Abstract
Various functions are ascribed to the HBx regulatory protein of the hepatitis B virus (HBV). Due to the low expression level of HBx, it has been difficult to correlate spatial and temporal HBx expression levels with specific functions. Based on a novel cell-permeable peptide, known as the translocation motif (TLM), cell-permeable HBx fusion proteins were generated. The TLM–HBx fusion protein is rapidly internalized from the medium into almost all cells, whereas no significant internalization was seen with wild-type HBx. The major fraction of internalized HBx protein moves from the cytoplasm to the nucleus. The cytosolic fraction, however, activates c-RAF1/extracellular-signal-related kinase 2 signalling and causes activation of activator protein 1 (AP1) and nuclear factor-κB. The TLM–HBx protein rescues HBV gene expression from an activator-deficient HBV genome. These results indicate that cell-permeable regulatory proteins provide a novel, efficient tool for a clearly defined, dose-dependent analysis of regulatory protein function, without affecting the integrity of the cell, and can be used for the safe reconstitution of virus production from a regulatory-protein-deficient virus genome.
Introduction
In addition to causing acute and chronic hepatitis, the hepatitis B virus (HBV) is considered to be a major aetiological factor in the development of human hepatocellular carcinoma (HCC). Almost all HBV-associated HCCs studied so far have chromosomally integrated HBV DNA (Buendia, 2000). The HBV genome encodes two transcriptional activators: the PreS2 activator LHBs (large hepatitis B virus surface protein; Hildt et al., 1996) and the HBx activator protein (Twu & Schloemer, 1987).
Various functions have been ascribed to HBx. The transcriptional activator function seems to depend on the activation of cytoplasmic signal transduction cascades, such as the RAS/RAF/extracellular-signal-regulated kinase (ERK) cascade (Klein et al., 1999; Klein & Schneider, 1997; Bouchard et al., 2001a). This activation can be mediated by an HBx-dependent modulation of intracellular calcium levels (Bouchard et al., 2001a). Furthermore, HBx triggers the activation of a variety of transcription factors, such as activator protein 1 (AP1; Kekule-Urchs et al., 1993; Benn et al., 1996), nuclear factor-κB (NF-κB; Lucito & Schneider, 1992) and ATF (activating transcription factor)–CREB (cAMP-responsive-element binding protein; Williams & Andrisani, 1995). In addition, a direct interaction of HBx with components of the transcriptional machinery, such as the TATA-binding protein (TBP), TFIIB, TF2H or the B5 subunit of RNA polymerase (Haviv et al., 1998) has been described. Finally, HBx can modulate pro-apoptotic as well as anti-apoptotic processes (Su & Schneider, 1997; Schuster et al., 2002; Elmore et al., 1997).
The function of HBx in HBV-associated carcinogenesis is of great interest (Andrisani & Barnabas, 1999). One possible mechanism is the induction of cell proliferation (Kekule-Urchs et al., 1993; Bouchard et al., 2001b; Madden et al., 2001). In addition, the protein might have a mutagenic effect by interfering with DNA repair (Elmore et al., 1997; Becker et al., 1998). Despite a well-established function of the X protein WHx in the woodchuck hepatitis virus (WHV) life cycle (Zoulim et al., 1994), there are conflicting reports about the relevance of HBx for the life cycle of HBV (Bouchard et al., 2001a; Reifenberg et al., 2002; Stoeckl et al., 2003).
This controversy might be due to different experimental approaches. In particular, due to low expression levels, detection of recombinant HBx after transient transfection is problematic. Conversely, strong, constitutive overexpression of the protein is unlikely to reflect the physiological situation. To overcome these problems, we established a novel experimental system that is based on cell-permeable HBx fusion proteins to investigate the biology of HBx. This system allows the defined, dose-dependent, direct correlation of the presence of HBx protein in almost all of the cells with the effects observed.
Cell permeability was achieved by the fusion of HBx to the TLM (translocation motif), a novel cell-permeable peptide (Oess & Hildt, 2000). The TLM is a 12 amino-acid, amphipathic, α-helical peptide, which is derived from the surface protein of HBV (Oess & Hildt, 2000). The TLM mediates the energy-independent and receptor-independent transfer of peptides, nucleic acids and proteins when fused to them, without affecting the integrity of the cell or interfering with intracellular signal transduction cascades (Saher & Hildt, 1999; Hildt et al., 2002), and is therefore a suitable tool for the analysis of HBx-dependent functions.
Results
TLM–HBx fusion proteins are cell permeable
TLM–HBx and wild-type HBx proteins (Fig. 1) were produced and purified as described in the Methods section. First, we investigated whether the highly purified and renatured TLM–HBx fusion proteins are cell permeable. Huh7 cells were grown for 20 min in the presence of purified TLM–HBx before fixation. Purified wild-type HBx was used as a control. Immunofluorescence microscopy showed an HBx-specific staining of almost all (>95%) of the cells grown in the presence of TLM–HBx (Fig. 2A–D), whereas in the case of cells incubated with wild-type HBx, no significant staining was seen.
Figure 1.

Structure of the HBx proteins and the eGFP fusion protein used in this study. The HBx proteins have an amino-terminal His6 tag followed by a V5 epitope to improve solubility. In the case of translocation motif (TLM)–HBx, the 12 amino-acid cell-permeable peptide is fused to the carboxyl terminus. eGFP, enhanced green fluorescent protein.
Figure 2.

TLM–HBx is a cell-permeable protein. (A–D) Immunofluorescence microscopy (200-fold magnification) of ethanol/DAPI (4′,6-diamidino-2-phenylindole)-fixed Huh7 cells that were grown in the presence of 0.5 μM TLM–HBx (A,C) or wild-type (WT) HBx (B,D) for 20 min. For detection, an HBx-specific antiserum and a Cy3-conjugated secondary antibody were used. Note that only in cells grown in the presence of TLM-HBx (A) was the HBx protein detected within the cells. DAPI staining was used to visualize nuclei (C,D). (E) Western blot analysis of lysosomal (lysos.), microsomal (micros.) and cytoplasmic (cyt.) fractions of Huh7 cells grown in the presence of 0.5 μM wild-type HBx (lanes 1, 3 and 5) or TLM–HBx (lanes 2, 4 and 6) for 20 min. For detection of HBx in these fractions, an HBx-specific antiserum was used (bottom panel). Detection of cathepsin b (a lysosomal marker), tumour necrosis factor receptor type I (TNFRI; a microsomal marker) and grb2 (a cytosolic marker) was performed to control for the purity of the subcellular fractions and as loading controls. The western blot analysis confirms the cell permeability of TLM–HBx. Aliquots of DMEM containing 0.5 μM TLM–HBx or wild-type HBx were analysed by western blotting using an HBx-specific serum to confirm that similar amounts of proteins were applied (input control). TLM, translocation motif.
To confirm the cell permeability of TLM–HBx by an independent experimental approach, Huh7 cells were subjected to subcellular fractionation. The lysosomal, microsomal and cytosolic fractions were analysed by western blotting using an HBx-specific antiserum. The blot showed that TLM–HBx, in contrast with wild-type HBx, is able to penetrate the plasma membrane and enter the cytoplasm (Fig. 2E). In the lysosomal and microsomal fractions, neither TLM–HBx nor wild-type HBx were detectable. These results indicate that the fusion of the TLM peptide to the HBx protein generates a cell-permeable protein.
TLM–HBx accumulates in the nucleus
The intracellular localization of HBx is still controversial (Bouchard et al., 2001a; Haviv et al., 1998; Sirma et al., 1998). Because the TLM, in contrast with other cell-permeable peptides (Elliot & O'Hare, 1997; Lin et al., 1995; Richard et al., 2003), does not target any particular subcellular compartment by itself (Oess & Hildt, 2000), fusion of HBx with this peptide is suitable for assessing this question.
Confocal immunofluorescence microscopy of Huh7 cells grown for 20, 40 or 60 min in the presence of TLM–HBx or wild-type HBx showed a nuclear accumulation of TLM–HBx. After 60 min, in almost all cells (>95%), the main fraction of TLM–HBx was seen in the nucleus (Fig. 3A). Incubation for a longer time did not result in a further nuclear accumulation of HBx. The nuclear accumulation of TLM–HBx was also demonstrated by z-stacking through cells grown for 60 min in the presence of TLM–HBx. From this, it was calculated that ∼85% of TLM–HBx is localized in the nucleus (Fig. 3B). This result was corroborated by an independent experimental approach. Preparation of nuclear and cytosolic fractions of cells grown for 30 or 60 min in the presence of TLM–HBx or wild-type HBx and subsequent western blot analysis using an HBx-specific antiserum (Fig. 3C) confirmed a nuclear accumulation of the main fraction of HBx.
Figure 3.

Nuclear accumulation of TLM–HBx. (A) Confocal laser-scanning immunofluorescence microscopy (1,000-fold magnification) of ethanol/DAPI (4′,6-diamidino-2-phenylindole)-fixed Huh7 cells grown for 20, 40 or 60 min in the presence of 0. 5 μM TLM–HBx or of wild-type (WT) HBx. The immunofluorescent staining was performed using an HBx-specific antiserum and a Cy3-conjugated secondary antibody. Note that a large fraction of TLM–HBx accumulates in the nucleus in a time-dependent manner. Transmission microscopy was used to visualize the cells. (B) Quantification of nuclear TLM–HBx in cells grown for 60 min in the presence of TLM–HBx by confocal scanning microscopy. The z-scans show that ∼85% of the TLM–HBx is localized in the nucleus. (C) Western blot analysis of the nuclear and cytosolic fractions of Huh7 cells grown in the presence of 0.5 μM TLM–HBx or WT HBx for 30 and 60 min using an HBx-specific antiserum. Histone H1 was used as marker for the nuclear fraction, and actin was used as a marker for the cytosolic fraction and as a loading control. Western blot analysis confirmed a time-dependent accumulation of TLM–HBx in the nucleus and a decrease in cytoplasmic TLM–HBx. The bottom right panel shows the input control. These two lanes were loaded with an aliquot of DMEM containing 0.5 μM TLM–HBx or WT HBx to verify that similar amounts of TLM–HBx and WT HBx were used. TLM, translocation motif.
TLM–HBx is a functional regulatory protein
The TLM does not interfere with intracellular signal transduction cascades. In the following set of experiments, we aimed to analyse the functionality of TLM–HBx. First, activation of c-RAF1 and ERK2 in cells treated with TLM–HBx was studied. Immunocomplex assays show that TLM–HBx is able to induce an ∼2.5-fold activation of c-RAF1 and ERK2 as compared with controls (buffer, wild-type HBx and TLM–eGFP (enhanced green fluorescent protein; Fig. 4A,B). Consistent with previous reports (Saher & Hildt, 1999; Oess & Hildt, 2000) that the TLM per se does not interfere with intracellular signal transduction cascades, TLM–eGFP does not activate c-RAF1/ERK2.
Figure 4.

Activation of the c-RAF1/ERK2 signal transduction cascade by TLM–HBx. (A) c-RAF1 activity was analysed by immunocomplex assays using MAPK/ERK kinase (MEK) [γ32P]ATP as substrates. The autoradiographs (upper panel; results of one representative experiment from three experiments are shown) show that, in case of the cells grown in the presence of 0.5 μM TLM–HBx (lane 4), a significant activation of c-RAF1 was observed as compared with the controls (buffer, lane 1; 0.5 μM TLM–enhanced-green-fluorescent protein (eGFP), lane 2; and 0.5 μM wild-type (WT) HBx, lane 3). To ensure that similar amounts of c-RAF1 were precipitated, an aliquot of the precipitate was analysed by western blotting (WB) using a c-RAF1-specific antiserum (lower panel). (B) ERK2 activity was determined by an immunocomplex assay using myelin basic protein (MBP) and [γ32P]ATP as substrates. The autoradiograph (upper panel; results of one representative experiment from three are shown) shows that in case of the cells grown in the presence of TLM–HBx (lane 4), a significant activation of ERK2 was observed as compared to the WT HBx control (lane 3). To ensure that similar amounts of ERK2 were precipitated, an aliquot of the precipitate was analysed by western blotting using an ERK2-specific antiserum (lower panel). (C) Western blot analysis of total cellular lysate derived from cells grown for 5 h in the presence of 0.5 μM TLM–eGFP (lane 1), TLM–HBx (lane 2), WT HBx (lane 3) and buffer (lane 4), using a heatshock protein 72 (HSP72)-specific antiserum. The blot shows that TLM-mediated transfer does not change the amount of HSP72. Actin was used as a loading control. ERK2, extracellular-signal-related kinase 2; TLM, translocation motif. Act., relative level of activation as compared with control.
To exclude the possibility that the effect observed is due to an unspecific stress, the amount of heatshock protein 72 (HSP72) was determined by western blot analysis. This showed that the presence of TLM–HBx or TLM–eGFP did not affect the level of the stress marker HSP72 as compared with controls (buffer and wild-type HBx), which confirms the specificity of the effects observed (Fig. 4C).
Next, the potential of TLM–HBx to activate the transcription factors AP1 and NF-κB was analysed by band-shift experiments. Nuclear extracts were prepared 30 min after the addition of TLM–HBx or wild-type HBx to the medium. The assays revealed significant activation of both transcription factors by TLM–HBx, as compared with the controls (Fig. 5A,B). These results show that passing across the plasma membrane does not affect the functionality of the HBx protein.
Figure 5.
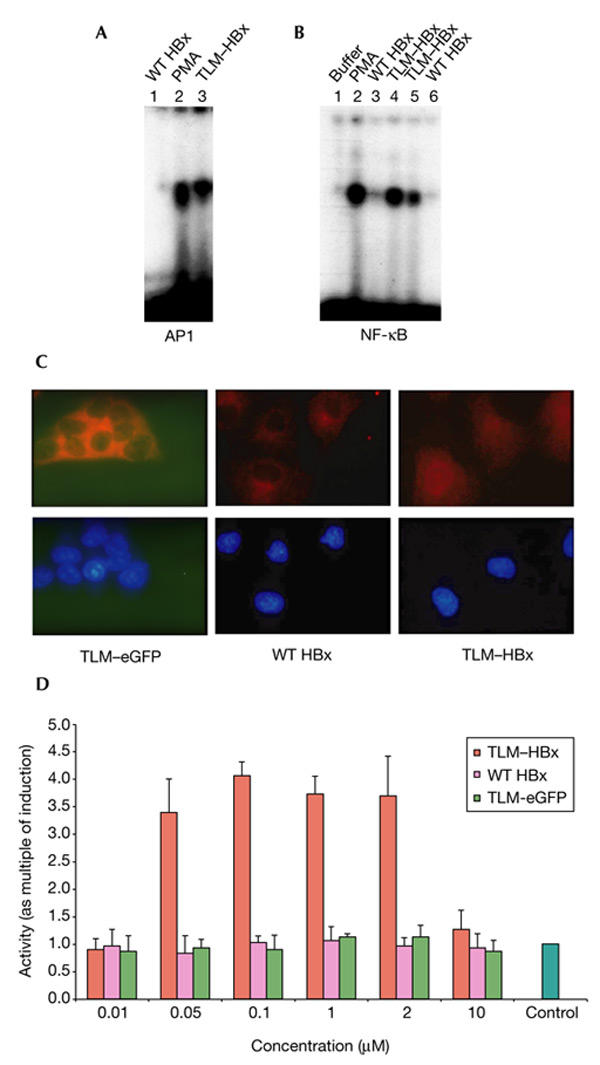
Activation of AP1 and NF-κB by TLM–HBx. (A) Elelctrophoretic mobility-shift assay (EMSA) using an AP1-specific consensus oligonucleotide of nuclear extracts from Huh7 cells grown in the presence of 0.5 μM TLM–HBx (lane 2) or 0.5 μM wild-type (WT) HBx (lane 1) for 30 min. Phorbol myristyl acetate (PMA) stimulation was used as a positive control (lane 3). For cells grown in the presence of TLM–HBx, but not for those grown in the presence of WT HBx, a significant induction of AP1-specific activity was seen. (B) EMSA using an NF-κB-specific consensus oligonucleotide of nuclear extracts from Huh7 cells grown in the presence of 0.5 μM TLM–HBx (lane 4), 0.5 μM WT HBx (lane 3), 0.05 μM TLM–HBx (lane 5) and 0.05 μM WT HBx (lane 6) for 30 min. PMA stimulation was used as a positive control (lane 2). Addition of buffer to the medium was used as a negative control (lane 1). For TLM–HBx, but not for WT HBx, a significant dose-dependent induction of the NF-κB-specific activity was seen. (C) Immunofluorescence microscopy (1,000-fold magnification) of ethanol/DAPI (4′,6-diamidino-2-phenylindole)-fixed Huh7 cells that were grown in the presence of 0. 5 μM WT HBx, TLM–eGFP (enhanced green fluoresent protein) or TLM–HBx for 40 min. Immuno-fluorescence was performed using an NF-κB-speciifc antiserum and a Cy3-conjugated secondary antibody. Note that TLM–HBx, in contrast with WT HBx, induces a nuclear accumulation of NF-κB, indicating its activation. (D) Reporter gene assay in Huh7 cells transfected with the reporter plasmid p2NF-κB–luc. Cells were grown in the presence of 0.01–10.00 μM TLM–HBx, TLM–eGFP or WT HBx for 6 h. Luciferase activity was determined using a commercial assay system. Activities, shown as multiples of induction, are mean values from three independent experiments. In the case of TLM–HBx, the assay revealed a dose-dependent induction of the reporter gene, whereas WT HBx or TLM–eGFP did not induce a significant induction of the reporter gene. AP1, activator protein 1; NF-κB, nuclear factor-κB; TLM, translocation motif.
Activation of NF-κB was also analysed on a single-cell level by immunofluorescence microscopy. In the case of cells that were grown for 40 min in the presence of TLM–HBx, translocation of NF-κB from the cytoplasm to the nucleus, which reflects activation, was seen in >95% of the cells analysed. In case of the controls (wild-type HBx or TLM–eGFP), no significant amount of NF-κB was detected in the nucleus (Fig. 5C).
To address the question of whether TLM–HBx is also able to induce long-term effects, cells were transfected with an NF-κB-dependent reporter construct and grown in the presence of various amounts of TLM–HBx, wild-type HBx or TLM–eGFP (as a control). Reporter gene assays showed a dose-dependent induction of the luciferase gene by the addition of TLM–HBx to the medium, but not by HBx. Interestingly, the presence of TLM–HBx in the medium at concentrations above 10 μM resulted in a loss of activator function. Under these conditions, no significant change in cell integrity was seen (Fig. 5D). These results show that passing across the plasma membrane does not affect the functionality of the HBx protein, and that TLM–HBx fusion proteins are functional activator proteins.
TLM–HBx rescues HBx/PreS2 deficiency
In a recent study, we found that functional knockout of both the PreS2 and HBx regulatory functions completely abolishes the production of viral particles (Stoeckl et al., 2003). To analyse the relevance of HBx-dependent transcriptional activator function to HBV gene expression, we investigated whether HBx/PreS2 deficiency can be rescued by the cell-permeable TLM–HBx fusion protein. To do this, Huh7 cells were transfected with a deficient HBV genome (pSPT1.2HBVdefXdef/PreS2; Stoeckl et al., 2003), which does not encode functional PreS2 and HBx regulatory proteins, and were grown in the presence or absence of TLM–HBx. Wild-type HBx and TLM–eGFP were used as negative controls. Expression of the viral genome was analysed by quantification of virus-specific DNA in the supernatant by TaqMan PCR.
After transfection of the activator-deficient HBV genome, no significant amount of viral DNA was detected in the supernatant. Subsequent addition of TLM–HBx, however, strongly enhanced the production of virus-specific DNA, producing levels up to 50% of those seen after transfection of the wild-type genome. Similar results were obtained from the application of cell-permeable, purified PreS2 protein, or by the simultaneous application of TLM–HBx and PreS2. Wild-type HBx and TLM–eGFP had no effect (Fig. 6).
Figure 6.

Rescue of hepatitis B virus gene expression from a PreS/HBx regulatory-protein-deficient hepatitis B virus genome by cell-permeable TLM–HBx. Huh7 cells were transfected with the plasmids pSPT1.2HBVadr (pHBV-WT, which carries a wild-type (WT) hepatitis B virus (HBV) genome) or pSPT1.2HBVdefXdef/PreS2 (pHBVdefX/PreS2, which lacks the HBx and PreS2 regulatory function). Virus production was quantified by TaqMan PCR. The number of viral particles derived from pSPT1.2HBVadr-transfected cells (∼1 × 106 genomes ml−1) was arbitrarily set at 100. TaqMan PCR showed that in the case of cells transfected with pSPT1.2HBVdefX/defPreS2, a complete loss of virus production is seen. However, in the case of pSPT1.2HBVdefX/defPreS2-transfected cells grown in the presence of 0.5 μM TLM–HBx for 8 h, a partial reconstitution of virus production was seen, whereas the presence of WT HBx or TLM–eGFP (enhanced green fluorescent protein; as an unrelated cell-permeable control) failed to have any effect on virus production. The presence of 0.5 μM purified PreS2 or 0.5 μM PreS2 and 0.5 μM TLM–HBx results in a partial reconstitution of virus production. TLM, translocation motif.
These results show that regulatory protein function is a prerequisite for HBV gene expression, that regulatory protein deficiency can be overcome by TLM–HBx, and that TLM–HBx can be used as a novel, efficient tool to analyse functions of the HBx protein in the viral life cycle.
Discussion
In this study, we show that by fusion of HBx to the TLM peptide, a functional, cell-permeable protein can be made. This cell-permeable HBx protein is a novel tool for the analysis of HBx, as it allows clear correlations to be made between the observed effects and the presence of the regulatory protein. TLM fusion proteins are characterized by the fact that almost all of the cells exposed to the cell-permeable protein internalize the fusion protein. The translocation of TLM fusion proteins into the cell neither affects the integrity of the cell nor interferes with intracellular signal transduction cascades—an important prerequisite for the analysis of HBx-specific regulatory functions (Oess & Hildt, 2000). In contrast to other cell-permeable peptides that target their cargo proteins to defined subcellular compartments (such as the nucleus in the case of VP22 (Elliott & O'Hare, 1997) and fibroblast growth factor 1/2 (Lin et al., 1995), or the endosomal compartment in the case of Tat-derived peptides (Richard et al., 2003), the TLM shows no preference for any subcellular compartment (Oess & Hildt, 2000; Saher & Hildt, 1999; Hildt et al., 2002). Due to the small size of the TLM (12 amino acids), the authentic localization of the cargo protein is not affected by fusion to the TLM tag.
In the case of the cell-permeable TLM–HBx protein, a time-dependent nuclear accumulation of the fusion protein is seen. However, a smaller but significant fraction can still be detected in the cytoplasm (∼15%). In the case of transfection experiments, it is possible that the transfer of plasmid DNA contributes to an increased expression of proteins that are involved in controlling DNA integrity, such as damaged-DNA binding protein 2 or p53 (Elmore et al., 1997), which results in a nuclear shift of their binding partner, HBx. However, even the small cytosolic fraction of TLM–HBx is able to trigger rapid activation of c-RAF1 and an induction of NF-κB, which results in its translocation from the cytoplasm to the nucleus. In parallel, the degradation of IκB (inhibitor of NF-κB) is detected (data not shown), showing that TLM–HBx-dependent activation of NF-κB follows the classic mechanism. In addition, it is possible that the nuclear fraction of HBx might be involved in general transcriptional coactivation by interference with components of the transcriptional machinery (Haviv et al., 1998).
We showed that, in contrast with the HIV activator protein tat, wild-type HBx is not cell permeable. It is therefore unlikely that the protein spreads from an infected cell to surrounding cells. This argues against a physiological role of HBx based on the spreading of HBx from an HBV-infected cell to the surrounding cells, thus preparing these cells for infection by HBV. Thus, the activator function seems to have another effect on HBV replication. Here, we show that the lack of HBV gene expression from an HBV double-mutant (defective with respect to the PreS2 and HBx regulatory functions) can be rescued by cell-permeable HBx. Virus production in HBV transgenic mice lacking the HBx protein is not affected by the HBx deficiency (Reifenberg et al., 2002). Consistent with this, transfection experiments using HBV genomes lacking the coding sequence for the PreS2 or HBx regulatory proteins showed that the loss of one regulatory protein (HBx or PreS2 activator) does not significantly affect HBV gene expression, indicating that the regulatory proteins can replace each other with respect to HBV genome expression (Stoeckl et al., 2003). Therefore, the observation that TLM–HBx and cell-permeable PreS2 induce only a partial reconstitution of viral gene expression from an activator-deficient HBV genome does not indicate that efficient HBV genome expression requires the simultaneous presence of both activator proteins. Simultaneous application of TLM–HBx and PreS2 protein resulted in a similar partial reconstitution of virus production from a defective genome (∼50% as compared with the wild-type genome). This probably reflects the time-limited effect induced by cell-permeable proteins that are subjected to proteolysis and are not regenerated. In the light of this, however, these experiments emphasize the relevance of the viral regulatory proteins for HBV gene expression.
The application of cell-permeable regulatory proteins, in contrast with transfection experiments, allows an easily adjustable amount of the activator protein in almost all of the cells exposed to it, without affecting the integrity of the cell. Therefore, cell-permeable regulatory proteins could be used as tools for basic research. Moreover, cell-permeable fusion proteins could be relevant for biotechnology. As the effect of cell-permeable regulatory proteins is regulated solely by the half-life of the respective protein, these fusion proteins, as shown paradigmatically in case of HBV, could be useful tools for the transient reconstitution of regulator-deficient viral genomes that are the foundation for many gene therapy approaches.
Methods
Construction of expression plasmids.
The sequence encoding HBx was amplified from plasmid pTKTHBV2 (which contains a tandem genome (subtype ayw); Will et al., 1985) by PCR, using a forward primer that introduced the coding sequence for the simian-virus-5-derived V5 epitope (Southern et al., 1991). In the case of TLM–HBx, the coding sequence for TLM was introduced by the reverse primer. The BglII-digested PCR product was inserted into the eubacterial expression vector pQe9, which encodes an amino-terminal His6 tag. The vectors encoding the PreS2 activator protein and TLM–eGFP were described recently (Oess & Hildt, 2000).
As a control, the sequence encoding the His6–V5–HBx and His6–V5–HBx–TLM fusion proteins were subcloned into the eukaryotic expression vector pCDNA3. These constructs were cotransfected with AP1-driven and NF-κB-driven reporter constructs to demonstrate that, under these conditions, both proteins have a similar function. The reporter gene assays showed that the insertion of the V5 or TLM peptides does not affect regulatory protein function (data not shown).
Protein purification.
Protein purification was performed under denaturing conditions, as described in Oess & Hildt (2000). Proteins were renaturated by stepwise dialysis against 30 mM Tris, pH 7.0, 5% glycerol, 25 mM NaCl, 1 mM dithiothreitol.
Protein analysis.
SDS–polyacrylamide gel electrophoresis was performed according to the method of Laemmli (1970). Before loading, samples were adjusted to identical protein concentrations. Gels were loaded with 20 μg of total protein per lane. For western blot analysis, an HBx-specific mouse-derived monoclonal antibody (Chemicon) was used. Sera specific for hexokinase, histone H1, cathepsin B, grb2 and TNFR1 (Santa Cruz) were used as controls for equal loading and for purity control of the subcellular fractions. Immunocomplex assays, subcellular fractionation and electrophoretic mobility-shift assays were performed as described in Oess & Hildt (2000).
Indirect immunofluorescence labelling.
Huh7 cells were grown on coverslides and incubated with 0.5 μM purified TLM–HBx or wild-type HBx diluted in DMEM. After two brief washes with PBS, cells were fixed with ice-cold DAPI (4',6-diamidino-2-phenylindole)/ethanol for 10 min. HBx was detected using a mouse-derived monoclonal antibody (Chemicon). Confocal immunofluorescence microscopy was performed using a Zeiss microscope (63×/1.23 objective).
Cell culture and transfection assays.
Huh7 cells were cultured in DMEM supplemented with 10% FCS. For reporter-gene assays, 0.8 × 106 cells were transfected with 1 μg of the reporter plasmid pNF-κB–luc by lipofection using DOTAP (Roche), in accordance with the manufacturer's instructions. Relative luciferase activity was determined using a commercial assay system in accordance with the manufacturer's instructions (Roche). For quantification of virus-specific DNA in the supernatant, DNAseI treatment was performed to eliminate transfected DNA. TaqMan PCR was performed as described in Stoeckl et al. (2003).
Acknowledgments
We are grateful to P.H. Hofschneider for his support, to B. Munz for many helpful discussions and critical reading of the manuscript, and to J. Tesch for excellent technical assistance.
References
- Andrisani O.M. & Barnabas S. ( 1999) The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int. J. Oncol., 15, 373–379. [DOI] [PubMed] [Google Scholar]
- Becker S.A., Lee T.H., Butel J.S. & Slagle B.L. ( 1998) Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol., 72, 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn J., Su F., Doria M. & Schneider R.J. ( 1996) Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol., 70, 4978–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M.J., Wang L.H. & Schneider R.J. ( 2001a) Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science, 294, 2376–2378. [DOI] [PubMed] [Google Scholar]
- Bouchard M.J., Giannakopoulos S., Wang E.H., Tanese N. & Schneider R.J. ( 2001b) Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol., 75, 4247–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia M.A. ( 2000) Genetics of hepatocellular carcinoma. Semin. Cancer Biol., 10, 185–200. [DOI] [PubMed] [Google Scholar]
- Elliott G. & O'Hare P. ( 1997) Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell, 88, 223–233. [DOI] [PubMed] [Google Scholar]
- Elmore L.W., Hancock A.R., Chang S.F., Wang X.W., Chang S., Callahan C.P., Geller D.A., Will H. & Harris C.C. ( 1997) Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl Acad. Sci. USA, 94, 14707–14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv I., Shamay M., Doitsh G. & Shaul Y. ( 1998) Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol. Cell. Biol., 18, 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildt E., Saher G., Bruss V. & Hofschneider P.H. ( 1996) The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology, 225, 235–239. [DOI] [PubMed] [Google Scholar]
- Hildt E., Munz B., Saher G., Reifenberg K. & Hofschneider P.H. ( 2002) The PreS2 activator MHBs(t) of hepatitis B virus activates c-raf-1/Erk2 signaling in transgenic mice. EMBO J., 21, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekule-Urchs A.S., Lauer U., Weiss L., Luber B. & Hofschneider P.H. ( 1993) Hepatitis B virus transactivator HBx uses a tumor promoter signalling pathway. Nature, 361, 742–745. [DOI] [PubMed] [Google Scholar]
- Klein N.P. & Schneider R.J. ( 1997) Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol., 17, 6427–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein N.P., Bouchard M.J., Wang L.H., Kobarg C. & Schneider R.J. ( 1999) Src kinases involved in hepatitis B virus replication. EMBO J., 18, 5019–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lin Y.Z., Yao S.Y., Veach R.A., Torgerson T.R. & Hawiger J. ( 1995) Inhibition of nuclear translocation of transcription factor NF-κB by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem., 270, 14255–14258. [DOI] [PubMed] [Google Scholar]
- Lucito R. & Schneider R.J. ( 1992) Hepatitis B virus X protein activates transcription factor NF-kB without a requirement for protein kinase C. J. Virol., 66, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden C.R., Finegold M.J. & Slagle B.L. ( 2001) Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol., 75, 3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oess S. & Hildt E. ( 2000) Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther., 7, 750–758. [DOI] [PubMed] [Google Scholar]
- Reifenberg K., Nusser P., Lohler J., Spindler G., Kuhn C., von Weizsacker F. & Kock J. ( 2002) Virus replication and virion export in X-deficient hepatitis B virus transgenic mice. J. Gen. Virol., 83, 991–996. [DOI] [PubMed] [Google Scholar]
- Richard J.P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M.J., Chernomordik L.V. & Lebleu B. ( 2003) Cell-penetrating peptides: a re-evaluation of the mechanism of cellular uptake. J. Biol. Chem., 278, 585–590. [DOI] [PubMed] [Google Scholar]
- Saher G. & Hildt E. ( 1999) Activation of c-Raf-1 kinase signal transduction pathway in α7 integrin-deficient mice. J. Biol. Chem., 274, 27651–27657. [DOI] [PubMed] [Google Scholar]
- Schuster R., Hildt E., Chang S.F., Terradillos O., Pollicino T., Lanford R., Gerlich W.H., Will H. & Schaefer S. ( 2002) Conserved transactivating and pro-apoptotic functions of hepadnaviral X protein in ortho- and avihepadnaviruses. Oncogene, 21, 6606–6613. [DOI] [PubMed] [Google Scholar]
- Sirma H., Weil R., Rosmorduc O., Urban S., Israel A., Kremsdorf D. & Brechot C. ( 1998) Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene, 16, 2051–2063. [DOI] [PubMed] [Google Scholar]
- Southern J.A., Young D.F., Heaney F., Baumgartner W.K. & Randall R.E. ( 1991) Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol., 72, 1551–1557. [DOI] [PubMed] [Google Scholar]
- Stoeckl L., Berting A., Malkowski B., Foerste R., Hofschneider P.H. & Hildt E. ( 2003) Integrity of c-Raf-1/MEK signal transduction cascade is essential for hepatitits B virus expression. Oncogene, 22, 2604–2610. [DOI] [PubMed] [Google Scholar]
- Su F. & Schneider R.J. ( 1997) Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc. Natl Acad. Sci. USA, 94, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu J.S. & Schloemer R.H. ( 1987) Transcriptional trans-activating function of hepatitis B virus. J. Virol., 61, 3448–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will H., Cattaneo R., Darai G., Deinhardt F., Schellekens H. & Schaller H. ( 1985) Infectious hepatitis B virus from cloned DNA of known nucleotide sequence. Proc. Natl Acad. Sci. USA, 82, 891–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.S. & Andrisani O.M. ( 1995) The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc. Natl Acad. Sci. USA, 92, 3819–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F., Saputelli J. & Seeger C. ( 1994) Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol., 68, 2026–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]


