998) Sequencespecific DNA recognition by the myb-like domain of the human telomere binding protein TRF1: a model for the protein–DNA complex. Nucleic Acids Res., 26, 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. (2000) Molecular biology. Telomeres keep on rappin'. Science, 288, 2141–2142. [DOI] [PubMed] [Google Scholar]
- Rubio M.A., Kim S.H. & Campisi J. (2002) Reversible manipulation of telomerase expression and telomere length. Implications for the ionizing radiation response and replicative senescence of human cells. J. Biol. Chem., 277, 28609–28617. [DOI] [PubMed] [Google Scholar]
- Shore D. (1997) Telomerase and telomere-binding proteins: controlling the endgame. Trends Biochem. Sci., 22, 233–235. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Shiue L., Kaplan S. & de Lange T. (1992) A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol., 12, 4834–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


