Abstract
Enzymes that use the cofactor pyridoxal phosphate (PLP) constitute a ubiquitous class of biocatalysts. Here, we analyse their variety and genomic distribution as an example of the current opportunities and challenges for the study of protein families. In many free-living prokaryotes, almost 1.5% of all genes code for PLP-dependent enzymes, but in higher eukaryotes the percentage is substantially lower, consistent with these catalysts being involved mainly in basic metabolism. Assigning the function of PLP-dependent enzymes simply on the basis of sequence criteria is not straightforward because, as a consequence of their common mechanistic features, these enzymes have intricate evolutionary relationships. Thus, many genes for PLP-dependent enzymes remain functionally unclassified, and several of them might encode undescribed catalytic activities. In addition, PLP-dependent enzymes often show catalytic promiscuity (that is, a single enzyme catalyses different reactions), implying that an organism can have more PLP-dependent activities than it has genes for PLP-dependent enzymes. This observation presumably applies to many other classes of protein-encoding genes.
Introduction
Pyridoxal phosphate (PLP; a vitamin B6 derivative) arguably represents the most versatile organic cofactor in biology, and is used by a variety of enzymes in all organisms (John, 1995; Jansonius, 1998; Mehta & Christen, 2000; Schneider et al., 2000; Christen & Mehta, 2001). Almost all PLP-dependent enzymes, with the exception of glycogen phosphorylases, are associated with biochemical pathways that involve amino compounds, mainly amino acids. The reactions carried out by the PLP-dependent enzymes that act on amino acids include the transfer of the amino group, decarboxylation, interconversion of L- and D-amino acids, and removal ('elimination') or replacement of chemical groups bound at the β- or γ-carbon. Such versatility arises from the ability of PLP to covalently bind the substrate and then to function as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates (John, 1995; Schneider et al., 2000).
The functional diversity of PLP-dependent enzymes is illustrated by the fact that more than 140 distinct enzymatic activities that are catalogued by the Enzyme Commission (EC; http://www.chem.qmul.ac.uk/iubmb/enzyme/) are PLP dependent, corresponding to ∼4% of all classified activities (Fig. 1). Thanks to the amount of genomic information that has accumulated over the past few years, it is now possible to ask more detailed questions about the importance, distribution and diversity of PLP-dependent enzymes. For example, what is the minimal set of such enzymes that is required by a free-living organism? How different are the number and variety of these enzymes in microorganisms and higher eukaryotes? Can we associate all known PLP-dependent activities with specific gene sequences? Finally, does genomic analysis suggest the existence of novel, as yet unclassified, PLP-dependent enzymes?
Figure 1.
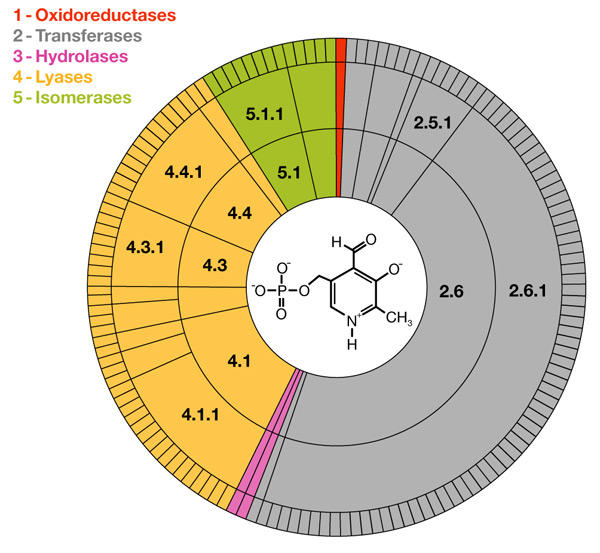
Catalytic versatility of pyridoxal-phosphate-dependent enzymes. The Enzyme Commission (EC) identifies an enzyme activity using a four-digit number, where the first digit indicates the general class of the catalysed reaction (1, oxidoreductases; 2, transferases; 3, hydrolases; 4, lyases; 5, isomerases; 6, synthetases). Of six general classes, five include pyridoxal-phosphate (PLP)-dependent enzymes. The catalytic diversity within enzyme families and superfamilies can be illustrated by a set of concentric pie charts (Thornton et al., 2000). Here, we show the distribution of the 145 PLP-dependent activities that have been classified so far. Sectors are coloured according to EC classes. The circles, from inner to outer, represent the first, second, third and fourth levels in the EC hierarchy. The angle subtended by any segment is proportional to the number of activities it contains, making it clear that aminotransferases (EC 2.6.1) constitute the largest group of PLP-dependent enzymes. A list of the EC-classified PLP-dependent activities is provided in the supplementary information online. The structure of PLP is shown in the centre of the graph.
This review addresses some of these points using a two-step approach: first, we survey a set of representative genomic sequences to obtain an outline of the inter- and intra-genomic distribution of PLP-dependent enzymes; second, we discuss several aspects of the resulting picture in the light of the most recent literature, relating them to more general issues that are relevant to the genomic analysis of protein families.
PLP-dependent enzymes in complete genomes
Despite their functional variety, all structurally characterized PLP-dependent enzymes belong to just five distinct structural groups (Grishin et al., 1995), which presumably correspond to five independent evolutionary lineages (Mehta & Christen, 2000; Christen & Mehta, 2001). The so-called fold-type I is the most common structure, and is found in a variety of aminotransferases and decarboxylases, as well as in enzymes that catalyse α-, β- or γ-eliminations. Fold-type II is found mainly in enzymes that catalyse β-elimination reactions. Fold-type III, which is characterized by a (β/α)8 barrel structure, is found in alanine racemase and in a subset of amino-acid decarboxylases. Fold-type IV enzymes include D-alanine aminotransferase and a few other enzymes. Finally, the fold-type V group includes glycogen and starch phosphorylases. This limited structural diversity facilitates the identification of PLP-dependent enzymes from genomic sequences using search methods that identify the structural conservation of protein families (Mehta & Christen, 2000).
We have used one such method (hidden Markov models) to describe the sequence conservation in families of PLP-dependent enzymes that are assigned to specific catalytic activities (see supplementary information online). Using these reference models, we have scanned a series of complete or near-complete genomes and have obtained a census of PLP-dependent enzymes in different organisms (Fig. 2), from which a few tentative conclusions can be drawn.
Figure 2.
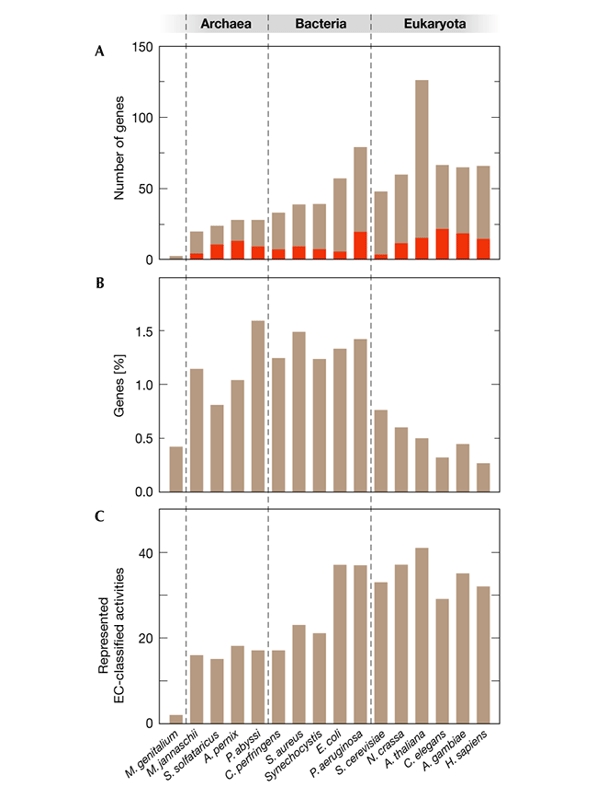
Distribution of genes that encode pyridoxal-phosphate-dependent enzymes in representative genomes. The organisms used as examples are: Mycoplasma genitalium (not a free-living organism), Methanococcus jannaschii, Sulfolobus solfataricus, Aeropyrum pernix, Pyrococcus abyssi, Clostridium perfringens, Staphylococcus aureus, Synechocystis sp. PCC 6803, Escherichia coli, Pseudomonas aeruginosa, Saccharomyces cerevisiae, Neurospora crassa, Arabidopsis thaliana, Caenorhabditis elegans, Anopheles gambiae and Homo sapiens. (A) Total number of pyridoxal-phosphate (PLP)-dependent genes per genome; the fraction of sequences that could not be classified in terms of function, on the basis of our search criteria, is shown in red. The relatively high number of PLP-dependent genes in A. thaliana can be explained in part by the occurrence, for many enzymes, of at least three isoforms (cytoplasmic, mitochondrial and plastidic) that are encoded by different genes. (B) The percentage of genes in each genome that encode PLP-dependent enzymes. (C) The number of Enzyme Commission (EC)-classified PLP-dependent activities for which sequences were found in each genome. Note that similar numbers of activities can be found in organisms with very different assortments of enzymes. For example, only about one-third of the PLP-dependent activities found in E. coli are also found in H. sapiens, despite the fact that the two organisms have a similar absolute number of activities.
The number of genes for PLP-dependent enzymes in extant, free-living microorganisms presumably depends on their adaptation to specific nutrient sources. The smallest number of genes, 20, is found in the archaeon Methanococcus jannaschii, an extremophile that is known to have peculiar nutrient requirements. Between 20 and 30 genes are present in several other Archaea, whereas slightly higher numbers are found in bacteria with small genomes, such as Clostridium perfringens (Fig. 2A). The fact that almost 1.5% of all genes in most prokaryotic genomes encode PLP-dependent enzymes attests to the importance of PLP as a biological cofactor in these species. This fraction decreases with expansion of the size and complexity of the genome (Fig. 2B), perhaps because PLP-dependent enzymes are mostly involved in basic metabolic pathways rather than in more specialized regulatory functions.
Although the absolute number of PLP-dependent enzymes is somewhat larger in higher eukaryotes than in microorganisms (Fig. 2A), the number of EC-classified activities that are represented is not (Fig. 2C). This can be explained in part by the occurrence of organelle-specific or tissue-specific isozymes, which are encoded by different genes but have the same enzymatic activity. However, the relatively modest number of classified activities that are found in higher eukaryotes might also imply that some gene products assigned to the same EC number on the basis of sequence homology actually have different activities (see below). Only two EC-classified activities are present in all of the available genomes of free-living organisms: aspartate aminotransferase (EC 2.6.1.1) and serine hydroxymethyltransferase (EC 2.1.2.1). Serine hydroxymethyltransferase (an enzyme that produces methylene tetrahydrofolate, which is required for the biosynthesis of nucleotides) was also one of just two PLP-dependent genes found in the obligatory intracellular parasite Mycoplasma genitalium.
Limitations to the identification of PLP enzymes
Although the above overview of the genomic distribution of PLP-dependent enzymes is informative, it is also preliminary, and to some degree incomplete. This is partly due to limits that are inherent to homology searches, which may fail to recognize PLP-dependent genes in at least two situations. First, it is possible that structurally similar enzymes escape detection if their sequence similarity has become negligible. A case in point is provided by Eswaramoorthy et al. (2003), who crystallized the yeast hypothetical protein YBL036C. This molecule was not predicted to be a PLP-dependent enzyme and was even suggested to have a novel protein fold. However, the solution of the crystal structure revealed a classic (β/α)8 barrel, with pyridoxal phosphate covalently bound at the bottom. Thus, YBL036C closely resembles PLP-dependent enzymes such as alanine racemase, and it has indeed been shown to have some amino-acid racemase activity (Eswaramoorthy et al., 2003).
A second situation in which homology searches can fail to identify PLP-dependent genes is when the enzymes that they encode do not fall into the five fold-type categories described above and do not resemble any of the known PLP-dependent enzymes. For example, some recently sequenced bacterial aminomutases (such as D-lysine-5,6-aminomutase and D-ornithine-4,5-aminomutase), which use vitamin B12 cofactors in addition to PLP, show no significant similarity to other PLP-dependent enzymes at the sequence level, but instead resemble some B12-containing proteins (Chang & Frey, 2000; Chen et al., 2001). One possibility is that these aminomutases have a fold type that is distinct from those of all other PLP-dependent enzymes. However, it will be necessary to obtain the three-dimensional structure for a representative of this group to clarify this issue.
Limitations to the functional assignment of PLP enzymes
A substantial fraction of the genes that have been identified as encoding PLP-dependent enzymes remain functionally unclassified or only tentatively classified; for example, we could not assign the catalytic activity of about 20% of the putative PLP-dependent enzymes encoded by the human genome (Fig. 2A). This number might even be an underestimate because, although bioinformatic tools are well suited to detecting structural similarities and determining phylogenetic relationships, they are much less reliable for establishing protein function solely based on homology, even when sequence identity is >50% (Thornton et al., 2000; Rost, 2002).
This problem is particularly severe for PLP-dependent enzymes, as their catalytic mechanisms almost invariably involve the formation of carbanionic intermediates that are stabilized by the cofactor. Such constant mechanistic features have presumably favoured the appearance of enzymes with identical or highly similar activities several times during the course of evolution (Mehta & Christen, 2000). In addition, PLP-dependent enzymes are involved in a surprising variety of cellular processes, so that even for an enzyme for which the catalytic activity can be assigned with high confidence on the basis of its sequence, the actual biological function may remain uncertain.
Enzymes that have the same catalytic activity but are evolutionarily unrelated and functionally diverse are exemplified by a group of PLP-dependent enzymes with desulphydrase activity, that is, enzymes that release H2S from thiol-containing amino acids. Bacteria use desulphydrases not only in amino-acid metabolism and in adaptation to new nutrient sources (Soutourina et al., 2001), but also sometimes as virulence factors (Krupka et al., 2000; Fukamachi et al., 2002). One Escherichia coli enzyme with desulphydrase activity is even known to act as a modulator of gene expression, although this function seems to be unrelated to catalysis (Clausen et al., 2000). Sulphide production by PLP-dependent enzymes is also important in vertebrates, in which H2S has been shown to function as a neuromodulator (Kimura, 2002).
PLP-dependent enzymes with desulphydrase activity come from several evolutionary lineages. For example, although most bacterial L-cysteine desulphydrases belong to the fold-type I group, cystathionine β-synthase (the enzyme responsible for H2S production in the mammalian brain) is a fold-type II enzyme. A recently identified L-cysteine desulphydrase from Fusobacterium nucleatum is also a fold-type II enzyme, and its closest sequence homologue is a cysteine synthase (Fukamachi et al., 2002). D-cysteine desulphydrase from E. coli is not closely related to other desulphydrases, and is most similar to 1-aminocyclopropane-1-carboxylate deaminase (Soutourina et al., 2001).
In search of novel PLP-dependent enzymes
The difficulties in establishing function on the basis of gene genealogies imply that much experimental work will be required to achieve a genome-wide classification of all PLP-dependent enzymes, even in the cases of model organisms. It is reasonable to anticipate that efforts in functional genomics will lead to the discovery of many 'novel' PLP-dependent enzymes that have activities that are not yet described or characterized. In fact, the recent literature supports this view.
One example is provided by the identification of a Salmonella typhimurium gene that encodes an enzyme with L-threonine-O-3-phosphate decarboxylase activity (Brushaber et al., 1998). This was the first enzyme described as having this activity, and its discovery shed new light on the pathway that leads to cobalamin biosynthesis in Salmonella. The sequence of this enzyme is only distantly related to those of other PLP-dependent decarboxylases, but is highly similar to those of histidinol-phosphate aminotransferases (Brushaber et al., 1998).
In another case, Wolosker et al. (1999) described a PLP-dependent serine racemase that is expressed in the human brain. The existence of this enzyme had been previously overlooked, as D-amino acids were not known to have any metabolic or physiological function in vertebrates. However, it is now recognized that D-serine is a neurotransmitter (Wolosker et al., 1999; De Miranda et al., 2000). The human serine racemase does not show sequence similarity to bacterial amino-acid racemases (fold-type III), but instead resembles threonine ammonia-lyases, which are fold-type II enzymes (De Miranda et al., 2000).
The discovery of new enzymes that produce chemical mediators in higher eukaryotes may have interesting implications for the recycling of certain proteins for new physiological functions. Indeed, plant and animal cells often use PLP-dependent enzymes for the synthesis of hormones and chemical messengers, and these enzymes can be close homologues of enzymes involved in basic metabolic pathways.
Another example of recycling is provided by the human homologue of 1-aminocyclopropane-1-carboxylate synthase (EC 4.4.1.14), a PLP-dependent enzyme that, in plants, is involved in the synthesis of the ripening hormone ethylene. The human gene product lacks 1-aminocyclopropane-1-carboxylate synthase activity, and although its function remains unknown (Koch et al., 2001), it is tempting to speculate that it might have a role in the synthesis of a chemical mediator.
Orphan PLP-dependent activities and catalytic promiscuity
It was recently noted that more than one-third of all PLP-dependent enzymes classified by the EC are still uncharacterized in terms of sequence (Christen & Mehta, 2001; supplementary information online). Why has it not been possible to identify the gene products that correspond to these catalytic functions? Some PLP-dependent enzymes may have remained uncharacterized at the sequence level because they occur only in relatively obscure organisms, or because their function is not deemed of sufficient scientific interest. Nevertheless, these reasons are unlikely to account for the large number of 'orphan' PLP-dependent activities.
A more basic explanation seems to lie in the phenomenon of 'catalytic promiscuity', that is, the ability of a single enzyme to catalyse different chemical reactions (for a review, see O'Brien & Herschlag, 1999). Catalytic promiscuity is particularly frequent among PLP-dependent enzymes, due to their common mechanistic features. For example, Strisovsky et al. (2003) recently reported that serine racemase catalyses the deamination of L-serine at a rate similar to that of serine racemization. In addition, PLP-dependent enzymes may have non-strict substrate specificity, as shown by Han et al. (2001), who described an aminotransferase that catalyses reactions corresponding to three different EC numbers. All this means that a single gene product can be responsible for several catalytic functions, which complicates functional classification and genomic annotation. In some instances, the same activity can even be carried out (possibly as a side reaction) by several different PLP-dependent enzymes. An extreme example of this is cysteine-S-conjugate β-lyase (EC 4.4.1.13); although the current databases do not contain any gene sequence that corresponds to this EC number, it is known that at least nine distinct PLP-dependent enzymes, including several aminotransferases, catalyse cysteine S-conjugate β-lyase reactions in mammals (Cooper et al., 2002).
The occurrence of catalytic promiscuity and loose substrate specificity also implies that an organism may have more PLP-dependent activities than it has genes encoding PLP-dependent enzymes. In higher eukaryotes, this may be compounded by the possibility of alternative splicing, a process that can increase the functional diversity of proteins in general (Graveley, 2001), and of enzymes in particular (for an example, see Christmas et al., 2001). It must be stressed, however, that although many alternatively spliced PLP-dependent genes have been described in animal cells, splice variants that differ significantly in terms of reaction or substrate specificity have not been reported. Moreover, in some cases it has been shown that certain splice variants cannot bind PLP and presumably do not function as enzymes (Bond et al., 1990; Liu et al., 2001).
Conclusions
The current flood of genomic information offers new opportunities for detecting and analysing the distribution of protein families within and between genomes. This, in turn, can provide a picture of the evolutionary appearance, loss and recycling of protein functions. PLP-dependent enzymes represent a particularly favourable case: structurally, they belong to a limited number of fold groups and are not, in general, found in larger, more complex multidomain proteins; and functionally, they have been studied for decades and are mostly involved in well-known metabolic pathways. Nevertheless, even for PLP-dependent enzymes, the assignment of function cannot rely solely on bioinformatics, and it will be of interest to see whether emerging high-throughput technologies can supply the necessary biochemical data.
Several of the factors that interfere with the functional classification of PLP-dependent genes may have a general relevance for genomic studies. In particular, the possibility of performing different biological functions is not specific to promiscuous enzymes, but occurs in a variety of proteins (Jeffery, 1999). This underlines the fact that a one-to-one correspondence should not be expected between functions (deduced from experimental studies) and genes (counted in genome scans). In genomic analyses, the recognition of proteins that have multiple biological functions is an issue that is only beginning to be appreciated (Copley, 2003), and represents a challenge for the coming years.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor914-s1.pdf).
Supplementary Material
Supplementary information online

Alessio Peracchi & Riccardo Percudani. Alessio Peracchi is the recipient of an EMBO Young Investigator Award
Acknowledgments
We thank G.-L. Rossi for support, A. Merli for discussions and F. Ravasini for technical assistance. We also thank S. Ottonello, A. Mozzarelli and D. Herschlag for comments on the manuscript.
References
- Bond R.W., Wyborski R.J. & Gottlieb D.I. ( 1990) Developmentally regulated expression of an exon containing a stop codon in the gene for glutamic acid decarboxylase. Proc. Natl Acad. Sci. USA, 87, 8771–8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushaber K.R., O'Toole G.A. & Escalante-Semerena J.C. ( 1998) CobD, a novel enzyme with L-threonine-O-3-phosphate decarboxylase activity, is responsible for the synthesis of (R)-1-amino-2-propanol-O-2-phosphate, a proposed new intermediate in cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem., 273, 2684–2691. [DOI] [PubMed] [Google Scholar]
- Chang C.H. & Frey P.A. ( 2000) Cloning, sequencing, heterologous expression, purification, and characterization of adenosylcobalamin-dependent D-lysine 5,6-aminomutase from Clostridium sticklandii. J. Biol. Chem., 275, 106–114. [DOI] [PubMed] [Google Scholar]
- Chen H.P., Wu S.H., Lin Y.L., Chen C.M. & Tsay S.S. ( 2001) Cloning, sequencing, heterologous expression, purification, and characterization of adenosylcobalamin-dependent D-ornithine aminomutase from Clostridium sticklandii. J. Biol. Chem., 276, 44744–44750. [DOI] [PubMed] [Google Scholar]
- Christen P. & Mehta P.K. ( 2001) From cofactor to enzymes. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Chem. Rec., 1, 436–447. [DOI] [PubMed] [Google Scholar]
- Christmas P. et al. ( 2001) Alternative splicing determines the function of CYP4F3 by switching substrate specificity. J. Biol. Chem., 276, 38166–38172. [DOI] [PubMed] [Google Scholar]
- Clausen T. et al. ( 2000) X-ray structure of MalY from Escherichia coli: a pyridoxal 5′-phosphate-dependent enzyme acting as a modulator in mal gene expression. EMBO J., 19, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.J., Bruschi S.A. & Anders M.W. ( 2002) Toxic, halogenated cysteine S-conjugates and targeting of mitochondrial enzymes of energy metabolism. Biochem. Pharmacol., 64, 553–564. [DOI] [PubMed] [Google Scholar]
- Copley S.D. ( 2003) Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol., 7, 265–272. [DOI] [PubMed] [Google Scholar]
- De Miranda J., Santoro A., Engelender S. & Wolosker H. ( 2000) Human serine racemase: moleular cloning, genomic organization and functional analysis. Gene, 256, 183–188. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy S., Gerchman S., Graziano V., Kycia H., Studier F.W. & Swaminathan S. ( 2003) Structure of a yeast hypothetical protein selected by a structural genomics approach. Acta Crystallogr. D Biol. Crystallogr., 59, 127–135. [DOI] [PubMed] [Google Scholar]
- Fukamachi H., Nakano Y., Yoshimura M. & Koga T. ( 2002) Cloning and characterization of the L-cysteine desulfhydrase gene of Fusobacterium nucleatum. FEMS Microbiol. Lett., 215, 75–80. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. ( 2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- Grishin N.V., Phillips M.A. & Goldsmith E.J. ( 1995) Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci., 4, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Fang J. & Li J. ( 2001) Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: identity with aspartate aminotransferase. Biochem. J., 360, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansonius J.N. ( 1998) Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol., 8, 759–769. [DOI] [PubMed] [Google Scholar]
- Jeffery C.J. ( 1999) Moonlighting proteins. Trends Biochem. Sci., 24, 8–11. [DOI] [PubMed] [Google Scholar]
- John R.A. ( 1995) Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta, 1248, 81–96. [DOI] [PubMed] [Google Scholar]
- Kimura H. ( 2002) Hydrogen sulfide as a neuromodulator. Mol. Neurobiol., 26, 13–19. [DOI] [PubMed] [Google Scholar]
- Koch K.A., Capitani G., Gruetter M.G. & Kirsch J.F. ( 2001) The human cDNA for a homologue of the plant enzyme 1-aminocyclopropane-1-carboxylate synthase encodes a protein lacking that activity. Gene, 272, 75–84. [DOI] [PubMed] [Google Scholar]
- Krupka H.I., Huber R., Holt S.C. & Clausen T. ( 2000) Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5′-phosphate-dependent protein acting as a haemolytic enzyme. EMBO J., 19, 3168–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Szebenyi D.M., Anguera M.C., Thiel D.J. & Stover P.J. ( 2001) Lack of catalytic activity of a murine mRNA cytoplasmic serine hydroxymethyltransferase splice variant: evidence against alternative splicing as a regulatory mechanism. Biochemistry, 40, 4932–4939. [DOI] [PubMed] [Google Scholar]
- Mehta P.K. & Christen P. ( 2000) The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv. Enzymol., 74, 129–184. [DOI] [PubMed] [Google Scholar]
- O'Brien P.J. & Herschlag D. ( 1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol., 6, R91–R105. [DOI] [PubMed] [Google Scholar]
- Rost B. ( 2002) Enzyme function less conserved than anticipated. J. Mol. Biol., 318, 595–608. [DOI] [PubMed] [Google Scholar]
- Schneider G., Kack H. & Lindqvist Y. ( 2000) The manifold of vitamin B6 dependent enzymes. Structure Fold. Des., 8, R1–R6. [DOI] [PubMed] [Google Scholar]
- Soutourina J., Blanquet S. & Plateau P. ( 2001) Role of D-cysteine desulfhydrase in the adaptation of Escherichia coli to D-cysteine. J. Biol. Chem., 276, 40864–40872. [DOI] [PubMed] [Google Scholar]
- Strisovsky K., Jiraskova J., Barinka C., Majer P., Rojas C., Slusher B.S. & Konvalinka J. ( 2003) Mouse brain serine racemase catalyzes specific elimination of L-serine to pyruvate. FEBS Lett., 535, 44–48. [DOI] [PubMed] [Google Scholar]
- Thornton J.M., Todd A.E., Milburn D., Borkakoti N. & Orengo C.A. ( 2000) From structure to function: approaches and limitations. Nature Struct. Biol., 7 (suppl.), 991–994. [DOI] [PubMed] [Google Scholar]
- Wolosker H., Sheth K.N., Takahashi M., Mothet J.P., Brady R.O., Ferris C.D. & Snyder S.H. ( 1999) Purification of serine racemase: biosynthesis of the neuromodulator D-serine. Proc. Natl Acad. Sci. USA, 96, 721–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information online


