Abstract
It has been suggested that DNA methylation/demethylation is involved in regulating V(D)J rearrangement. Although methylated DNA is thought to induce an inaccessible chromatin structure, it is unclear whether DNA methylation can directly control V(D)J recombination independently of chromatin structure. In this study, we tested whether DNA methylation directly affects the reactivity of the RAG1/RAG2 complex. Specific methylation within the heptamer of the recombination signal sequences (RSS) markedly reduced V(D)J cleavage without inhibiting RAG1/RAG2–DNA complex formation. By contrast, methylation at other positions around the RSS did not affect the reactivity of the RAG proteins. The presence of a methyl-CpG binding-domain protein inhibited the binding of the RAG1/RAG2 complex to all the methylated CpG sites that were tested. Our findings suggest that DNA methylation around the RSS may have a previously unexpected function in regulating V(D)J recombination by directly inhibiting V(D)J cleavage, in addition to its general function of inducing an inaccessible chromatin configuration.
Introduction
During lymphocyte development, antigen receptor genes are assembled from V, D and J gene segments to form functional exons of their variable region. This process, termed V(D)J recombination, is initiated by the lymphoid-specific proteins, RAG1 and RAG2 (Schatz et al., 1989; Oettinger et al., 1990). RAG1 and RAG2 form a stable complex to recognize the well-conserved recombination signal sequences (RSSs) that flank each antigen-receptor gene segment. RSSs consist of highly conserved heptamer and nonamer regions that are separated by a less-conserved 12-bp or 23-bp spacer (12-RSS and 23-RSS, respectively). Efficient recombination occurs only between 12-RSSs and 23-RSSs, and this is known as the 12/23 rule, because a double-stranded break (DSB) is catalysed preferentially within the paired complex (PC), in which a 12-RSS and a 23-RSS are synapsed (coupled cleavage). The RAG1/RAG2 complex mediates DSB formation in a two-step process. First, a nick is introduced at the border between the coding sequence and the RSS heptamer on its sense strand; next, the 3′ OH of the nick attacks the opposite (antisense) strand to form a hairpin coding end and a blunt signal end. Purified RAG1 and RAG2 proteins are sufficient to obey the 12/23 rule to form the PC and perform coupled cleavage, supported by the high-mobility group protein HMG1, or HMG2. These RAG-mediated reactions are the first step in the several processes that are involved in V(D)J recombination (Gellert, 2002).
V(D)J recombination is highly ordered and is controlled at several levels, including the levels of tissue- and site-specificity, lymphocyte developmental stage and allelic exclusion (Bassing et al., 2002). To account for the different levels of regulation, an accessibility model has been proposed whereby germline RSS sites, which are inaccessible to the recombination machinery due to chromatin structure or modification, must be actively opened before recombination takes place (Blackwell & Alt, 1989). One of the potential mechanisms of controlling accessibility is DNA methylation. It has been shown that CpG methylation has an important function in regulating transcription and chromatin structure in several genes (Bird & Wolffe, 1999; Wade, 2001). DNA methylation is known to repress gene expression directly by impeding the binding of trans-acting factors, and indirectly by the recruitment of histone deacetylases (HDACs) through methyl CpG (mCpG) binding-domain (MBD) family proteins (Bird & Wolffe, 1999). Current studies suggest a model in which MBD family proteins function as anchors on methylated DNA, recruiting accessory proteins such as HDACs that are able to modulate chromatin structure and the transcriptional activity of genes (Wade, 2001).
Differences in methylation status are also correlated with antigen-receptor gene rearrangement and expression. For example, immunoglobulin and T-cell receptor (TCR) loci are highly methylated before V(D)J recombination and undergo demethylation concomitantly with gene rearrangement (Mostoslavsky & Bergman, 1997). The demethylation at the Igκ locus is monoallelic, suggesting a function in the maintenance of allelic exclusion (Mostoslavsky et al., 1998). At the TCR-β locus, deletion of the Dβ1 promoter enhances methylation at proximal sites and results in reduced recombination (Whitehurst et al., 2000). Although demethylation alone is not sufficient to activate V(D)J rearrangement (Cherry et al., 2000), these studies strongly suggest that DNA methylation is important in the regulation of antigen-receptor genes in vivo.
It has been shown that a CpG site in the heptamer of broken signal ends derived from the 3′ Dβ1 RSS are completely demethylated in mouse, suggesting that methylation of this site is incompatible with V(D)J cleavage (Whitehurst et al., 2000). This heptamer sequence is present in the 12-RSSs of the DQ52, Vκ24C, Dδ2 and Jδ2 gene segments in mouse antigen-receptor genes, and is also commonly found in the DH genes of chicken and rabbit. As potential methylation sites are localized in many RSSs, as well as in their flanking coding sequences (Ramsden et al., 1994), the 3′ Dβ1 RSS may not be a special case. Although DNA methylation may regulate gene expression by altering chromatin structure to an inaccessible configuration, the possibility of a direct effect on the recombination machinery cannot be excluded.
In this study, we have found that DNA methylation inhibits the cleavage activity of the RAG1/RAG2 complex by two different mechanisms, depending on the position of the mCpG around the RSS. A novel mechanism that may directly control the regional accessibility for V(D)J recombination through DNA methylation will be discussed.
Results
To investigate the effect of CpG methylation on the initiation of V(D)J recombination, we tested synthetic oligonucleotide substrates containing mCpGs at various positions in a V(D)J cleavage assay. First, the effect of methylation within the heptamer was analysed using the sequence CACGGTG as a heptamer in place of the consensus CACAGTG (see Table 1 for a list of all oligonucleotide sequences used). This sequence was chosen because it contains a naturally occurring, unique CpG site (Ramsden et al., 1994). The singlesite cleavage assay was performed in the presence of Mn2+, using purified core RAG1 and RAG2 proteins (Cuomo et al., 1996). Little difference in the cleavage efficiency was seen on changing the fourth nucleotide in the consensus heptamer from adenine to guanine (Fig. 1A, lanes 2–5), consistent with previous observations (Ramsden et al., 1996). Interestingly, hemimethylation on the top strand (the coding strand in our standard RSS orientation) markedly decreased the amounts of nick and hairpin products compared with the unmethylated control (Fig. 1A, lanes 6 and 7), indicating that a single methyl group at this position interfered with the reaction. Conversely, hemimethylation on the bottom strand (the noncoding strand) had almost no effect on V(D)J cleavage (Fig. 1A, lanes 8 and 9). Methylation on both strands markedly reduced cleavage efficiency (Fig. 1A, lanes 10 and 11) to the levels seen with top-strand methylation (<10% of the unmethylated control). These results suggest that the mCpG on the top strand is responsible for this direct inhibition.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| 12-RSS standard | |
| HN100 | 5′-CTGCAGGTCAACCTGCACAGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN101 | 3′-GACGTCCAGTTGGACGTGTCACGATGTCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| Methylation at the heptamer1 | 1234567 |
| HN102 | 5′-CTGCAGGTCAACCTGCACGGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN103 | 3′-GACGTCCAGTTGGACGTGCCACGATGTCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| HN104 | 5′-CTGCAGGTCAACCTGCAmCGGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN105 | 3′-GACGTCCAGTTGGACGTGmCCACGATGTCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| Methylation at the spacer2 | 123456789 |
| HN110 | 5′-CTGCAGGTCAACCTGCACAGTGCTACGGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN111 | 3′-GACGTCCAGTTGGACGTGTCACGATGCCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| HN112 | 5′-CTGCAGGTCAACCTGCACAGTGCTAmCGGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN113 | 3′-GACGTCCAGTTGGACGTGTCACGATGmCCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| Methylation at the coding region3 | -6-5-4-3-2-1 |
| HN140 | 5′-CTGCAGGTCCGCCTGCACAGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN141 | 3′-GACGTCCAGGCGGACGTGTCACGATGTCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| HN142 | 5′-CTGCAGGTCmCGCCTGCACAGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ |
| HN143 | 3′-GACGTCCAGGmCGGACGTGTCACGATGTCTGACCTTGTTTTTGGGTCCAGAG-5′ |
| 23-RSS4 | |
| HN124 | 5′-GTGGGGACAGGGGGCCACAGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN125 | 3′-CACCCCTGTCCCCCGGTGTCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
| Methylation at the heptamer5 | 1234567 |
| HN120 | 5′-GTGGGGACAGGGGGCCACGGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN121 | 3′-CACCCCTGTCCCCCGGTGCCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
| HN122 | 5′-GTGGGGACAGGGGGCCAmCGGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN123 | 3′-CACCCCTGTCCCCCGGTGmCCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
| 23-RSS standard6 | |
| HN138 | 5′-GTGGGGACAGGGGTACACAGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN139 | 3′-CACCCCTGTCCCCATGTGTCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
| Methylation at the heptamer7 | 1234567 |
| HN134 | 5′-GTGGGGACAGGGGTACACGGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN135 | 3′-CACCCCTGTCCCCATGTGCCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
| HN136 | 5′-GTGGGGACAGGGGTACAmCGGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCATTCT-3′ |
| HN137 | 3′-CACCCCTGTCCCCATGTGmCCACTAAGTTAAGATACCCTTCGGAAATGTTTTTGGTAAGA-5′ |
Positions of the heptamer and nonamer are underlined.
1Hemimethylation on the top strand, HN104/103; on the bottom strand, HN102/105.
2Hemimethylation on the top strand, HN112/111; on the bottom strand, HN110/113.
3Hemimethylation on the top strand, HN142/141; on the bottom strand, HN140/143.
4Based on the mouse 3′ Dβ1 recombination signal sequence (RSS), with a consensus heptamer.
5Natural 3′ Dβ1 RSS, HN120/121; hemimethylation on the top strand, HN122/121; on the bottom strand, HN120/123.
6Good flank, with a consensus heptamer.
7Hemimethylation on the top strand, HN136/135; on the bottom strand, HN134/137.
Figure 1.
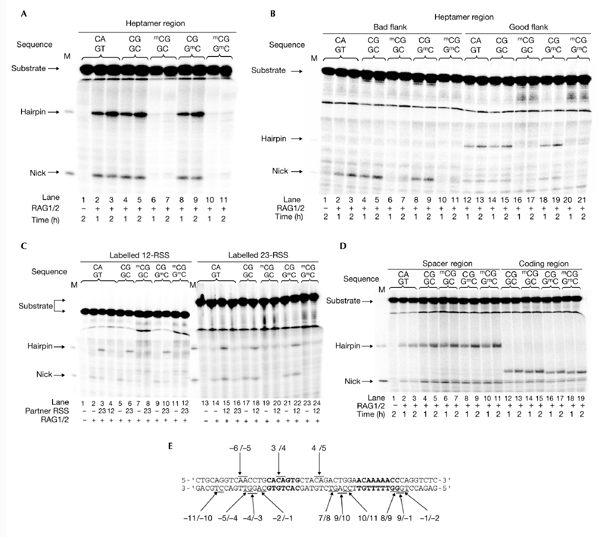
V(D)J cleavage is sensitive to the methylation at the heptamer, but not at the spacer or the coding sequence. Cleavage reactions were carried out in a mixture containing RAG1/RAG2 proteins and 32P-labelled recombination signal sequences 12-RSS or 23-RSS, with or without methyl-CpG (mCpG). The sequences of the target dinucleotides (for example, the fourth and fifth of the heptamer) are indicated at the top of the each lane. Products were separated using a super-denaturing gel. Positions of hairpins and nicks are indicated (M). (A) Effect of methylation within the heptamer of 12-RSS. Lanes 1–3, HN100/101; lanes 4 and 5, HN102/103; lanes 6 and 7, HN104/103; lanes 8 and 9, HN102/105; lanes 10 and 11, HN104/105. (B) Effect of methylation within the heptamer of 23-RSS. Lanes 1–3, HN124/125; lanes 4 and 5, HN120/121; lanes 6 and 7, HN122/121; lanes 8 and 9, HN120/123; lanes 10 and 11, HN122/123; lanes 12 and 13, HN138/139; lanes 14 and 15, HN134/135; lanes 16 and 17, HN136/135; lanes 18 and 19, HN134/137; lanes 20 and 21, HN136/137. (C) Effect of methylation within the heptamer under the conditions of coupled cleavage. Left half, 32P-labelled 12-RSS (HN100/101-HN104/105) with or without unlabelled HN138/139; right half, 32P-labelled 23-RSS (HN138/139-136/137) with or without unlabelled HN100/101. (D) Effect of methylation within the spacer (lanes 1–11) and the coding region (lanes 12–19) in the single-site cleavage. Lanes 1–3, HN100/101; lanes 4 and 5, HN110/111; lanes 6 and 7, HN112/111; lanes 8 and 9, HN110/113; lanes 10 and 11, HN112/113; lanes 12 and 13, HN140/141; lanes 14 and 15, HN142/141; lanes 16 and 17, HN140/143; lanes 18 and 19, HN142/143. (E) Summary of the positions of mCpG tested in this study. Each dinucleotide pair tested for methylation sensitivity is indicated. Heptamer and nonamer sequences are shown in bold. Ten other mutations that were tested, but for which the results are not shown, are indicated under the nucleotide sequences.
We also examined the effect of heptamer methylation in the 23-RSS. Because the mouse 3′ Dβ1 RSS is the only RSS that has been shown to be methylated at the heptamer in vivo, substrates were designed based on this naturally occurring 23-RSS (HN120/121). The hairpin product was barely detected in all the 23-RSS substrates tested, consistent with the finding that certain sequences that flank cleavage sites (GC in this case) do not form hairpins easily (so-called 'bad flanks'; Cuomo et al., 1996; Ramsden et al., 1996; Fig. 1B, lanes 2–11). In fact, hairpin formation was restored when it was changed to a 'good' flanking sequence, TA (Fig. 1B, lanes 12–21). Similar to the results obtained with the 12-RSS, hemimethylation on the top strand limited cleavage (Fig. 1B, lanes 6, 7, 16 and 17), whereas hemimethylation on the bottom strand did not (Fig. 1B, lanes 8, 9, 18 and 19). Again, methylation on both strands decreased the amounts of cleavage product to the levels seen with top-strand methylation (Fig. 1B, lanes 10, 11, 20 and 21). These results show that methylation at the third cytosine of the heptamer severely affected RAG-mediated single-site cleavage.
Next, we tested the possibility that the inhibition of single-site cleavage could be overcome under coupled-cleavage reaction conditions. Thus, we analysed further the effects of heptamer methylation in the presence of Mg2+, under which condition hairpin formation requires a 12/23-RSS pair (Fig. 1C, lanes 1–4 and 13–16), as shown previously (Hiom & Gellert, 1998). We used the 23-RSS with a good flank for these experiments. A base exchange at the fourth nucleotide of the heptamer that introduce a CpG site had no effect on the cleavage reaction (Fig. 1C, lanes 5, 6, 17 and 18). Similar to the results seen for the single-site cleavage, the production of both nicks and hairpins was severely reduced by the presence of mCpG in the top strand (Fig. 1C, lanes 7, 8, 19 and 20). Furthermore, mCpG in the bottom strand did not affect the cleavage efficiency (Fig. 1C, lanes 9, 10, 21 and 22), whereas it was markedly affected when both strands were methylated (Fig. 1C, lanes 11, 12, 23 and 24). These results indicate that heptamer methylation inhibited V(D)J cleavage even under coupled-cleavage reaction conditions, suggesting that methylation at this site might control V(D)J rearrangement under physiological conditions.
We carried out further cleavage assays on substrates containing a methylation site either within the spacer or the coding sequence, in the presence of Mn2+. For the spacer methylation, we introduced a CpG sequence at the 4/5 position of the 12-bp spacer, as this is the most common CpG site in mouse and human 12-RSSs (Ramsden et al., 1994). For the coding-region methylation, a CpG sequence was introduced at the −6/−5 position from the cleavage site, which is also a common CpG site in the RSSs of the mouse Tcr gene. A base exchange of adenine to guanine at the fourth nucleotide of the 12-bp spacer that created a methylation site did not affect cleavage efficiency (Fig. 1D, lanes 2–5). In contrast with the results of heptamer methylation, spacer and coding-region methylation had no apparent effect on cleavage efficiency (Fig. 1D, lanes 6–11 and 14–19, respectively). The hairpin products derived from the substrates that contained a coding-region methylation migrated faster (Fig. 1D, lanes 12–19) due to nucleotide sequence influences on migration (Yu et al., 2002). In fact, when the substrate was labelled at the 5′ end of the bottom strand, the mobility of the DSB product was the same as that of the product formed from the standard substrate, HN100/101, indicating that those hairpins were formed at the expected position (data not shown). We also examined positions 7/8, 9/10 and 10/11 of the spacer and −2/−1, −4/−3, −5/−4 and −11/−10 of the coding region for effects of methylation, but these also had no effect on V(D)J cleavage (data not shown).
Potential methylation sites at position 8/9 of the nonamer ACAAAAAcg, at the border of position 9 of the nonamer and the intron region ACAAAAACCG, and at the first two bases of the intron region (−1/−2) flanking the nonamer, were also tested. For these substrates, no clear difference was seen in the amounts of nick and hairpin products between methylated and unmethylated substrates, indicating that methylation of these sites has no effect on the reactivity of RAG1/RAG2 proteins (data not shown).
In summary, these results show that a unique mCpG in the heptamer region severely inhibits V(D)J cleavage and this inhibition is specifically caused by a methyl group at the third cytosine of the top strand. By contrast, mCpG at the other sites did not directly affect the V(D)J cleavage reaction. All the positions tested for the effect of the presence of an mCpG are summarized in Fig. 1E.
The reduced V(D)J cleavage due to the mCpG in the heptamer region could be caused by poor binding of the RAG proteins to the substrate. Thus, we next analysed the effect of RSS methylation on the binding of RAG proteins. Reactions were carried out at 37 °C for 30 min in the presence of Ca2+, which supports RAG–DNA complex formation but not RAG-mediated cleavage. Incubation of the 12-RSS standard substrate (HN100/101) with RAG1 alone or with both RAG1 and RAG2 generated a single band (Fig. 2A, lane 2), or two additional bands (Fig. 2A, lane 3; single complexes, SC1 and SC2,) , respectively. When unlabelled standard 23-RSS was added with HMG1, a larger complex was seen, which is referred to as the PC (Fig. 2A, lane 4). Under this experimental condition, we tested the effect of RSS methylation on the formation of all the RAG1/RAG2–DNA complexes.
Figure 2.
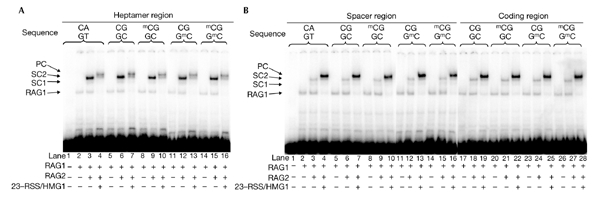
Methyl-CpG has little effect on the binding activities of RAG1 and RAG2. Binding reactions were carried out in buffer containing RAG1/RAG2 and 32P-labelled recombination signal sequence 12-RSS, with or without mCpG. To detect the paired complex (PC), unlabelled 23-RSS (HN138/139) and high-mobility group 1 (HMG1) were added to the reaction mixtures. The complexes were separated on a native gel. Positions of each complex are indicated with arrows. Combinations of the oligonucleotide pairs are as described in the legend for Fig. 1. (A) Effect of methylation within the heptamer region. (B) Effect of methylation within the spacer region (lanes 1–16) and coding region (lanes 17–28). SC, single complex.
The use of a substrate containing an unmethylated CpG sequence at the heptamer (HN102/103) gave similar results as when the standard substrate was used (Fig. 2A, lanes 5–7). When the RSS was methylated at the heptamer region, all the expected complexes were detected, and the intensities of each band were similar to that of the unmethylated substrate (Fig. 2A, lanes 8–16). Similarly, no difference was seen between complex formation on 23-RSS with an mCpG at the heptamer compared with the unmethylated control (data not shown). These results indicate that an mCpG at the heptamer region does not affect the binding of RAG1 and RAG2, although it severely inhibits the cleavage reaction. Furthermore, in the case of methylation at the spacer or coding regions, the binding features of the methylated substrate were the same as the unmethylated controls (Fig. 2B), consistent with no effect being shown in the cleavage assay. In conclusion, RSS methylation has little effect on the DNA binding of RAG1 and RAG2. Importantly, the reduction in V(D)J cleavage of heptamer-methylated substrate is not due to the reduced binding of RAG proteins.
The mCpG is specifically recognized by MBD family proteins that recruit HDAC complexes, which results in the generation of an inaccessible chromatin structure. To study the function of MBD proteins in RAG-mediated cleavage of methylated substrates, the effect of the MBD protein was tested in a binding assay (Fig. 3). The results shown were obtained in the presence of Mg2+; however, similar results were obtained with Ca2+ as the divalent cation (data not shown). As it is not known which MBD family protein is involved in the regulation of the antigen-receptor genes, we used MBD core protein derived from MBD1, which is highly conserved in all the MBD family members (Ohki et al., 2001). MBD core protein contains a mCpG-binding domain only, and specifically recognizes a fully methylated CpG (Ohki et al., 2001). More than 90% of fully methylated substrates bound to MBD (Fig. 3A, lane 2). When the RSS containing a methylated heptamer was tested, the bands representing SC1, SC2 and PC were reduced in the presence of MBD (Fig. 3A, lanes 6–8). The interference to RAG1/RAG2–DNA complex formation was not caused by non-specific binding of MBD, as MBD had no effects on RAG complexes when an unmethylated substrate was used (Fig. 3A, lanes 14–16). The MBD band seen with the unmethylated substrates disappeared in the presence of poly(dI-dC), indicating that it was a result of nonspecific binding (data not shown). These results show that MBD inhibits RAG1/RAG2 complex binding to methylated substrates.
Figure 3.
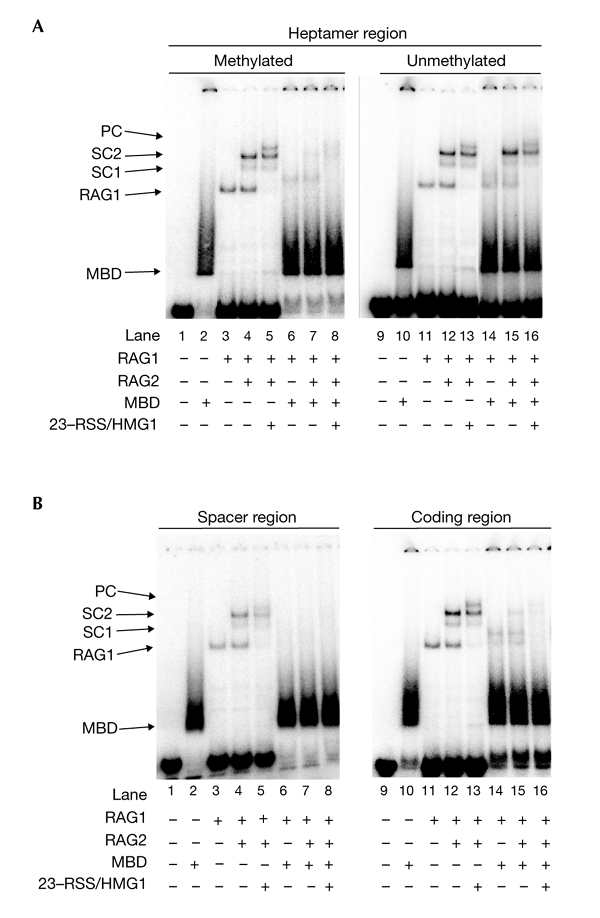
Binding of RAG proteins to the methylated substrate was inhibited by the methyl-CpG binding-domain protein. The binding assay was performed in the presence of the methyl-CpG binding domain (MBD) of MBD1. (A) Effect of MBD on the binding of RAG proteins to methylated or unmethylated substrates at the heptamer. Lanes 1–8, HN104/105; lanes 9–16, HN102/103. (B) Effect of MBD on the binding of RAG proteins to methylated substrates at the spacer or the coding regions. Lanes 1–8, HN112/113; 9–16, HN142/143. HMG1, high-mobility group protein 1; PC, paired complex; SC, single complex; RSS, recombination signal sequence.
Similar effects were seen for the substrate containing an mCpG in the spacer or coding regions (Fig. 3B). The levels of all of the RAG1/RAG2–DNA complexes were severely reduced in the presence of MBD (Fig. 3B, lanes 6–8 and 14–16). We also confirmed that no V(D)J cleavage was seen in the presence of MBD for all the methylated substrates used in this study, including those methylated at ten other positions, shown in Fig. 1E (data not shown). In conclusion, MBD proteins inhibit the initiation of V(D)J recombination by interfering with the DNA binding of the RAG1/RAG2 complex when the mCpG is located in or near the RSS.
Discussion
In this study, we showed that DNA methylation negatively controls V(D)J recombination in vitro, although there may be some cases not tested in this study in which RSS methylation has no effect. There are three possible mechanisms by which a CpG site could regulate V(D)J recombination. The simplest mechanism is by directly preventing RAG proteins from binding to the RSS by methylation. There are several examples in which an mCpG around the recognition sequence inhibits the function of a transcription factor by sterically hindering its binding. These include the binding of the transcription factors E2F, cyclic AMP response element binding protein (CREB), major late transcription factor (MLTF), AP2, c-MYC/MYN, nuclear factor-κB (NF-κB), c-MYB and CCAAT-binding factor (CBF; for a review, see Mostoslavsky & Bergman, 1997). This may therefore be a common mechanism for regulating the functions of biological factors by DNA methylation. Interestingly, none of our methylated substrates inhibited RAG proteins from binding DNA. As the RAG1/RAG2 complex makes contacts throughout the entire RSS (Fugmann et al., 2000), a small modification, such as a single mCpG, may be tolerated.
The second mechanism is the indirect inhibition of RAG proteins from binding to the RSS by recruiting a protein factor. We showed that binding of the MBD core protein to the methylated substrate interfered with the formation of the RAG1/RAG2–DNA complex. This indicates that methylation can indirectly control the reactivity of RAG proteins through binding of an MBD protein, independently of chromatin structure. This may be a major mechanism for regulation by methylation, as the presence of the MBD core protein affected all the methylated substrates tested. Because the size of a full-length MBD family protein is larger than the MBD core protein used in this study, the area around the RSS that is rendered inaccessible to RAG protein binding may span a wider region.
The third mechanism is the direct inhibition of the V(D)J cleavage reaction. We found that an mCpG within the heptamer markedly inhibited the cleavage reaction, which was caused by a direct effect on the catalytic mechanism, without affecting the initial recognition of the site. Furthermore, we found that a single methyl group on the top strand is responsible for this effect. Figure 4 shows a predicted three-dimensional structure of the consensus heptamer, on the basis of the results of Schwabe et al. (1993). In this model, the predicted position of the methyl group of methyl-cytosine on the top strand is facing the nicking surface (Fig. 4, shown in green), which may explain why the catalytic reaction was severely affected. In fact, the third nucleotide of the heptamer has been shown to make direct contact with RAG1, as determined by ultraviolet-light cross-linking experiments (Eastman et al., 1999), which strongly suggests that this cytosine is located near the catalytic centre.
Figure 4.
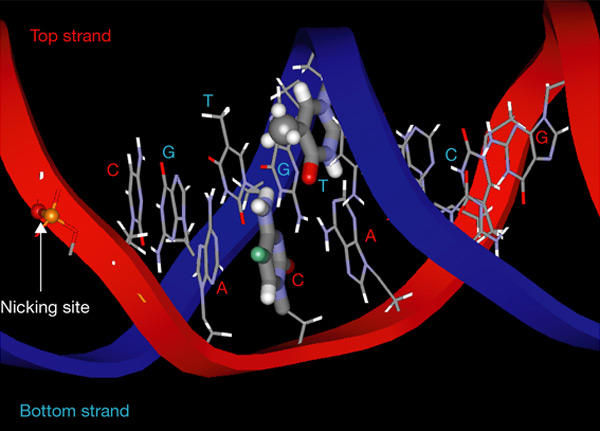
Three-dimensional structure of the consensus heptamer sequence CACAGTG. The bases of the heptamer region are shown by a stick model. The phosphodiester bond where RAG1/RAG2 introduces a nick is indicated. The third cytosine of the top strand and the fourth thymidine of the bottom strand are shown by thick lines. The site of methylation on the third cytosine is shown in green. The methyl group of the thymidine base on the bottom strand is shown as a ball model.
The cleavage reaction is apparently very sensitive to structural changes caused by the methylation of the top strand, but not of the bottom strand. Consensus and methylated heptamer sequences of the bottom strand are 5′-CACTGTG-3′ and 5′-CACmCGTG-3′, respectively. The position of the methyl group in methyl-cytosine occupies a spatial position similar to the thymine base of the consensus sequence. Therefore, methylation on the bottom strand would not be expected to have an effect, and our results support this view.
CpG sequences are common and appear naturally throughout the RSS. However, those in the heptamer are limited to the third cytosine of the 12-RSS. As the fourth nucleotide of the heptamer has been shown to tolerate nucleotide exchanges, such mutations might have been well conserved, as they may provide certain advantages for regulation. For example, a CpG site of mouse 3′ Dβ1 RSS is always demethylated when V(D)J cleavage occurs (Whitehurst et al., 2000), perhaps because demethylation is required for V(D)J cleavage to proceed. The authors of this study also pointed out that the methylated allele is accessible to the V(D)J recombinase, suggesting that demethylation does not always occur before chromatin configuration changes. Thus, the CpG sequences in or near the RSS may in some cases have an additional function that is independent of controlling accessibility though chromatin structure. Although our results, in conjunction with observations from other laboratories, suggest that specific methylation can regulate V(D)J recombination beyond chromatin regulation, the biological relevance of having this type of control remains to be explored in vivo.
Methods
Proteins and oligonucleotide substrates.
The core RAG1 protein (amino acids 352–1,040) was purified from Escherichia coli, as described previously (Kim & Oettinger, 1998). The core RAG2 protein (amino acids 1–384) was purified from HeLa cells infected by recombinant vaccinia virus (McBlane et al., 1995). Bacterially expressed, purified HMG1 (amino acids 1–162) and the core MBD protein (amino acids 1–77) were provided by M.A. Oettinger and M. Shirakawa, respectively. Synthetic oligonucleotides containing 5-methyl-cytidine were purchased from Sigma. The standard HN100/101 oligonucleotides were based on the sequence of VDJ100/101 (Cuomo et al., 1996), with a base exchange to remove a pre-existing CpG sequence.
Oligonucleotide cleavage assays.
Single-site and coupled cleavage reactions were carried out following methods described previously (Cuomo et al., 1996; Hiom & Gellert, 1998). Briefly, for coupled cleavage, combinations of labelled and unlabelled RSSs were first mixed with RAG and HMG1 proteins at 37 °C for 10 min in the presence of 5 mM CaCl2. After the pre-incubation, MgCl2 was added to a final concentration of 5 mM, and the reactions were continued at 37 °C for 30 min. For both single-site and coupled cleavage, products were separated on a 10% polyacrylamide/0.67 × TBE denaturing gel containing 7 M urea, 30% formamide and 12.5 mM MOPS–KOH (pH 7.0). Gels were run at 1,800 V for 2 h and visualized using a Bio-imaging analyser, BAS-2500 (Fuji). The scanned images were quantified using Image Gauge software.
DNA binding assay.
Binding assays were performed as described previously (Mundy et al., 2002). Binding reactions were carried out for 30 min at 37 °C in the presence of Ca2+. Binding assays were also performed in the presence of Mg2+, but no difference was seen in the results obtained (data not shown). To elucidate the effect of MBD, 30 ng was added to the binding reactions. Samples were separated uising 5% polyacrylamide/0.25 × TBE gels containing 6% glycerol. Gels were run at 200 V at 4 °C using 0.25 × TBE buffer containing 6% glycerol and were visualized as described above.
Acknowledgments
We thank M. Shirakawa for providing reagents and M.A. Oettinger for her comments on the manuscript and for providing reagents, and P. Sieh and K.B. Morshead for careful reading of the manuscript. We are grateful to T. Korenaga for his encouragement. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (to Y.A. and Y.T.).
References
- Bassing C.H., Swat W. & Alt F.W. ( 2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109, S15–S55. [DOI] [PubMed] [Google Scholar]
- Bird A.P. & Wolffe A.P. ( 1999) Methylation-induced repression—belts, braces, and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K. & Alt F.W. ( 1989) Mechanism and developmental program of immunoglobulin gene rearrangement in mammals. Ann. Rev. Genet., 23, 605–636. [DOI] [PubMed] [Google Scholar]
- Cherry S.R., Beard C., Jaenisch R. & Baltimore D. ( 2000) V(D)J recombination is not activated by demethylation of the kappa locus. Proc. Natl Acad. Sci. USA, 97, 8467–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C.A., Mundy C.L. & Oettinger M.A. ( 1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., Villey I.J. & Schatz D.G. ( 1999) Detection of RAG protein-V(D)J recombination signal interacts near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol., 19, 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J. & Schatz D.G. ( 2000) The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- Gellert M. ( 2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- Hiom K. & Gellert M. ( 1998) Assembly of a 12/23 paired signal complex: A critical point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- Kim D.R. & Oettinger M.A. ( 1998) Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol., 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane J.F., van Gent D.C., Ramsden D.A., Romeo C., Cuomo C.A., Gellert M. & Oettinger M.A. ( 1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R. & Bergman Y. ( 1997) DNA methylation: regulation of gene expression and role in the immune system. Biochim. Biophys. Acta, 1333, F29–F50. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh N., Kirillov A., Pelanda R., Cedar H., Chess A. & Bergman Y. ( 1998) κ chain monoalleic demethylation and the establishment of allelic exclusion. Genes Dev., 12, 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy C.L., Patenge N., Matthews A.G.W. & Oettinger M.A. ( 2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz D.G. & Baltimore D. ( 1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- Ohki I., Shimotake N., Fujita N., Jee J.-G., Ikegami T., Nakao M. & Shirakawa M. ( 2001) Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell, 105, 487–497. [DOI] [PubMed] [Google Scholar]
- Ramsden D.A., Baetz K. & Wu G.E. ( 1994) Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res., 22, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden D.A., McBlane J.F., van Gent D.C. & Gellert M. ( 1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger M.A. & Baltimore D. ( 1989) The V(D)J recombination activating gene, RAG-1. Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Schwabe J.W., Chapman L., Finch J.T. & Rhodes D. ( 1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell, 75, 567–578. [DOI] [PubMed] [Google Scholar]
- Wade P.A. ( 2001) Methyl CpG-binding proteins and transcriptional repression. Bioessays, 23, 1131–1137. [DOI] [PubMed] [Google Scholar]
- Whitehurst C.E., Schlissel M.S. & Chen J. ( 2000) Deletion of germline promoter PDβ1 from the TCRβ locus causes hypermethylation that impairs Dβ1 recombination by multiple mechanisms. Immunity, 13, 703–714. [DOI] [PubMed] [Google Scholar]
- Yu K., Taghva A. & Lieber M.R. ( 2002) The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex. J. Biol. Chem., 277, 5040–5046. [DOI] [PubMed] [Google Scholar]


